Abstract
The MSI2 RNA binding protein is a potent oncogene playing key roles in hematopoietic stem cell homeostasis and malignant hematopoiesis. Here we demonstrate that MSI2 is expressed in the intestinal stem cell compartment, that its expression is elevated in colorectal adenocarcinomas, and that MSI2 loss of function abrogates colorectal cancer cell growth. MSI2 gain of function in the intestinal epithelium in a drug inducible mouse model is sufficient to phenocopy many of the morphological and molecular consequences of acute loss of the APC tumor suppressor in the intestinal epithelium in a Wnt-independent manner. Transcriptome-wide RNA-binding analysis indicates that MSI2 acts as a pleiotropic inhibitor of known intestinal tumor suppressors including Lrig1, Bmpr1a, Cdkn1a, and Pten. Finally, we demonstrate that inhibition of the PDK-AKT-mTORC1 axis rescues oncogenic consequences of MSI2 induction. Taken together, our findings identify MSI2 as a central component in an unappreciated oncogenic pathway promoting intestinal transformation.
INTRODUCTION
The Drosophila melanogaster RNA binding protein Musashi contributes to asymmetric stem cell division and cell fate determination in the neuroblast1. In mammals, there are two Musashi orthologs, MSI1/Msi1 and MSI2/Msi22,3. Recently, Msi2 has been implicated as a critical regulator of hematopoietic stem cell self-renewal and fate determination and MSI2 is a potent cooperative oncogene in human leukemias4–6. The role of MSI2 in leukemia progression was recently revealed by two groups who independently observed increased MSI2 expression during disease progression in patients with CML blast crisis and in acute myeloid leukemias4,5. Forced MSI2 expression drove a more aggressive myeloid disease in a transplantation model utilizing the BCR-ABL oncogene. In contrast, MSI2 abrogation in myeloid leukemia cells leads increases differentiation, decreases proliferation and increases apoptosis4. These studies demonstrate that MSI2 cooperates with known oncogenes in hematopoietic malignancies. In addition, high MSI2 expression is observed in a variety of other cancers, including hepatocellular carcinoma and lung cancer7,8, suggesting an important role for MSI2 in a variety of epithelial-derived carcinomas.
Aggressive leukemias are characterized by the prevalence of an increasingly hematopoietic stem cell (HSC)-like transcriptional profile. Consistent to the role of MSI2 in leukemia, MSI2 also plays an important role in HSC homeostasis. MSI2 is highly expressed in the most primitive HSCs, including long-term hematopoietic stem cells (LT-HSC) and short-term hematopoietic stem cells (ST-HSC), but not in more committed hematopoietic lineages. Inactivation of Msi2 in HSCs impairs their competitive repopulation ability upon transplantation4,6,9. Thus, although the functions of MSI2 in normal and malignant hematopoiesis are well established, little is known regarding the role MSI2 plays in stem cells and cancers in other organ systems.
In contrast to reports of MSI2 function in the hematopoietic system, several reports have suggested a role for the second Musashi family member, MSI1 in colorectal cancer. Msi1 is expressed in the putative intestinal stem cell (ISC) compartment10 and overexpressed in colorectal adenocarcinoma, where higher expression level of MSI1 is correlated to increased metastatic risk and poorer survival11,12. The putative role of MSI1 in colorectal cancer and ISCs, coupled with our past observations of MSI2 function in the HSC and hematopoietic cancers prompted us to investigate a role of MSI2 in intestinal transformation.
Colorectal cancer (CRC) is one the leading causes of cancer-related deaths globally. Genetic inactivation of the APC tumor suppressor is believed to initiate the majority of human colorectal cancers, and elegant genetic studies suggest that APC loss only initiates tumorigenesis when it occurs in ISCs with self-renewal capacity13. APC loss drives constitutive activity of the canonical Wnt signaling pathway by preventing the degradation of its downstream transcriptional effector β-catenin. Thus, constitutive β-catenin activity is thought to be a primary initiator of intestinal stem cell transformation. Genetic inactivation of APC is found in approximately 80% of human patients with CRC, and families harboring a germline mutation in one APC allele suffer from Familial adenomatous polyposis (FAP), a disease characterized by the formation numerous intestinal polyps resulting from stochastic loss of heterozygosity at the APC locus, some of which will invariably progress to CRC14–16. The role of MSI2 in this process and its potential interaction with the Wnt signaling pathway remains entirely unknown.
In this study, we find that MSI2 is overexpressed in human colorectal adenocarcinomas, as well as in early stage adenomas arising in the APCmin/+ mouse model of intestinal tumorigenesis. Using both loss- and gain-of-function approaches we demonstrate that constitutive MSI2 activation is sufficient to phenocopy many histological and molecular aspects of APC loss in the absence of canonical Wnt pathway induction. Transcriptome-wide RNA binding analysis reveals that MSI2 binds several known intestinal tumor suppressor genes, including Pten, a potent tumor suppressor frequently silenced in colorectal cancers. Our findings identify MSI2 as an intestinal oncogene and suggest that MSI2 inhibition may offer a novel point for therapeutic intervention in gastrointestinal cancers.
RESULTS
MSI2 is overexpressed in intestinal cancers
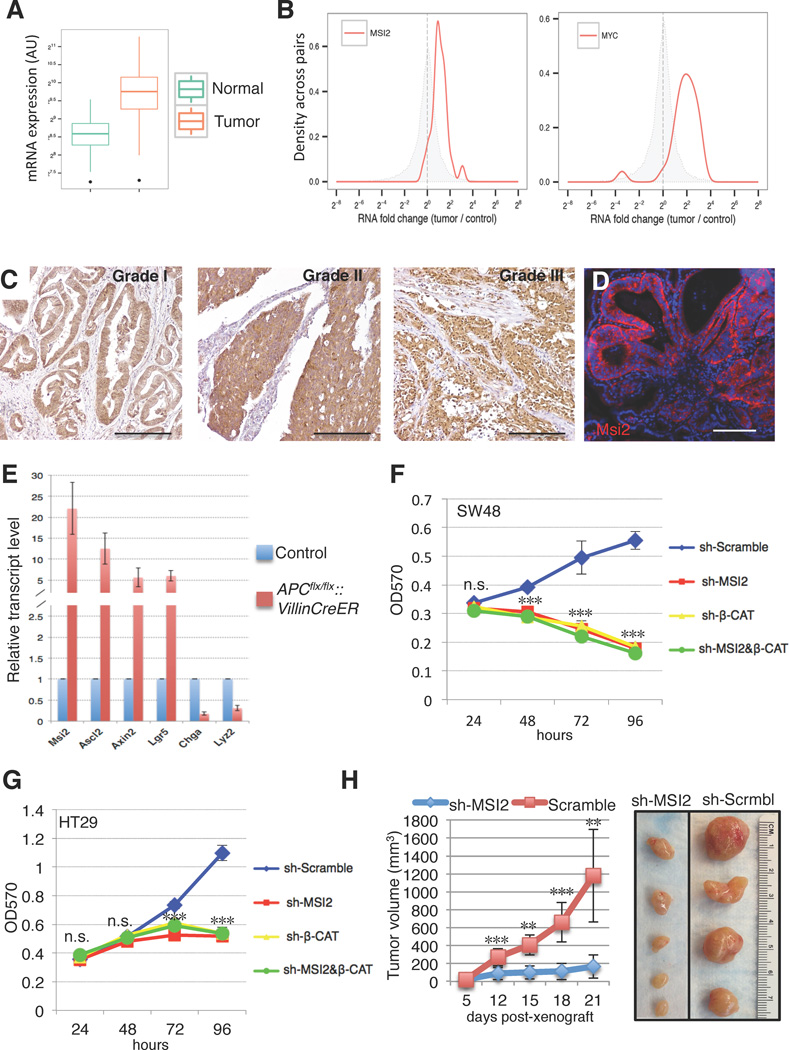
To begin understanding the role of MSI2 in colorectal cancer, we examined MSI2 expression in human gastrointestinal (GI) cancers, and colorectal adenocarcinomas in particular. Analysis of transcriptome profiles from a number of GI cancers reveals elevated MSI2 in all analyzed tumors, including adenocarcinomas of the cecum, colon, and rectum (Supplementary Figure 1A). Focusing on colorectal adenocarcinoma, analysis of cancer genome atlas data (TCGA) derived form 314 healthy control individuals and 314 colon adenocarcinomas patient tumors revealed consistent MSI2 overexpression in cancer tissue (Figure 1A). Similarly, analysis of 26 colon adenocarcinoma/adjacent normal tissue pairs from individual patients revealed uniform overexpression of MSI2 in tumors (Figure 1B). Strikingly, MSI2 overexpression was more consistent in these matched samples than that of the well-established colorectal oncogene c-MYC, which was underexpressed in a small fraction of these tumors (Figure 1B). Consistent with the upregulation of MSI2 transcripts, immunohistochemistry analysis of a CRC tissue array confirmed high MSI2 immunoreactivity in all grades of CRC, with an increased MSI2-immunoreactive area correlating with tumor grade, suggesting that MSI2 activation may be an early event in the ontogeny of cancer (Figure 1C and Supplementary Figure 1B). We also examined Msi2 expression in intestinal adenomas resulting from loss of heterozygosity (LOH) of the APC tumor suppressor in the APCmin/+ mouse model of intestinal transformation. APC loss occurs in the vast majority of human colorectal cancers, where it is believed to be the initiating oncogenic event. Consistent with our observation in human tumors, adenomas in APCmin/+ mice exhibited strong Msi2 immunoreactivity, suggesting a potential role for Msi2 early in intestinal transformation and tumorigenesis (Figure 1D). In order to determine whether Msi2 activation was an immediate early event downstream of APC loss, we deleted APC throughout the epithelium using Villin-CreER17 and observed a striking, upregulation of Msi2 mRNA along with the activation of Wnt target genes (Axin2, Ascl2, Lgr5) and suppression of differentiation markers Chromogranin-A and Lysozyme characteristic of acute APC loss (Sansom et al. 2004) (Figure 1E). Thus, Msi2 activation is a direct consequence of APC loss and is characteristic of all grades of colorectal adenocarcinoma.
Figure 1. MSI2 contributes to human colorectal cancer cell growth.
(A) MSI2 mRNA expression levels from RNA-Seq on TCGA (the cancer genome atlas) COAD (colorectal adenocarcinoma) tumor samples (314 solid tissue samples from healthy individuals, 314 tumor samples). Expression values were obtained by TMM normalization of read counts in genes. (B) Fold changes for MSI2 (left) and MYC (right) in matched tumor/control RNA-Seq sample pairs from TCGA COAD (total of 26 patients). The distribution of MSI2/MYC fold changes in tumor/control pairs for 26 individuals is plotted in red (intra-individual comparison). The distribution of MSI2/MYC fold changes between control/control comparisons for 26 pairs of healthy individuals is plotted in grey (inter-individual comparison). (C) MSI2 immunochemistry of graded human colorectal adenocarcinomas (scale bar=200µM). (D) Msi2 immunofluorescence showing broad expression in adenomas arising in APCmin/+ mice (scale bar=200µM). (E) Expression analysis for Msi2, Wnt target genes (Ascl2, Axin2, Lgr5) and differentiation-related genes (Chga, Lyz) in the intestinal epithelium after acute ablation of APC in APCflox/flox::Villin-CreER mice (error bars denotes s.d., n=3). (F, G) MTT proliferation assays of human colorectal cancer cell lines SW48 (F) and HT29 (G) infected with lentiviral knockdown vectors for MSI2, β-CATENIN, or scramble controls. Error bars indicated s.e.m. from six independent experiments per cell line per condition. ***: p<0.0005, Student’s t-test. (H) Tumor growth curves of HT29 colon cancer cell xenografts infected with lentivirus expressing scramble control hairpin or sh-MSI2. Error bars denote the s.d. derived from 4 tumors in each of 2 mice per group. Representative images of xenograft tumors are shown on the right. **: p<0.005, ***: p<0.0005, Student’s t-test.
MSI2 promotes cancer cell proliferation and tumor growth
To investigate a potential oncogenic role for MSI2 in colorectal cancer, we initially performed MSI2 loss of function experiments in 2 human CRC cell lines, SW48 and HT29. Numerous studies have implicated constitutive Wnt pathway activity as a driving force in colorectal cancer onset and progression, and these cancers are dependent on the transcription activity of the Wnt effector β-CATENIN18. We therefore knocked down MSI2 alone or in combination with β-CATENIN and confirmed loss of MSI2 protein by Western blot (Supplementary Figure 1C). Surprisingly, MSI2 knockdown completely abrogated growth of SW48 and HT29 cells, and was as potent as loss of β-CATENIN in growth inhibition (Figure 1F, G). This finding strongly indicates an important role for MSI2 in promoting cancer cell proliferation. We confirmed these in vitro observations by performing xenograft assays upon MSI2 knockdown in HT29 cells. Twenty-one days after xenografting, tumor volume was significantly reduced in MSI2 knockdown tumors, with some xenografts exhibiting no growth after transplantation (Figure 1H).
MSI2 overexpression largely phenocopies APC loss
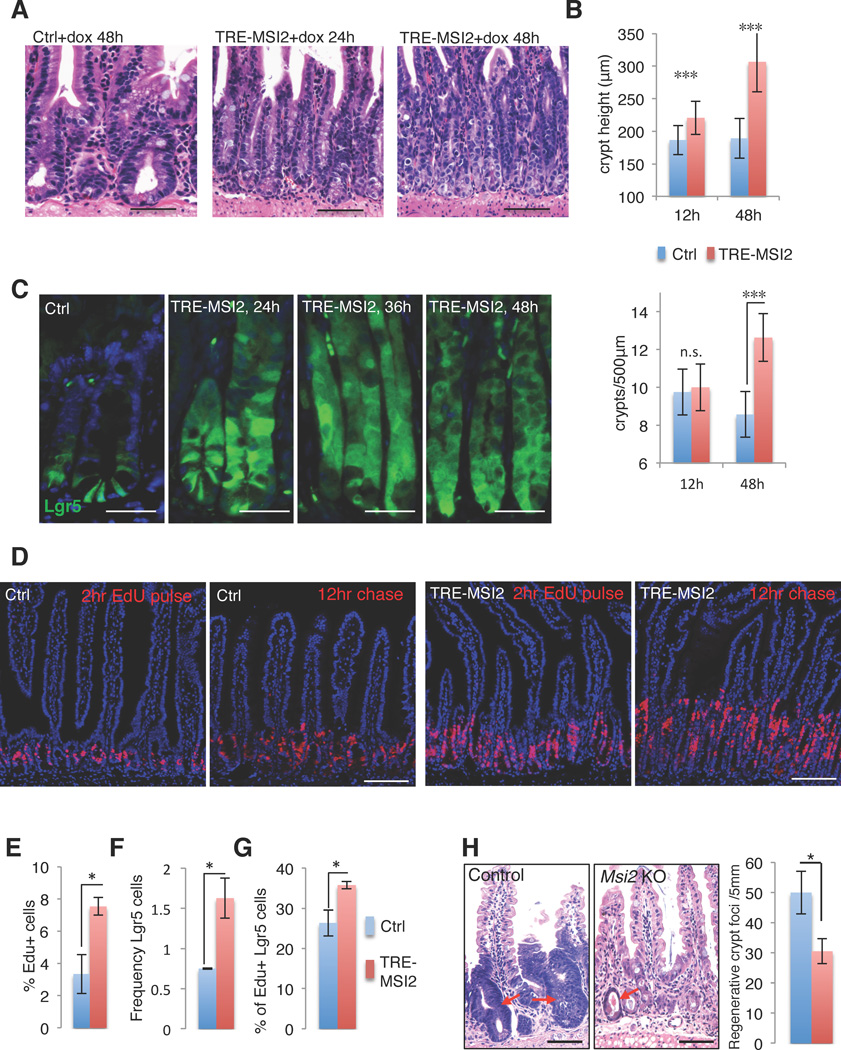
To gain insight into the potential function of Msi2 in intestinal homeostasis and transformation, we examined Msi2 expression in wild type mice and found it to be largely restricted to cells of the crypt in the small intestine. This includes crypt base columnar (CBC) stem cells marked by expression of the Wnt co-receptor Lgr519, Paneth cells, and the rapidly proliferating cells of the transit-amplifying zone (Figure 2A,B, and Supplementary Figure 2A). In addition, rare Msi2-positive cells were observed in the differentiated villi, possibly marking enteroendocrine or tuft cells based on frequency and position (Supplementary Figure 2A). We next sought to determine the functional consequences of ectopic MSI2 activity in the intestinal epithelium. We therefore employed a Doxycycline (Dox)–inducible, site-specific, single copy human MSI2 transgenic mouse model (TRE-MSI2) that we previously utilized to study the oncogenic functions of MSI2 in the hematopoietic system (Supplementary Figure 2A)4. Addition of Dox to the drinking water of TRE-MSI2 mice resulted in activation of MSI2 expression throughout the intestinal epithelium within 12 hours (Figure 2B and Supplementary 2C). Strikingly, MSI2 induction resulted in a dramatic expansion of the proliferative zone and an increased frequency of apoptotic cells at the tips of villi, suggesting that cells were being pushed from the crypt to the villus and shed into the lumen at an increased rate (Figure 2C, D). We further observed that differentiated cells of intestinal epithelium including goblet, enteroendocrine and enterocyte cells were obviously absent in the TRE-MSI2 mice (Figure 2E, F and Supplementary Figure 2D). However, lysozyme-positive Paneth cells persisted, likely due to their long lifespan and stable positioning at the base of the crypt (Supplementary Figure 2E). This block in differentiation resulted in severe dehydration and morbidity within 3–4 days of Dox induction.
Figure 2. MSI2 induction drives intestinal epithelial cell hyperproliferation and blocks differentiation.
(A) Immunofluorescence for Msi2 (red) and eGFP (Green) in the intestinal crypts of Lgr5-eGFP-IRES-CreER knockin mice. Msi2 expression is observed in Lgr5+ cells and in adjacent Paneth cells (arrowheads) (scale bar=50µM). (B–F) Immunofluorescence for MSI2 (B), Ki-67 marking proliferating cells (C), cleaved Caspase 3 (Casp3) marking apoptotic cells in (D), Chromagranin-A marking enteroendocrine cells (E), and Alcian blue marking Goblet cells (F) in control (M2rtTA) and TRE-MSI2 intestines 48 hours after 2mg/mL Dox induction. Brackets in C indicate the crypt proliferative zone. Scale bars in B–F=100µM.
To further investigate the effect of MSI2 induction on proliferation and the crypt stem/progenitor cell compartment, we quantified crypt height and crypt fission (frequency of crypts per linear unit) in TRE-MSI2 mice and observed a significant increase in both of these parameters (Figure 3A, B). Interestingly, the increased crypt proliferation, crypt fission, block in differentiation, and rapid morbidity in TRE-MSI2 mice are all phenotypes entirely consistent with the consequences downstream of acute APC loss in the intestinal epithelium20,21. Taken together, these observations suggest that MSI2 induction drives proliferative expansion of an undifferentiated stem/progenitor cell and may act downstream of APC loss to promote intestinal transformation.
Figure 3. MSI2 expression induces crypt fission and expansion of intestinal stem cells.
(A) H&E staining of control (M2rtTA) and TRE-MSI2 small intestine treated with 2mg/mL Dox for 24 and 48 hours showing expanded crypt height and increased crypt fission (scale bar=100µM). (B) Quantification of crypt length (top panel) and crypt fission (lower panel) in control (M2rtTA) and TRE-MSI2 intestines at indicated time points after Dox treatment. Error bars indicate the s.d. derived from n=3 mice per group per time point. ***: p<0.0005, Student’s t-test. (C) Immunofluorescence for eGFP in control (Lgr5-eGFP-IRES-CreER::M2rtTA) and TRE-MSI2::Lgr5-eGFP-IRES-CreER intestinal crypts at the indicated time points after Dox induction (scale bar=50µM). (D) Histological staining for EdU incorporation 2 hours or 12 hours after an EdU pulse in control (M2rtTA) and TRE-MSI2 mice. Images were taken 48hrs after initiation of 2mg/mL Dox treatment (scale bar=100µM). (E) Flow cytometric analysis of EdU incorporation in control (M2rtTA) and TRE-MSI2 mice 48hrs after 2mg/mL Dox induction. Error bars denote s.d. (n=3 mice per group), *: p<0.05, Student’s t-test. (F–G) Flow cytometric analysis of Lgr5-eGFP+ cell frequency (F) and 2hr-EdU incorporation in Lgr5-eGFP+ cells (G) from control (Lgr5-eGFP-IRES-CreER::M2rtTA) and TRE-MSI2::Lgr5-eGFP-IRES-CreER mice 48 hours after 2mg/mL Dox. Error bars denote s.d. (n=3 mice per group), *: p<0.05, Student’s t-test. (H) Histological analysis and quantification of regenerative crypt foci (arrows) in control (Msi2wt/wt::Villin-CreER) or Msi2flox/flox::Villin-CreER mice three days after exposure to 12Gy gamma-irradiation (scale bar=50µM). Error bars denote s.d. (n=3 mice per group). *: p<0.05, Student’s t-test.
APC loss drives intestinal tumorigenesis only when it occurs in an intestinal stem cell13. We therefore sought to assess the effects of MSI2 induction specifically in the intestinal stem cell compartment. We bred TRE-MSI2 mice to Lgr5-eGFP-IRES-CreER reporter mice that mark CBC stem cells19 and observed a dramatic expansion of the Lgr5+ zone within the crypts of TRE-MSI2 mice (Figure 3C). This expansion was accompanied by an overall increase in the proliferation rate of intestinal epithelial cells (Figure 3D, E), as well as an increase in the absolute frequency and proliferation rate of Lgr5+ CBCs in TRE-MSI2 mice relative to controls (Figure 3F, G). To test whether the increased apoptotic events at the villus tip in TRE-MSI2 might be a result of increased turnover of the epithelium and more rapid replacement of the differentiated villus compartment with proliferative cells from the crypts, we assessed the migration of Edu labeled cells after 12 hours in the presence of ectopic MSI2 and found that indeed, proliferative cells in TRE-MSI2 mice migrated higher than their control counterparts (Figure 3D). Taken together these findings are consistent with a model where MSI2 increases the shedding of differentiated cells and promotes their replacement by undifferentiated cells.
To determine whether the effects of MSI2 induction are epithelial cell-autonomous, we cultured crypts from TRE-MSI2::Lgr5-eGFP-IRES-CreER in vitro and observed an increase in crypt budding and Lgr5+ cell expansion, consistent with an epithelial cell-autonomous phenotype (Supplementary Figure 3A, B). Thus, MSI2 induction drives the proliferative expansion of undifferentiated intestinal stem cells. This suggests that endogenous Msi2 may play a role in intestinal epithelial proliferation and regeneration. To test this, we first examined regenerative crypt foci three days after exposure of wildtype mice to high dose (12Gy) gamma-irradiation and observed high Msi2 expression in proliferative cells (Supplementary Figure 3C). We next ablated Msi2 expression throughout the intestinal epithelium using Msi2flox/flox::Villin-CreER mice6 and observed that in the absence of Msi2 activity there was a significant decrease in the formation of regenerative crypt foci (Figure 3H), indicating that endogenous Msi2 plays a role in supporting proliferation in the epithelium.
MSI2-driven transformation is Msi1-independent
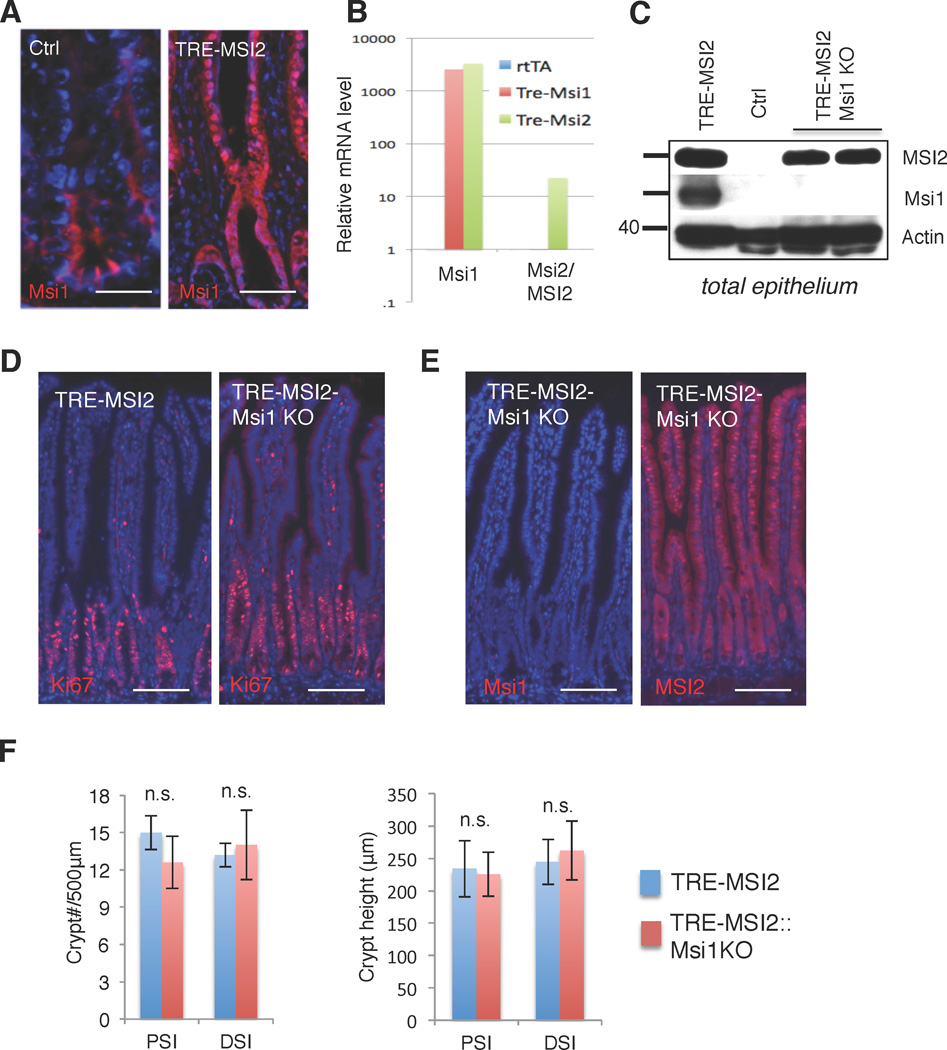
Msi2/MSI2 is one of two mammalian orthologs of Drosophila melanogaster Musashi. The second mammalian protein, Msi1/MSI1, is expressed in intestinal stem cells and has been implicated as an oncogene in intestinal cancers and elsewhere11,22–24. We therefore examined Msi1 expression in the TRE-MSI2 intestinal epithelium and observed a striking activation of Msi1 throughout the epithelium in response to MSI2 induction (Figure 4, A–C). RNA analysis demonstrates that the activation of Msi1 in response to MSI2 induction occurs at the transcript level, and that the activation is not reciprocal: Msi1 induction in an analogous mouse model (TRE-Msi1) did not result in Msi2 activation (Figure 4B).
Figure 4. Msi1 is dispensable for MSI2-driven intestinal transformation.
(A) Immunofluorescence for Msi1 in control (M2rtTA) and TRE-MSI2 mice (scale bar=50µM). (B) Quantitative RT-PCR for Msi1 and Msi2 in control (M2rtTA), TRE-Msi1, or TRE-MSI2 mice treated with 2mg/mL Dox for 24 hours. Msi1 is induced in TRE-MSI2 mice whereas Msi2 expression is not reciprocally effected by TRE-Msi1 induction. (C) Western blot showing Msi1 protein induction upon dox treatment of TRE-MSI2 mice. Msi1 induction is prevented in TRE-MSI2::Msi1flox/flox:: VillinCreER mice (TRE-MSI2-Msi1KO) after Tamoxifen-mediated inactivation of Msi1 alleles followed by Dox treatment. (D) Immunostaining for Ki67 in the intestinal crypts of TRE-MSI2 and TRE-MSI2::Msi1KO mice (scale bar=100µM). (E) Immunofluorescence for Msi1 (left) and Msi2/MSI2 (right) in TRE-MSI2-Msi1KO intestine (scale bar=100µM). (F) Quantification of crypt fission (left) and crypt height (right) in TRE-MSI2 and TRE-MSI2::Msi1KO mice. Error bars indicate s.d. derived from at least 20 fields per slide from each of 2 mice per condition. PSI/DSI: proximal and distal small intestine.
We therefore tested whether the phenotype resulting from MSI2 induction was Msi1-dependent. We generated Msi1flox/flox::Villin-CreER mice25 enabling the deletion of Msi1 within all cells of the intestinal epithelium, deleted Msi1, and induced MSI2 with Dox. Msi1 deletion with Villin-CreER resulted in no morphological phenotype during homeostasis consistent with the viability and lack of any overt GI phenotype in germline Msi1 knockout mice26 Deletion of Msi1 was confirmed at the protein level (Figure 4C, E). We observed that the phenotype resulting from MSI2 induction was fully penetrant in the absence of Msi1, with increased crypt proliferation and fission, and decreased differentiation being unaffected in TRE-MSI2::Msi1flox/flox::Villin-CreER mice (Figure 4D–F and Supplementary Figure 3D). Thus, Msi1 activity is not required for MSI2-driven transformation of the intestinal epithelium.
The oncogenic effects of MSI2 are β-catenin-independent
To gain mechanistic insights into the molecular events underlying the effects of MSI2 induction, we analyzed transcriptome profiles of intestinal epithelia 24 hours after Dox administration to TRE-MSI2 or control (M2rtTA) mice, prior to overt phenotypic changes. MSI2 induction drove rapid and robust changes in the intestinal epithelial transcriptome (Figure 5A and Supplementary Table I). Unbiased Gene Set Enrichment Analysis (GSEA)27 of these changes revealed that the APC-loss gene signature was among the most significantly enriched in the TRE-MSI2 epithelium (Figure 5B and Supplementary Figure 4A and B). Thus, genes that are activated upon APC loss are also activated upon MSI2 induction, and genes suppressed upon APC loss are similarly suppressed upon MSI2 induction, with a false discovery rate (FDR) approaching zero.
Figure 5. MSI2 activation imposes an APC-loss gene signature in the absence of Wnt pathway activation.
(A) Hierarchical clustering of transcriptome profiles from intestinal epithelium of 3 control (M2rtTA) and 3 TRE-MSI2 mice 24 hours after 2mg/mL Dox induction. (B) Gene Set Enrichment Analysis (GSEA) of the transcriptome profiles showing a highly significant enrichment of APC loss gene signatures in TRE-MSI2 mice. Top panel: Genes activated by both APC loss and MSI2 induction, Bottom panel: Genes suppressed upon APC loss and MSI2 induction. FDR: False Discover Rate (C) Western blot for APC in lysates of intestinal epithelium of control (M2rtTA), TRE-MSI2 and TRE-MSI2-Msi1KO mice 48hrs after 2mg/mL Dox induction. (D) Immunohistochemical staining for β-catenin in control (M2rtTA) and TRE-MSI2 intestinal crypts 48 hours after 2mg/mL dox induction, with number of β-catenin+ nuclei per crypt section quantified at right (PSI/DSI: proximal and distal small intestine). Error bars indicate the S.D. derived from at least 50 crypts from each mouse (n=3 mice per group, Student’s t-test). (E) QRT-PCR analysis of Wnt target gene and differentiation-related gene expression in purified crypts from control (M2rtTA) and TRE-MSI2 intestinal crypts 48 hours after 2mg/mL dox induction. Error bars indicate the S.D., n=3. (F) β-Catenin/TCF transcriptional reporter activity (TOPFLash) normalized to activity of the reporter with a mutated binding element (FOPFlash) in HEK293 cells upon MSI2 knockdown (sh-MSI2), overexpression (pcDNA-MSI2), or β-CATENIN knockdown (sh-β-CAT) as a control Error bars indicate the S.D. (n=4). ***: p<0.0005, Student’s t-test. (G) Frequency of adenomas in Apcmin/+ (n=4) and TRE-MSI2::Apcmin/+ (n=4) mice treated with low dose Dox (0.1mg/mL) in drinking water for 3 weeks. Error bars denote the SD. **: p<0.005, Student’s t-test.
In addition to the APC loss gene signature, MSI2 induction significantly enriched gene signatures associated with RNA processing, RAS oncogenic signalling, and a common stem cell gene signature, suggesting that MSI2 drives proliferative expansion of an undifferentiated stem/progenitor cell, consistent with the observed increase in Lgr5+ cell frequency and cycling (Supplementary Figure 4C). Further, MSI2 activity strongly suppressed an oxidative phosphorylation expression signature, indicating that a metabolic shift consistent with the Warburg effect may occur in response to MSI2 induction (Supplementary Figure 4C).
Given the ability of MSI2 induction to phenocopy many aspects of APC at both the molecular and morphological level, we tested whether MSI2 activation resulted in loss of APC protein, as MSI proteins are known to function as translational repressors3. We were unable to detect any decrease in APC upon MSI2 induction (Figure 5C and Supplementary Figure 4D). Since APC loss is believed to drive transformation of the intestinal epithelium primarily through constitutive activation of the Wnt effector β-catenin and, subsequently, hyperactivation of the canonical Wnt pathway, it was important to address the consequences of MSI2 induction for transcriptionally active (nuclear) β-catenin in TRE-MSI2 crypts. Normally, nuclear β-catenin is limited to the Lgr5+ CBCs and Paneth cells, rapidly disappearing above the +4 position of the crypt (Figure 5D). We observed no change in the pattern or frequency of nuclear β-catenin after MSI2 induction (Figure 5D), or in the expression levels of known direct β-catenin target genes in the transcriptome profiles (Supplementary Figure 4E). We also observed that canonical Wnt target genes such as Ascl2, Axin2, Lgr5, and Sox9 were not among the genes commonly upregulated upon APC loss and MSI2 induction in the GSEA (Supplementary Figure 4B).
To further explore the effect of MSI2 on activity of the canonical Wnt pathway, we analyzed the expression of Wnt target genes Ascl2, Axin2, Lgr5, and Sox9, as well as differentiation markers Lysozyme and Chromogranin-A upon MSI2 induction both in crypts in vivo and in organoid cultures. MSI2 had identical effects on the expression of these genes both in vivo and in vitro. Remarkably, not only were Wnt target genes Ascl2, Axin2, and Sox9 not activated, they exhibited suppression upon MSI2 induction, providing further evidence that the effects of MSI2 are not mediated through canonical Wnt signaling (Figure 5E and Supplementary Figure 3E). One exception was Lgr5, which was activated by MSI2. Differentiation markers Lysozyme and Chromogranin-A were suppressed upon MSI2 induction in vivo and in vitro, consistent with the effect of APC loss on differentiation (Figure 1E), and with the effects of MSI2 being epithelial cell autonomous. Further, in vitro analysis of β-Catenin/TCR reporter activity (TOPFlash) confirmed that MSI2 does not activate canonical Wnt signaling (Figure 5F).
We also directly compared the effects of β-Catenin transcriptional activation to MSI2 induction in intestinal organoid cultures from TRE-BcatS33 mice29. In these mice, a non-degradable, transcriptionally active β-Catenin is targeted to the identical chromosomal location as MSI2 in the TRE-MSI2 mouse model in the same genetic background, enabling direct comparison of the effects of these two proteins. As expected, induction of β-CateninS33 resulted in the growth of large, spherical, cystic organoids consistent with the known effects of Wnt pathway hyperactivation (Supplementary Figure 3F). In contrast, MSI2 induction resulted in large organoids with prolific crypt budding (Supplementary Figure 3F), consistent with the in vivo phenotype.
Consistent with our earlier observation of elevated Msi2 expression in adenomas resulting from LOH in APCmin/+ mice and upon acute APC deletion in APCflox/flox mice, these findings support a model in which MSI2 acts downstream of APC loss in a pathway parallel to and independent of β-catenin activation. Thus, MSI2 may cooperate with Wnt/β-catenin signalling to drive tumorigenesis. We therefore crossed TRE-MSI2 mice to APCmin/+ mice and administered a low dose of Dox for 3 weeks (0.1mg/mL, or 5% of the standard dose). TRE-MSI2::APCmin/+ mice treated with low dose Dox exhibited an increase in tumor burden resulting from APC LOH, while adjacent tissue that had not undergone LOH remained phenotypically normal (Figure 5G and Supplementary Figure 4F). This finding provides additional support to the model in which MSI2 promotes tumorigenesis in a pathway parallel to canonical Wnt signaling.
Transcriptome-wide MSI2 RNA-binding analysis
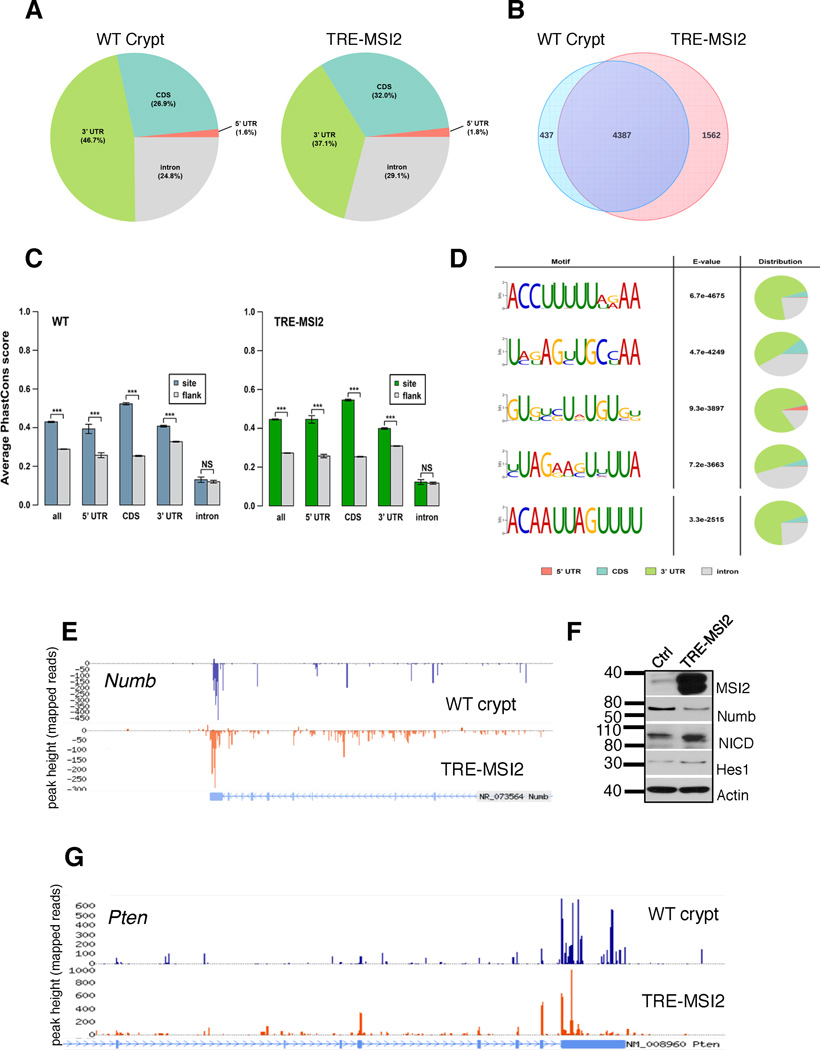
MSI2 is an RNA binding protein that, similar to MSI1, harbors two RNA recognition motifs (RRMs) that mediate RNA binding3. Binding of RNA targets by MSI1 results in their translational suppression without affecting overall mRNA stability30,31. Therefore, if we assume that MSI2 (which is highly homologous to MSI1, particularly in the RRMs3) acts in a manner similar to MSI1, we expect that the transcriptional changes associated with MSI2 induction are not direct effects of its RNA binding activity. To identify direct MSI2 binding targets within the intestinal epithelium, we performed crosslinking and immunoprecipitation of Msi2/MSI2-bound RNA in intestinal crypts isolated from wildtype mice and in TRE-MSI2-transformed intestinal epithelium, followed by RNase-digestion of unprotected RNA and massively parallel sequencing (CLIP-Seq). Msi2/MSI2 primarily bound sites in 3’ untranslated regions (UTRs), consistent with the established binding activity of Msi130,31, and consistent with what we previously observed for MSI2 in hematopoietic cancers6 (Figure 6A). The vast majority of transcripts bound by Msi2 in wildtype crypts were also bound by ectopic MSI2 (4387/4824, or 90%) (Figure 6B). Conservation analysis of Msi2/MSI2 binding sites revealed that sites in coding sequences and UTRs were significantly conserved relative to flanking sequences, however this was not true of intronic binding events, suggesting that Msi2/MSI2 binding of introns may represent promiscuous events. (Figure 6C and Supplementary Figure 5A). Limiting analysis specifically to 3’UTR-bound transcripts reveals a similar overlap between Msi2-targets in wildtype crypts and MSI2-bound targets in TRE-MSI2 epithelium (91% of Msi2 targets also bound by ectopic MSI2, and 76% of TRE-MSI2 targets also bound by Msi2 in wildtype crypts) (Supplementary Figure 5B).
Figure 6. Transcriptome-wide MSI2 RNA binding analysis.
(A) RNA Crosslinking, Immunoprecipitation, and massively parallel Sequencing (CLIP-Seq) shows distribution of Msi2/MSI2 RNA binding events in wildtype intestinal crypts and TRE-MSI2 epithelium. (B) Venn diagrams show overlap between Msi2/MSI2 bound transcripts in wildtype crypts (left) and TRE-MSI2 epithelium (right) 24 hours after 2mg/mL Dox induction. (C) PhastCons conservation analysis of Msi2/MSI2 binding quantifying evolutionary conservation of binding sites relative to flanking sequences in all categories with the exception of intronic binding sites (these were not conserved relative to flanking sequences). Error bars represent 95% confidence intervals. ***: p<0.0005, two-sample Kolmogorov-Smirnov test. (D) Msi2 binding motif identification and distribution in WT crypts. The previously identified Msi1 binding motive is the final motif pictured. (E) CLIP-Seq track showing Msi2/MSI2 binding to the 3’UTR of the mRNA encoding Numb (F) Western blot analysis of Numb, activated Notch (Notch intracellular domain, NICD) and the downstream Notch target gene Hes1 in control (M2rtTA), and TRE-MSI2 intestinal epithelium. (G) CLIP-Seq track showing Msi2/MSI2 binding to the 3’UTR of the mRNA encoding Pten.
Msi2/MSI2 binding was, overall, not selective for the most highly expressed transcripts, and binding did not globally affect overall mRNA levels, as transcripts newly bound upon MSI2 induction in TRE-MSI2 epithelium exhibited no change in mRNA levels (Supplementary Figure 5C–E). This is consistent with MSI2-RNA interaction promoting neither mRNA degradation nor stabilization. Binding motif analysis for Msi2 in WT crypts revealed that Msi2 bound a number of U-rich motifs, and the previously published Msi1 binding motif [(G/A)U(n)AGU (n = 1–3)]30, while not among the most significant, was still highly enriched and significant in the data (Figure 6D). Electrophoretic mobility shift assays confirmed interaction of recombinant human MSI2 with the dominant 3’UTR motif ACCUUUUUAGAA (Supplementary Figure 5F). Global analysis of Msi2 RNA-binding targets revealed an enrichment in transcripts encoding proteins involved in protein localization and transport, ribosome biology, and nucleotide/ribonucleotide binding, consistent with what was observed previously for MSI2 in hematopoietic cancers6 (Supplementary Figure 6A). Interestingly, mRNA targets involved in neurodegenerative diseases including Parkinson’s, Alzheimer’s and Huntington’s were also significantly enriched, possibly reflecting a role for MSI2 in these disorders (Supplementary Figure 6A).
We next examined binding of Msi2 to transcripts that have previously been identified as Msi1-binding targets in vitro, including Cdkn1a (encoding p21) and Numb30,31. Msi2/MSI2 bound both Cdkn1a and Numb mRNAs in their 3’UTRs (Figure 6E and Supplementary Figure 6B). Interestingly, MSI2 induction had only modest effects reducing Numb protein and activating Notch signaling, and no enrichment for a Notch expression signature was observed in our transcriptome profiles, indicating that Notch activation is not a major consequence of MSI2 induction.
These data validate our CLIP-Seq approach and indicate that Msi2 has an RNA-binding activity similar to the second mammalian Musashi family member, Msi1. Given the observed binding of MSI2 to the transcripts encoding the tumor suppressors p21 and Numb, we examined the CLIP dataset for other interactions between MSI2 and tumor suppressors. Remarkably, MSI2 bound several well-established intestinal tumor suppressions including Lrig132, Bmpr1a33 and Pten34,35 (Figure 6F and Supplementary Figure 6B). Interestingly, PTEN protein is lost in many colorectal cancers, however the PTEN gene is rarely the target of genetic inactivation36. Thus, it is tempting to speculate that binding of PTEN mRNA by MSI2 may account for the loss of PTEN protein in PTEN-wildtype cancers.
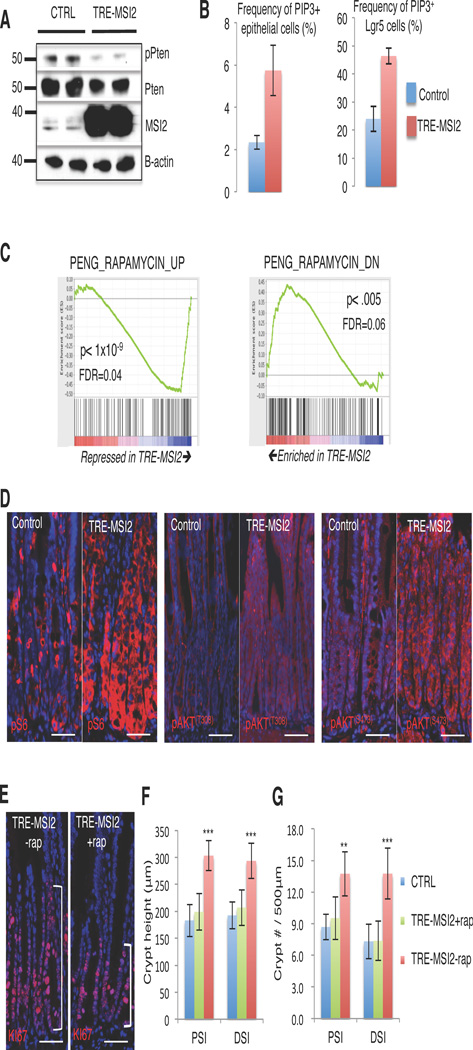
MSI2 inhibits Pten and activates the mTORC1 pathway
PTEN is a well-known tumor suppressor in colorectal cancer and has been implicated in the regulation of intestinal stem cells34. Deletion of Pten in the context of APC deficiency accelerates adenoma formation and activates the PDK-AKT-mTORC1 pathway34,35,37. To investigate the possibility that MSI2-mediated transformation of the intestinal epithelium is a result of Pten inhibition, we examined Pten protein levels and observed a reduction upon TRE-MSI2 induction (Figure 7B). Pten is a phosphatase that dephosphorylates phosphatidylinositol (3,4,5)-triphosphate (PIP3), converting it to PIP2. Loss of Pten thus blocks conversion of PIP3 to PIP2, leading to an accumulation of PIP3. Analysis of PIP3 in both total intestinal epithelial cells and specifically in Lgr5+ crypt base columnar stem cells reveals an increase in PIP3-positive cells after TRE-MSI2 induction, consistent with an inhibition of Pten activity (Figure 7B).
Figure 7. Activation of the Pten-AKT-mTORC1 axis is necessary for MSI2-transformation of the intestinal epithelium.
(A) Western blotting showing reduction of Pten protein in TRE-MSI2 crypts vs. controls (M2rtTA) 48 hours after 2mg/mL Dox induction. (B) Flow cytometric quantification of PIP3+ cell frequency in total intestinal epithelial cells (left) and Lgr5-eGFP+ crypt base columnar stem cells in (right) between control (M2rtTA) and TRE-MSI2 mice 48 hours after Dox induction (n=3 mice per group), error bars indicate the S.D. (C) Gene set enrichment analysis demonstrating an inverse correlation between the TRE-MSI2 transcriptome and Rapamycin-regulated genes. Genes upregulated by Rapamycin treatment are suppressed by MSI2 (left), and genes suppressed by Rapamycin treatment are upregulated by MSI2 induction. (D) Immunofluorescence for pS6, pAKT (T308) and pAKT (s473) showing activation AKT/mTORC1 signaling axis in TRE-MSI2 mice, (scale bar=50µM). (E) Immunofluorescence staining for Ki67 in Dox-induced TRE-MSI2 mice treated with or without Rapamycin (scale bar=50µM). (F and G) Qualification of crypt height in F and crypt frequency in G in control (M2rtTA), TRE-MSI2, and Rapamycin treated TRE-MSI2 mice. Error bars denote S.D. Crypt height was measured based on 40 crypts in each of 3 independent pairs of mice. Crypt frequency was scored in 20 randomly selected areas in proximal and distal intestine (PSI and DSI, respectively) in each of 3 independent pairs of mice. **: p<0.005, ***: p<0.0005.
Accumulating PIP3 binds to PDK1, in turn activating the AKT-mTorc1 pathway38. Consistent with Pten suppression and mTORC1 activation downstream of MSI2 induction, GSEA revealed an inverse correlation between the TRE-MSI2 gene signature and the Rapamycin gene signature (Figure 7C). Rapamycin is a potent inhibitor of the mTORC1 complex, and thus these findings indicate that the mTORC1 complex is being activated by MSI2 induction. We further examined activity of the pathway downstream of Pten and observed increased phosphorylation of AKT (Phospo-AKT(T308) and Phospo-AKT(S473), Figure 7D), as well as increased mTORC1 activity, assessed by phosphorylation of S6 by S6 kinase and phosphorylation of 4EBP1 (Figure 7D and Supplementary Figure 7A).
Ultimately, we asked whether MSI2-mediated mTORC1 activation functionally contributes to intestinal transformation in TRE-MSI2 mice. Since it is well established that mTORC1 activity is dispensable for intestinal homeostasis39,40, we blocked mTORC1 signaling in TRE-MSI2 mice with Rapamycin for two days prior to Dox induction of MSI2 for 48 hours. Phosphorylation of S6 was markedly reduced in Rapamycin-treated TRE-MSI2 mice (Supplementary Figure 7B), confirming that mTORC1 activity was effectively inhibited. Strikingly, we found that the expanded proliferative zone within the crypts marked by Ki67 in TRE-MSI2 mice was fully rescued by the Rapamycin treatment (Figure 7E, F). Furthermore, we observed that the increase in crypt density/crypt fission was also rescued in the Rapamycin treated TRE-MSI2 mice, demonstrating that activation of mTORC1 functionally contributes the increased crypt fission and hyperplasia observed in TRE-MSI2 mice (Figure 7G). In summary, these findings demonstrate that MSI2 is a potent oncogene capable of recapitulating phenotypes resulting from APC-loss through the activation of mTORC1 in an unappreciated, Wnt-independent oncogenic pathway.
DISCUSSION
Here we identify MSI2 as a potent oncogene in colorectal cancer. The current dogma of colorectal cancer initiation suggests that loss of the APC tumor suppressor results in the failure to degrade β-CATENIN, consequently resulting in constitutive activation of the Wnt signaling pathway. This process is believed to result in tumorigenesis only when it occurs in intestinal stem cells, as genetic ablation of APC in other cell types of the intestine does not initiate tumorigenesis13. In the current study we observe high levels of MSI2 in primary human colorectal cancers, along with Msi2 induction upon loss of APC in mouse models. Further, we observe that loss of MSI2 abrogates growth of colorectal cancer cells (both APC-wildtype and APC-negative), where its loss is as effective as loss of β-CATENIN in inhibiting CRC cell growth. These findings, coupled with our previous findings in acute and chronic myelogenous leukemias4 indicate that MSI2 is upregulated in a number of cancers (possibly through a variety of mechanisms, as elevated MSI2 levels are found in cancers with distinct underlying genetic mutations), and that these cancers become dependent on oncogenic MSI2 activity.
In the current study we sought to model the consequences of MSI2 induction on the intestinal epithelium. Using a single-copy, drug-inducible MSI2 transgenic mouse model we are able to rapidly and robustly induce MSI2 activity throughout the intestinal epithelium. Strikingly, MSI2 activity alone was sufficient to phenocopy many aspects of APC loss in the intestinal epithelium (the exception being Paneth cell mislocalization), resulting in a dramatic expansion of the crypt proliferative zone, a block in differentiation, and an increase in crypt fission20. This phenotype was also reflected at the molecular level, with unbiased Gene Set Enrichment Analysis of MSI2-expressing epithelium identifying the APC loss gene signature among the most significantly enriched.
Given the dogma that APC loss drives intestinal cancer through constitutive Wnt pathway activation, we examined whether Wnt hyperactivation could contribute to the TRE-MSI2 phenotype and, surprisingly, found no evidence of Wnt pathway activation, either at the level of APC loss, nuclear β-catenin localization, or β-catenin target gene activity. This result prompts a reassessment of the dogma that constitutive β-catenin activity is the sole oncogenic consequence of APC loss, a notion that is supported by recent evidence that constitutive β-catenin activation is not sufficient to recapitulate many phenotypes associated with APC loss28,29. Taken together, our findings suggest that MSI2 activation represents an oncogenic pathway downstream of APC loss and parallel to β-CATENIN activation.
This concept runs contrary to recent in vitro reports that the second mammalian MSI family member, MSI1 activates canonical Wnt signaling24,41,42, possibly reflecting differences between the in vivo setting used in the current study and the in vitro approaches taken in published studies, as well as tissue specific differences, as only one of these studies examined colorectal cancer cells in vitro41. Given the evidence for functional redundancy between the two mammalian MSI proteins26, and that MSI1 appears to have an oncogenic role in colorectal cancer cell lines23, we also examined Msi1 expression in our TRE-MSI2 mouse model and observed a striking induction of Msi1 at the mRNA level, however we conclude, through Msi1 loss of function studies, that the oncogenic properties of MSI2 are not dependent on Msi1, although this certainly does not rule out an oncogenic role for Msi1/MSI1 independently of Msi2/MSI2.
The co-expression of both Msi proteins in stem cells of the intestinal crypt (this study and10, the lack of any major intestinal phenotype described in either germline Msi1 knockout mice26 and our conditional Msi1flox/flox::VillinCreER mice (this study), along with the aforementioned functional redundancy of Msi proteins26 raises the intriguing possibility that Msi proteins act redundantly to maintain stem cell self renewal in the intestinal stem cell compartment, as we have previously observed for Msi2 in hematopoietic stem cells6. It will be important to address this possibility in future studies utilizing dual-Msi conditional knockout mice.
Transcriptome-wide RNA-binding analysis of endogenous Msi2 in intestinal crypts, ectopic MSI2 in the intestinal epithelium, and of MSI2 in human leukemic cell lines offers insight into the tissue-specific versus common functions of MSI2 in these tissues. In the intestinal transcriptome analyzed in this study and the leukemic transcriptome analyzed in6, MSI2 bound to transcripts involved in RNA biogenesis and metabolism, suggesting that MSI regulation of these fundamental biological processes may be conserved across tissues. Pathway analysis in hematopoietic tissues indicates MSI2 primarily acts to modulate TGF-β signalling, whereas in the intestinal epithelium, the Pten-AKT-mTORC1 axis was identified as being a major target of MSI2 activity, indicating that MSI2 may have cell type-specific functions (although the mTOR pathway was also identified as an MSI2 target in the hematopoietic system, however it appears to not be as dramatically effected as in the intestinal epithelium).
Conceptually, this phenomenon is not unexpected as RNA binding protein activity must, by definition, be restricted to the repertoire of transcripts expressed in a particular cell type. This is in contrast to transcription factors, whose DNA targets are identical in all cell types, epigenetic-regulated accessibility and cofactor binding notwithstanding. Thus, MSI2 and other RNA binding proteins might serve as hubs of signal integration by acting on messenger RNAs in the cytoplasm, with the possibility of cell type-specific function depending on the mRNA repertoire available for binding in any given cell type. In the current study, we identify Pten-AKT-mTORC1 axis as a major MSI2 target with functional relevance to the observed phenotype, however we note that MSI2 binds numerous additional tumor suppressors with known roles in colorectal cancer, including Cdkn1a, Lrig1, Bmpr1a and Numb. At least two of these targets (Bmpr1a and Lrig1, in addition to Pten) are also known repressors of mTorc1, and thus the effects of MSI2 on mTorc1 activity may be the result of pleiotropic inhibition of these tumor suppressors. While such pleiotropic inactivation of tumor suppressors may make MSI2 a potent oncogene, it also represents an attractive point for therapeutic intervention. In principle, MSI2 inhibition may allow for the concerted re-activation of this cadre of colorectal tumor suppressors. Thus, the viability of MSI2 as a drug target will be of great interest moving forward in the fight against colorectal cancer, and likely a variety of other cancers.
METHODS
Mouse strains
All procedures involving mice were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania (Animal Welfare Assurance Reference Number #A3079-01, approved protocol #803415 granted to Dr. Lengner) and were in accordance with the guidelines set forth in the Guide for the Care and Use of Laboratory Animals of the National Research Council of the National Institutes of Health.
The generation of TRE-MSI2 mice is described in4. Briefly, the human MSI2 cDNA (open biosystems clone ID#5942 Accession CV027635) under control of the tetracycline operator with a minimal CMV promoter was targeted to safe-haven chromatin downstream of the Collagen1a1 locus in KH2 embryonic stem cells also harboring the modified reverse tetracycline transactivator (M2rtTA) targeted to and under transcriptional control of the ROSA26 locus43. Lgr5-eGFP-CreER mice were obtained from Jackson Laboratories (stock #008875). Conditional loss of function Msi1flox/flox mice will be described in detail elsewhere. Briefly the two exons, including the transcriptional start site of Msi1 were targeted with flanking LoxP sites resulting in the 2lox, or floxed locus. Deletion of this region results in the loss of the transcriptional start site and a frame shift in the Msi1 coding sequence. Proper recombination at both the 5’ and 3’ end of the arms of homology was confirmed by Southern blotting. Mice generated from 2lox ES cells were bred to homozygosity and crossed with Villin-CreER mice17. Msi1 was conditionally deleted by administration of 5 consecutive doses of Tamoxifen (50mg/kg) in corn oil (Sigma), and loss of Msi1/Msi1 confirmed by Southern blotting, qRT-PCR, and immunostaining. Dox induction of MSI2 in TRE-MSI2::Msi1flox/flox::VillinCreER mice was initiated 5 days after the final dose of Tamoxifen. TRE-B-CatS3329 mice were a kind gift of Dr. Konrad Hochedlinger.
For ablation of APC, APCflox/flox mice44 were crossed with Villin-CreER mice and APC was deleted with 4 consecutive daily doses of Tamoxifen (50mg/kg) in corn oil (Sigma). Epithelial RNA was isolated 5 days after the initial Tamoxifen dose and processed for qRT-PCR as described below.
The generation of Msi2flox/flox mice is described in6. Gamma-irradiation after Msi2 deletion in Msi2flox/flox::Villin-CreER mice was performed on at least 4 mice in each of knockout and control groups, treated with 5 consecutive doses of Tamoxifen (50mg/kg) in corn oil (Sigma) followed by 5 days of recovery and exposure to 12Gy whole body γ-irradiation. Controls and experimental mice were sex-matched littermates. Intestines were washed with PBS, fixed in 4% paraformaldehyde, paraffin-embedded and sectioned. Surviving crypts were defined as 10 or more adjacent chromophilic cells and a lumen in hematoxylin and eosin stained sections from intestinal epithelium.
Doxycycline induction and isolation of intestinal epithelium
For Dox induction experiments of control (M2rtTA+/+) and TRE-MSI2+/−::M2rtTA+/+ mice, 2mg/ml Dox (Doxycycline hyclate, Sigma) was added to the drinking water along with 1% w/v sucrose of mice 8–10 weeks of age. All experiments except tumor formation in TRE-MSI2::APCmin/+ mice utilized the 2mg/ml Dox dosage in mice homozygous for M2rtTA and heterozygous for the TRE-MSI2 allele. Most experiments were conducted after 48 hours of exposure to Dox-containing drinking water, with the exception of transcriptome profiling which was carried out 24 hours after initiation of Dox treatment. For APCmin/+ experiments, TRE-MSI2+/−::APCmin/+::M2rtTA+/− or APCmin/+::M2rtTA+/− (in these experiments the M2rtTA was heterozygous to reduce MSI2 dosage) were administered 0.1 mg/ml of Dox in the drinking water for 3 weeks starting at 2 months of age while being fed low protein/high fat diet (Research Diets, D12079B). For isolation of whole intestinal epithelial cells, mouse intestine was cut longitudinally and washed 2–3 times with ice-cold DPBS, then cut into small pieces (3–7mm long) and incubated for 1 hour at 4 °C in DPBS containing 2 mM EDTA and 0.2 mM DTT on a rotating platform, and collecting suspended cells after gentle vortexing. For isolation of intestinal crypts, wash mouse small intestine with DPBS as above. The villi were scraped using a hemocytometer coverslip. The crypts were released by incubation for 30 min at 4 °C in DPBS containing 2 mM EDTA and gentle vortexing. Isolated crypts were counted and pelleted as described in45.
Flow cytometry
The intestine was cut open longitudinally and incubated with 5mM EDTA-HBSS solution at 4°c for 30min. Single cell suspension was generated with Dispase (BD Biosciences, San Jose, CA). Flow cytometry analysis was performed with BD LSR Fortessa cell analyzer (BD Biosciences, San Jose, CA). DAPI negative cells were selected, then gated for single cell based on Forward-scatter height versus forward-scatter width (FSC-H vs. FSC-W) and side-scatter height vs. side-scatter width (SSC-H vs. SSC-W) profiles. The size of the nozzle for all sorting is 100µm (20 psi). Lgr5+ stem cells were quantified by flow cytometry in TRE-MSI2::Lgr5-eGFP-IRES-CreER and M2rtTA::Lgr5-eGFP-IRES-CreER control mice after 48 hours of Dox treatment.
EdU labeling and flow cytometry
TRE-MSI2::Lgr5-eGFP-IRES-CreER and M2rtTA::Lgr5-eGFP-IRES-CreER control mice were treated with 2mg/ml doxycycline (Sigma) and were injected with 0.15 mg of 5-ethynyl-2´-deoxyuridine (Edu) (Life technologies) 2 hours or 12 hours before euthanasia and intestinal isolation. The intestine was cut open longitudinally and washed twice with ice-cold PBS. The intestine was incubated with 30 mM EDTA (EDTA, Sigma) and 1.5 mM DTT (Sigma) in HBSS at 4°C for 20 minutes. Then it was moved to 30 mM EDTA in HBSS at 37°C for 10 minutes and dissociated by pipetting vigorously. Single cell suspension was generated with 0.8 mg/ml Dispase (GIBCO)46. Cells were fixed with Click-iT fixative (Life technologies) for 15 minutes at room temperature and stained with FITC-anti-GFP (Abcam) for 30 minutes on ice and washed twice with Click-iT permeabilization and wash reagent. For Click-it labeling cells were incubated with CuSO4 and Alexa fluor 647 azide dye per the user manual and cells were analyzed using flow cytometry. Flow cytometry was performed on an LSR Fortessa and Flowjo software was used for data analysis.
PIP3 flow cytometry
Intestinal epithelial cells were suspended in PBS containing 1 uL/ml of fixable viability dye eFluor 450(Affymetrix ebioscience). Cells were incubated on ice for 30 minutes and were washed two times with PBS. Then, cells were fixed and permeabilized using Cytofix/ Cytoperm solution (BD PharMingen), stained with FITC-anti-GFP (Abcam) and biotinylated anti-PIP3 (Echelon inc.) for 30 min on ice and washed twice. They were incubated with Streptavidin-Allophycocyanin (APC) (biolegend) for 30 min on ice and washed twice. Flow cytometry was performed on an LSR Fortessa and Flowjo software was used for data analysis.
Administration of Rapamycin
Rapamycin (LC Laboratories) was administered by daily intraperitoneal injection (4 mg per kg of body weight) for 5 days. It was reconstituted in absolute ethanol at 10 mg/ml and diluted in 5% Tween-80 (Sigma) and 5% PEG-400 (Hampton Research) before injection. The final volume of all injections was 200 ul. Dox was administered to TRE-MSI2 mice as above for 48 hours after the third dose of Rapamycin.
Crypt organoid culture and qRT-PCR from intestinal epithelium and organoids
Crypt culture was performed as previously described in45. After intestinal crypt isolation, a total of 500 crypts were mixed with 50 µl of Matrigel (BD Bioscience) and plated in 24-well plates. After polymerization of Matrigel, 500 µl of crypt culture medium (Advanced DMEM/F12 containing growth factors (50 ng/ml EGF (Invitrogen), 1 µg/ml R-spondin 1 (Wistar Institute protein production facility), 100 ng/ml Noggin (Peprotech) and 3 µM GSK-3 inhibitor (CHIR99021, Stemgent) was added. After 2 days culture at 37 °C incubator, the organoids were treated with 2 µg/ml Dox and harvested for QRT-PCR analysis 72 hrs later.
RNA was isolated from organoids, crypts, or total epithelium, using QIAGEN RNeasy mini plus kits. 1 µg total RNA was used for cDNA synthesis. Primers used in qRT-PCR are as follows:
| Forward | Reverse | |
|---|---|---|
| Ascl2 | ATCTGCCCGGAGCATGGAA | CACTGCTGCAGGACTCCCTA |
| Axin2 | GATCCACGGAAACAGCTGAA | AGCCGGAACCTACGTGATAA |
| Chga | CCCGAAGTGACTTTGAGGAA | ATGGCTGACAGGCTCTCTA |
| Gapdh | AGACGGCCGCATCTTCTT | TTCACACCGACCTTCACCAT |
| Lgr5 | ACGCCTTTGGAAACCTCTCC | CAGGCTGTGGAGTCCATCAAA |
| Lyz2 | GTGACAGTCAGCCAACACAA | TGGCTGAAGAACTGACCTACA |
| Msi1 | GCCATGCTGATGTTCGACAA | CTACGATGTCCTCGCTCTCAA |
| Msi2 | GCGATGCTGATGTTCGACAA | TCTCCACAACGTCTTCATTCTCA |
| Sox9 | AGTACCCGCATCTGCACAA | GTCTCTTCTCGCTCTCGTTCA |
Histological analyses
Intestines were washed with DPBS, fixed in 10% Formalin, paraffin-embedded and sectioned. Hematoxylin, eosin, Alcian blue, and Alkaline Phosphatase staining was performed in the Morphology Core of the Penn Center for Molecular Studies in Digestive and Liver Diseases. For immunohistochemistry staining, antigen-retrieval was performed by heating slides in 0.01 M citrate buffer (pH 6) with a pressure cooker. The sections were then immunostained by the ABC peroxidase method (Vector labs) with diaminobenzidine (DAB) as the enzyme substrate and hematoxylin as a counterstain. For detection of nuclear β-catenin, antibody clone 15B8 (Sigma; 1:1000) was used in combination with the MOM kit (Vector Laboratories). For immunofluorescence staining, paraffin sections were pretreated in 0.01 M citrate buffer (pH 6) with a pressure cooker, and incubated in primary antibodies, then incubated with Cy2- or Cy3- conjugated fluorescent secondary antibodies (Jackson Laboratory) and counterstained with DAPI in mounting media (Vector labs). For amplification of signal from Msi1 and Msi2 we used a Biotin-Streptavidin system. The following antibodies were used: Ki67 (1:1250,Leica), Msi1 (1:400, MBL), Msi2 (1:200, Novus Biologicals), Lysozyme (1:50, Santa Cruz), Chromogramin A (1:1000, Abcam), GFP(1:200,Abcam), pS6 (1:200; Cell Signaling), p-AKT (T308) (1:200; Cell Signaling), p-AKT (S473) (1:200; Cell Signaling), p-4EBP1 (T37/46) (1:200; Cell Signaling).
CLIP-Seq
Clip libraries were made as previously described in47 with minor modification. Total intestinal epithelial cells from TRE-MSI2 mice treated with Dox for 24 hours were isolated as above, and wildtype intestinal crypts were isolated as above. For crosslinking, cell suspension was exposed to 2 pulses of 265nm UV light at 400mJ/cm2 in a Stratalinker (Model 2400, Stratagene). Epithelial cells were then lysed using PXL buffer (PBS, 01% SDS, 0.5% deoxycholate, 0.5% NP-40, plus protease inhibitor and RNAsin). The lysates were sequentially treated with DNaseI and RNase, and spin in ultra-microcentrifuge at 40,000g for 20 min. The supernatant was added to protein A Dynabeads (Dynal, 100.02) conjugated with MSI2 antibody (EMD Millipore 03-115) and incubated for 4 hours at 4 °C. 32P-γ-ATP labeled 3’ RNA (RL-3) linker was ligated to the RNA fragment on beads overnight at 16°C. The beads were re-suspended in 30 µl of Novex loading buffer (without reducing agent), and separated with Novex NuPAGE 10% Bis-Tris gel and transferred to S&S BA-85 nitrocellulose membrane. After overnight exposure, around 50 KD band was visualized and the corresponding membrane was cut to small pieces. The RNA was released by proteinase K digestion and isolated using RNA phenol and CHCl3 solution. 5’ RNA (RL-5) linker was ligated into the RNA fragments. The RNA was transcribed into complementary DNA using RT-PCR and amplified using Re-PCR with Solexa fusion primers. The CLIP library underwent single-end sequencing on an Illumina hiSeq2000 at the University of Pennsylvania Functional Genomics Core. RL-3: 5’-OH GUG UCA GUC ACU UCC AGC GG 3’ –puromycin; RL-5: 5’ –OH AGG GAG GAC GAU GCG G 3’-OH. Adapter sequences were removed from the 3' end using cutadapt (http://journal.embnet.org/index.php/embnetjournal/article/view/200) with options '-a GTGTCAGTCACTTCCAGCG -e 0.06 -O 6 -m 12'. Reads were then mapped to the mouse (mm9) transcriptome and genome using Tophat48 with options '--read-mismatches 1 --read-gap-length 1 --read-edit-dist 1 --max-multihits 100 --b2-very-sensitive --transcriptome-max-hits 100 --no-coverage-search --no-novel-juncs'. Peaks were called using Piranha49 with a window size of 200bp. Motifs were identified using the MEME software suite50.
Transcriptome profiling
Total RNA was isolated from total mouse small intestinal epithelial cells from 3 M2rtTA and 3 TRE-MSI2 mice administered Dox for 24 hours in the drinking water (as above) using TRIzol Reagent (Life Technologies) according to the manufacturer’s instructions. Total RNA was DNAse treated with an RNAse-free DNAse kit (Zymo Research). Purified RNA was submitted to the University of Pennsylvania Molecular Profiling Core, where samples were labeled and hybridized to Affymetrix Mouse Gene 1.0ST arrays. Microarray data was analyzed using Partek® Genomics SuiteTM software. Following RMA background subtraction and normalization, a 1-way-ANOVA analysis between controls (M2rtTA) and MSI2 induced (TRE-MSI2) was run to compute p-values of significance and F-statistic for each probeset. q-value, a measure of false discovery rate (FDR) was computed within the SAM (Significance Analysis of Microarrays) software for each probeset by running an unpaired t-test. The FDR values were integrated with the 1-way-ANOVA results. Genes that were significant at FDR cut-off of 5% and changed at least 2-fold in either direction in the MSI2 induction group when compared to the control group were selected as the set of differentially expressed genes. This set of 836 unique genes and 6 samples were subjected to agglomerative hierarchical clustering analysis. Log2 intensities were median-centered across samples. Euclidean distance was used as the dissimilarity metric and average linkage method as clustering strategy. Results were visualized as intensity heatmap (Figure 5A).
For analysis of direct β-catenin target gene expression in Figure 5F, genes with published evidence for direct regulation by β-catenin demonstrated by chromatin immunoprecipitation were selected. These genes include Edn1, Fra-1 (Fosl1), c-Myc, Met, c-Jun, Pinx1, Tcf4, Egfr, Klf5, Cyclin D (Ccnd1), Mycbp, Mmp7, Cdkn2a, Vegfa, SNAI1, Fgf18, Gja1 (connexin-43), Axin2, Claudin1, Runx2, Lef1, Bglap, ENPP2, Nrcam, DLK1, Vcan, Fgf4, Fn1, Tcf1, (Hnf1a), Lgr5, Ppard, Sp5, Ovol1, Id2, L1CAM, Cdh1 (E-cadherin), Fst, Tnfrsf19, Neurod1, Ctla4, Wisp1, Fzd7, Mitf, Pou3f2, Gbx2, Nkx2-2, Neurog1, Eda, T, Pitx2, Btrc, Cdx1, Birc5 (survivin), Pkd1, Pml, and Lect2 (chemotaxin2).
Oncomine and TCGA
Using Oncomine analysis, MSI2 expression level was analyzed in the Kaiser colon database. The database includes full transcriptome profiles of 105 samples: Control (5); Cecum Adenocarcinoma (17); Colon Adenocarcinoma (41); Colon Mucinous Adenocarcinoma (13); Colon Signet Ring Cell Adenocarcinoma (2); Colon Small Cell Carcinoma (2); Rectal Adenocarinoma (8); Rectal Mucinous Adenocarcinoma (4); Rectal Signet Ring Cell Adenocarcinoma (1); Rectosigmoid Adenocarcinoma (10); Rectosigmoid Mucinous Adenocarcinoma (2).51 For TCGA analysis, mRNA expression levels from RNA-Seq on TCGA COAD tumor samples (314 solid tissue samples from healthy individuals, 314 tumor samples). Expression values were obtained by TMM normalization of read counts in genes. Fold changes for MSI2 in matched tumor/control RNA-Seq sample pairs from TCGA COAD were calculated (total of 26 patients.) Distribution of MSI2 fold changes in tumor/control pairs for 26 individuals plotted are in red (intra-individual comparison). Distribution of MSI2 fold changes between control/control comparisons for 26 distinct individuals plotted in grey (inter-individual comparison).
Cell proliferation assays
SW48 (ATCC stock number CCL-231) and HT29 (ATCC stock number HTB-38) were seeded in 96-well plates at a density of 1500 and 2000 cell and cultured in DMEM supplemented with 10% FBS, 1% sodium pyruvate, 1% penn/strep, 1%L-glutamine 24hrs before infection. For infection, medium was removed, and cells gently washed with PBS. 100 ul viral supernatant containing pSICO-eGFP-shScramble, -sh-MSI2, -sh-BCAT, and -sh-MSI2 & -sh-BCAT. Cell proliferation assays were performed using MTT (3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide) assays. Absorbance was measured at 570nm. All data ware expressed as a mean from at least three independent biological experiments with n=4 per condition per experiment.
Xenograft Assays
6-week-old female nude mice were obtained from the Stem Cell and Xenograft Core at UPENN. Stably infected colorectal cancer cells were trypsinized and suspended in phosphate-buffered saline (PBS). A total volume of 0.2 ml containing 2–5 × 106 cells and 25% volume MatriGEL was injected subcutaneously into the mouse flank. Tumor size was measured using a Vernier caliper. Tumor volumes were calculated using the formula V=1/2 (L×W2), where L is length (longest dimension) and W is width (shortest dimension) of the tumor. Moribund animals were euthanized according to the protocols of the University of Pennsylvania. Tumor growth rates in the xenograft experiment were evaluated by fitting a linear mixed effects model on the log10-transformed tumor volume with days, experiment indicator (MSI shRNA versus control shRNA), and interaction between days and experiment included as independent variables.
TOPFlash Assays
TOP/FOPFlash assays were carried out using the Dual-Luciferase Reporter Assay System (Promega), 293 cells were seeded in 24 well plate 12hrs before cotransfection with 1200ng scrambled hairpin, Sh-MSI2, Sh-B-CAT, or pcDNA-MSI2 and 400ng M50 Super 8×TOPflash or M50 Super 8×FOPflash reporter plasmids per well (Addgene plasmids 12456 and 12457). FugeneHD was used to mediate co-transfection, and the media contained 3nM CHIR99021 GSK3β inhibitor to insure Wnt pathway activation. The Renilla luciferase reporter vector pRL-SV40 (200 ng) was simultaneously transfected as the control for Firefly luciferase in TOP/FOP vectors. Cells were harvested 48 hrs after transfection. Experiments were performed at least twice with n=4 wells as technical replicates in each experiment.
Western Blots
Cells were lysed in RIPA buffer with protease inhibitors (Roche). After quantification using a BCA protein assay kit (Pierce), 40 µg of total protein was separated by 10% SDS-PAGE under denaturing conditions and transferred to PVDF membranes (GE Healthcare). Membranes were blocked in 5% BSA (Sigma) and then incubated with a Msi2 primary antibody (1:1000; Novus Biologicals), Msi1 (1:1,000; MBL), NICD (1:1,000; Abcam), Hes1 (1:1,000; Abcam), β-actin (1:10000, Abcam), or Pten (1:1000, Cell Signaling) APC (1:200, Santa Cruz sc-896) followed by incubation with a secondary antibody conjugated with horseradish peroxidase (HRP) (1:2,000; Cell Signaling) together with an HRP-conjugated primary antibody for β-actin (1:10,000; Sigma). Immunoreactive proteins were visualized using LumiGLO chemiluminescent substrate (Pierce). Raw scans of Western blots in Figures 4C, 5C, 6F, and 7A can be found in the supplementary information accompanying the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to acknowledge members of the University of Pennsylvania P01 Center for Molecular Studies in Digestive and Liver Diseases for technical support and thoughtful discussions, particularly the Drs. Anil Rustgi and John Lynch and members of their laboratories, as well as the Center’s Molecular Pathology and Imaging core for histological processing, microscopy, and analysis. GM was supported by Mr. William H. and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center. MY was supported by a Howard Hughes Medical Institute International Student Fellowship. MGK was supported by the US National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Career Development Award and NIDDK NIH R01-DK101989-01A1, Louis V Gerstner Young Investigator Award and the American Society of Hematology Junior Scholar Award, Kimmel Scholar Award and V-Scholar Award. ZY was supported by the National Natural Science Foundation of China (NSFC, 31271584), the National Basic Research program of China (973 program, 2011CB944103) and the National Transgenic Breeding Project of China (2011ZX08009-001-003). CJL was supported by a pilot award from the Penn P01 Center for Molecular Studies in Digestive and Liver Diseases, a grant from the State of Pennsylvania Health Research Formula Fund, a Fellowship from the W.W. Smith Charitable Trust, and R01 CA16865 from the NCI. This work was supported in part by, the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30DK050306) and its core facilities.
Footnotes
AUTHOR CONTRIBUTION
CJL and MGK designed and developed the Msi mouse models used in the study. SW, NL, ZY, and CJL designed and performed all experiments unless noted otherwise below. MY performed mTOR and PIP3 analysis, irradiation, and stem cell flow cytometry, AND and KP performed mouse husbandry, genotype analyses, and technical support. FL and BDG performed analysis of CLIP-Seq data. SR performed gene expression analyses. GM performed electrophoretic mobility shift assays. YK performed TCGA data analysis. CJL and SW wrote the manuscript with editorial support from NL, ZY, and MGK.
COMPETING FINANCIAL INTEREST STATEMENT
None of the authors have competing financial interests.
REFERENCES
- 1.Okano H, et al. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 2.Sakakibara S, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Developmental biology. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 3.Sakakibara S, Nakamura Y, Satoh H, Okano H. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2001;21:8091–8107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kharas MG, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nature medicine. 2010;16:903–908. doi: 10.1038/nm.2187. PCMID:3090658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito T, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. PCMID:2918284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SM, et al. Musashi-2 controls cell fate, lineage bias, and TGF-beta signaling in HSCs. The Journal of experimental medicine. 2014;211:71–87. doi: 10.1084/jem.20130736. PCMID:3892968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, et al. Musashi2 predicts poor prognosis and invasion in hepatocellular carcinoma by driving epithelial-mesenchymal transition. Journal of cellular and molecular medicine. 2014;18:49–58. doi: 10.1111/jcmm.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, et al. Expression of seven stem-cell-associated markers in human airway biopsy specimens obtained via fiberoptic bronchoscopy. Journal of experimental & clinical cancer research : CR. 2013;32:28. doi: 10.1186/1756-9966-32-28. PCMID:3689624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hope KJ, et al. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell stem cell. 2010;7:101–113. doi: 10.1016/j.stem.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Potten CS, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation; research in biological diversity. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 11.Li D, et al. Msi-1 is a predictor of survival and a novel therapeutic target in colon cancer. Annals of surgical oncology. 2011;18:2074–2083. doi: 10.1245/s10434-011-1567-9. [DOI] [PubMed] [Google Scholar]
- 12.Fan LF, et al. Expression of putative stem cell genes Musashi-1 and beta1-integrin in human colorectal adenomas and adenocarcinomas. Int J Colorectal Dis. 2010;25:17–23. doi: 10.1007/s00384-009-0791-2. [DOI] [PubMed] [Google Scholar]
- 13.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi Y, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Human molecular genetics. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 15.Nagase H, et al. Screening for germ-line mutations in familial adenomatous polyposis patients: 61 new patients and a summary of 150 unrelated patients. Human mutation. 1992;1:467–473. doi: 10.1002/humu.1380010603. [DOI] [PubMed] [Google Scholar]
- 16.Kinzler KW, et al. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 17.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 18.van de Wetering M, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 19.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 20.Sansom OJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes & development. 2004;18:1385–1390. doi: 10.1101/gad.287404. PCMID:423189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasan HS, et al. APC in the regulation of intestinal crypt fission. The Journal of pathology. 1998;185:246–255. doi: 10.1002/(SICI)1096-9896(199807)185:3<246::AID-PATH90>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 22.Asai R, Okano H, Yasugi S. Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Development, growth & differentiation. 2005;47:501–510. doi: 10.1111/j.1440-169X.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- 23.Sureban SM, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 24.Rezza A, et al. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. Journal of cell science. 2010;123:3256–3265. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 25.Katz Y, et al. Musashi proteins are post-transcriptional regulators of the epithelial-luminal cell state. eLife. 2014;3 doi: 10.7554/eLife.03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakakibara S, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. PCMID:1239896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elyada E, et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470:409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 29.Hirata A, et al. Dose-dependent roles for canonical Wnt signalling in de novo crypt formation and cell cycle properties of the colonic epithelium. Development. 2013;140:66–75. doi: 10.1242/dev.084103. PCMID:3513993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imai T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Molecular and cellular biology. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. doi: 10.1016/j.mcn.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Powell AE, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. PCMID:3563328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howe JR, et al. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nature genetics. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 34.He XC, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nature genetics. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goel A, et al. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer research. 2004;64:3014–3021. doi: 10.1158/0008-5472.can-2401-2. [DOI] [PubMed] [Google Scholar]
- 36.Naguib A, et al. Alterations in PTEN and PIK3CA in colorectal cancers in the EPIC Norfolk study: associations with clinicopathological and dietary factors. BMC cancer. 2011;11:123. doi: 10.1186/1471-2407-11-123. PCMID:3080834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marsh V, et al. Epithelial Pten is dispensable for intestinal homeostasis but suppresses adenoma development and progression after Apc mutation. Nature genetics. 2008;40:1436–1444. doi: 10.1038/ng.256. [DOI] [PubMed] [Google Scholar]
- 38.Stocker H, et al. Living with lethal PIP3 levels: viability of flies lacking PTEN restored by a PH domain mutation in Akt/PKB. Science. 2002;295:2088–2091. doi: 10.1126/science.1068094. [DOI] [PubMed] [Google Scholar]
- 39.Ashton GH, et al. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Developmental cell. 2010;19:259–269. doi: 10.1016/j.devcel.2010.07.015. PCMID:3291717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujishita T, Aoki K, Lane HA, Aoki M, Taketo MM. Inhibition of the mTORC1 pathway suppresses intestinal polyp formation and reduces mortality in ApcDelta716 mice. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13544–13549. doi: 10.1073/pnas.0800041105. PCMID:2533226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spears E, Neufeld KL. Novel double-negative feedback loop between adenomatous polyposis coli and Musashi1 in colon epithelia. The Journal of biological chemistry. 2011;286:4946–4950. doi: 10.1074/jbc.C110.205922. PCMID:3037606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang XY, et al. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Molecular and cellular biology. 2008;28:3589–3599. doi: 10.1128/MCB.00040-08. PCMID:2423292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 44.Shibata H, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 45.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 46.Gracz AD, Puthoff BJ, Magness ST. Identification, isolation, and culture of intestinal epithelial stem cells from murine intestine. Methods Mol Biol. 879:89–107. doi: 10.1007/978-1-61779-815-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. doi:nature08170 [pii] 10.1038/nature08170 PCMID:2733940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. PCMID:2672628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uren PJ, et al. Site identification in high-throughput RNA-protein interaction data. Bioinformatics. 2012;28:3013–3020. doi: 10.1093/bioinformatics/bts569. PCMID:3509493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. PCMID:2703892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. PCMID:1813932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.