Abstract
Viral reactivation from latently infected cells has become a promising therapeutic approach to eradicate HIV. Due to the complexity of the viral latency, combinations of efficient and available drugs targeting different pathways of latency are needed. In this work, we evaluated the effect of various combinations of bryostatin-1 (BRY) and novel histone deacetylase inhibitors (HDACIs) on HIV-reactivation and on cellular phenotype. The lymphocyte (J89GFP) or monocyte/macrophage (THP89GFP) latently infected cell lines were treated with BRY, panobinostat (PNB) and romidepsin (RMD) either alone or in combination. Thus, the effect on the viral reactivation was evaluated. We calculated the combination index for each drug combination; the BRY/HDACIs showed a synergistic HIV-reactivation profile in the majority of the combinations tested, whereas non-synergistic effects were observed when PNB was mixed with RMD. Indeed, the 75% effective concentrations of BRY, PNB and RMD were reduced in these combinations. Moreover, primary CD4 T cells treated with such drug combinations presented similar activation and proliferation profiles in comparison with single drug treated cells. Summing up, combinations between BRY, PNB and/or RMD presented a synergistic profile by inducing virus expression in HIV-latently infected cells, rendering these combinations an attractive novel and safe option for future clinical trials.
Effective combination antiretroviral therapy (cART) has improved the quality of life and the life expectancy of HIV-infected patients. Nevertheless, achieving the cure of HIV is still an unattainable challenge for the scientific community. Although cART achieves undetectable plasma viral RNA and the normalization of CD4 T cell levels in the majority of patients, several studies have shown that HIV remains incurable owing to the persistence of latently infected cells1,2,3,4. Most of these cells are resting memory or naïve CD4 T cells and other cells belonging to the monocyte/macrophage lineage that contain integrated provirus within their genome5,6. This latent infection escapes from the cART effect and remains undetectable to the immune system.
Several therapeutic interventions to eradicate HIV focus on the stimulation of viral production from latently infected cells. This is followed by a “kill” phase that permits the elimination of infected cells through existing immune responses or cytotoxic drugs under the aim to purge and clear HIV reservoirs. This strategy involves the use of a wide range of small molecules called latency-reversing agents (LRAs)7. Such drugs include: (1) histone deacetylase inhibitors (HDACIs)8,9 (2) disulfiram, postulated to involve the nuclear factor κB (NF-κB) activation10,11 (3) bromodomain-containing protein 4 (BRD4) inhibitor JQ1, which elicits effects through positive transcription elongation factor (P-TEFb)12 and (4) protein kinase C (PKC) activators such as ingenols13, prostratin14, 1,2-diacylglycerol analogues15 and bryostatin-1 (BRY)16,17. Not only the interest in these drugs has grown greatly, but also the number of on-going clinical trials about the safety and the effect of LRAs as disruptors of HIV latency have increased. HDACIs are the most advanced HIV-1 anti-latency agents in current clinical testing, mainly due to the synthesis in recent years of novel and more specific pan-HDACIs, such as givinostat, belinostat and panobinostat (PNB)18,19 and newly synthesized class I selective HDACIs that include oxamflatin20, NCH-5121 and romidepsin (RMD)22. Recently, published results validate the safety and the effect of PNB on HIV expression in patients on suppressive cART in a clinical trial, and postulated this compound as a promising reactivator of HIV viral latency. However, this study reveals that PNB did not reduce the number of latently infected cells and must be combined with other drugs in order to significantly affect the size of HIV reservoirs23. Consistent with these results Bullen, C. K., et al. have shown a comparative ex vivo evaluation of different LRAs demonstrating that none of the leading candidate drugs can singly disrupt the latent HIV reservoir. Thereby, the combination of several LRAs can be the best strategy for HIV latency reactivation and precludes possible synergisms24. Indeed, some published results described possible synergisms between the classic HDACIs (valproic acid, vorinostat and sodium butyrate) and either prostratin25 or BRY26 in HIV expression activation, possibly due to the other role attributed to HDACIs as stimuli promoting NF-κB activity27. Many other combinatorial approaches have been postulated as good strategies to induce the reactivation of latent reservoirs24,28,29. Therefore, we study the possible synergism between the new promising HDACIs PNB or RMD and BRY as non-carcinogenic PKC activator, in order to reveal new insights in LRA combinatorial strategies that could be useful for future clinical trials design.
Results
Enhanced reactivation profile of bryostatin-1 and HDACIs combinations in latently HIV-1 infected cells
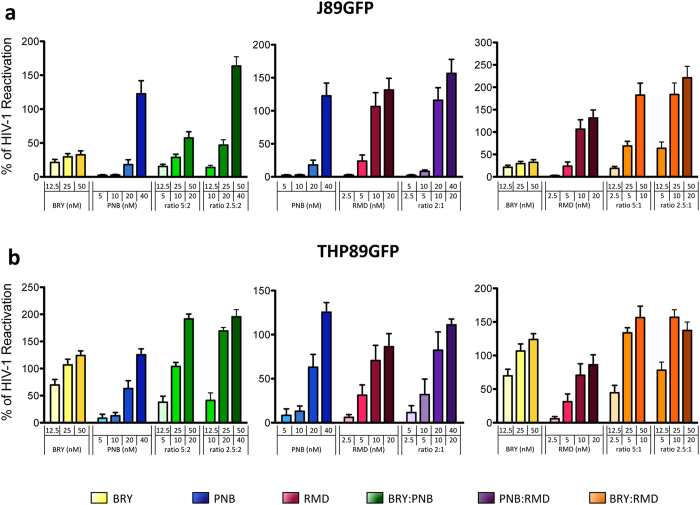
As an experimentally tractable and relevant model to study post-integration HIV latency and reactivation30, we employed J89GFP and THP89GFP, which are respectively lymphocyte and monocyte-derived cell lines latently infected by HIV that carry out a copy of latent EGFP under the control of HIV promoter. The HIV reactivation effect of BRY, PNB and RMD was tested as individual drugs or in combination at different ratios. The election of the ratios was performed taking into account the non-toxic concentration for each drug (BRY: 12.5–50 nM, PNB: 5–40 nM, and RMD: 2.5–20 nM) (see Supplementary Figs S1 and S2 online). After 1 day of exposure, the reactivation effect was measured by flow cytometry and expressed as EGFP (integrated MFI or iMFI, see Supplementary Fig. S3 online). Moreover, flow-cytometry profiles of a representative experiment are depicted in Supplementary Fig. S4 online, showing both cell viability and HIV-reactivation by 7AAD staining and EGFP detection, respectively. Nevertheless, to avoid experimental deviations, results were normalized according to the EGFP expression profile of TNF-α treated cells (Fig. 1).
Figure 1. Reactivation-effect of drugs alone or in double combinations.
J89GFP (a) and THP89GFP (b) cell lines were treated with BRY (yellow), PNB (blue) and RMD (red) at the indicated concentrations, alone or in the following double combinations: BRY:PNB (green), PNB:RMD (purple) and BRY:RMD (orange) at the specified ratios. After 24 hours, HIV reactivation was analysed by flow cytometry as EGFP expression (iMFI). Percentage of HIV reactivation was normalized to TNF-induced viral reactivation. Results represent the arithmetic mean + SEM of at least three independent experiments.
To research the reactivation activity of each individual drug in these HIV latency models, we analysed the dose-effect curves in the two cell lines (see Supplementary Fig. S5 online), and we determined the effective concentration (EC50, EC75, EC90, and EC95) of each drug (Table 1). The effective concentration of the drugs in J89GFP cells was higher than in THP89GFP and BRY. The doses needed to obtain an effective reactivation in J89GFP cells were extremely high (micromolar range), exceeding the commonly used nanomolar range. As shown in Fig. 1 and Supplementary Fig. S3 online, all of the tested combinations containing BRY and HDACIs resulted in a more efficient reactivation profile in either J89GFP and THP89GFP cell lines compared to the effects of the individual drugs. On the contrary, the combination of PNB and RMD, slightly improved the single drug effect.
Table 1. Dose-effect values of single drugs.
| Cell type | Individual Drug | Dose-effect values |
|||
|---|---|---|---|---|---|
| EC50 | EC75 | EC90 | EC95 | ||
| J89GFP | BRY (μM) | 0.5 ± 0.18 | 2.02 ± 0.76 | 8.23 ± 3.41 | 21.48 ± 9.58 |
| PNB (nM) | 49.85 ± 12.65 | 80.31 ± 25.32 | 130.36 ± 50.69 | 182.04 ± 80.92 | |
| RMD (nM) | 12.88 ± 2.88 | 20.17 ± 5.17 | 31.87 ± 10.13 | 43.72 ± 16.42 | |
| THP89GFP | BRY (nM) | 17.3 ± 8.7 | 31.4 ± 9.8 | 62.6 ± 23.3 | 105.5 ± 55.6 |
| PNB (nM) | 19.34 ± 6.43 | 28.87 ± 10.41 | 44.19 ± 18.99 | 60.02 ± 30.38 | |
| RMD (nM) | 10.55 ± 4.09 | 19.07 ± 6.9 | 35.83 ± 14.04 | 56.7 ± 27.87 | |
The effective concentration (EC) for each drug after a 24-hour treatment of J89GFP and THP89GFP cells normalized to TNF-induced viral reactivation. Data are mean ± SD from at least six independent experiments performed in triplicate.
To evaluate the type of drug interactions, we calculated the combination index (CI) values (Table 2). For this purpose, the CalcuSyn software based on the median effect principle was used31,32. The obtained CI values demonstrated the positive interactions between BRY and HDACIs, and synergy was observed for the majority of the BRY+PNB and BRY+RMD combinations in both HIV latently infected cell lines. The only exception was the EC50 values in THP89GFP cell line, which showed non-synergism. Nevertheless, very strong or strong synergistic interactions were observed at higher effective doses. On the other hand, non-synergistic or additive effects were observed when PNB was mixed with RMD. Since enhanced reactivation effects and generally synergistic profiles were observed when BRY was combined with the HDACIs, the use of both HDACIs must be challenged for future utilization. Consequently, even though slight or no synergism was observed between PNB and RMD, we wondered whether the triple combination BRY+PNB+RMD could render a positive interaction. The ratios assessed in the double combinations were maintained, resulting in two different triple combination ratios 2.5:2:1 and 5:2:1. As depicted in Table 3, a potent synergistic profile (CI values between 0.22–0.76) was found at calculated EC50, EC75, EC90, and EC95 for both ratios in J89GFP cells, whereas a moderate to strong synergistic effect (CI values between 0.4–0.84) tended to appear only at the highest calculated effective doses (EC90, and EC95) in THP89GFP cells. Summing up, we demonstrated that BRY is highly efficient in combination with HDACIs and that PNB and RMD can be used efficiently when mixed with BRY to reactivate HIV from latency in our cell line models.
Table 2. Combination Index values (CI) of double combinations.
| Double combination | Ratio | J89GFP |
THP89GFP |
||||||
|---|---|---|---|---|---|---|---|---|---|
| EC50 | EC75 | EC90 | EC95 | EC50 | EC75 | EC90 | EC95 | ||
| BRY:PNB | 2.5:2 | 0.56 ± 0.11 (+++) | 0.48 ± 0.1 (+++) | 0.42 ± 0.09 (+++) | 0.38 ± 0.09 (+++) | 1.16 ± 0.24 (−) | 0.65 ± 0.11 (+++) | 0.44 ± 0.12 (+++) | 0.35 ± 0.11 (+++) |
| 5:2 | 0.41 ± 0.3 (+++) | 0.34 ± 0.21 (+++) | 0.33 ± 0.19 (+++) | 0.34 ± 0.2 (+++) | 1.27 ± 0.29 (−) | 0.69 ± 0.32 (+++) | 0.39 ± 0.2 (+++) | 0.29 ± 0.15 (++++) | |
| PNB:RMD | 2:1 | 1.11 ± 0.1 (−) | 0.92 ± 0.08 (±) | 0.76 ± 0.06 (++) | 0.67 ± 0.05 (+++) | 2.05 ± 0.58 (−) | 2.3 ± 0.37 (−) | 2.71 ± 0.45 (−) | 3.09 ± 0.79 (−) |
| BRY:RMD | 2.5:1 | 0.47 ± 0.04 (+++) | 0.26 ± 0.07 (++++) | 0.16 ± 0.07 (++++) | 0.11 ± 0.06 (++++) | 0.88 ± 0.2 (+) | 0.73 ± 0.29 (++) | 0.64 ± 0.3 (+++) | 0.63 ± 0.26 (+++) |
| 5:2 | 0.46 ± 0.04 (+++) | 0.38 ± 0.05 (+++) | 0.32 ± 0.05 (+++) | 0.28 ± 0.05 (++++) | 1.34 ± 0.42 (−) | 0.87 ± 0.28 (+) | 0.58 ± 0.18 (+++) | 0.46 ± 0.15 (+++) | |
CI calculated at the EC50, EC75, and EC90. CI < 0.9 indicates synergism; 0.9 < CI < 1.1 indicates additive effects, and CI > 1.1 indicates antagonism (−). Synergy level: ± indicates additive effects; 0.85 < CI < 0.9 + (slight synergism); 0.7 < CI < 0.85 ++ (moderate synergism); 0.3 < CI < 0.7 +++ (synergism); CI < 0.1 < 0.3 ++++ (potent synergism). Compound range concentration: BRY (12.5–50 nM), PNB (5–40 nM) and RMD (2.5–20 nM). Each experiment was performed in duplicate. Data are represented as the mean ± SD of at least three independent experiments. Abbreviations: CI, combination index; EC50, 50% effective concentration; EC75, 75% effective concentration; EC90, 90% effective concentration; EC95, 95% effective concentration; SD, standard deviation; BRY, bryostatin-1; PNB, panobinostat; RMD, romidepsin.
Table 3. Combination Index values (CI) of triple combinations BRY:PNB:RMD.
| Cell type | ratio 2.5:2:1 | ratio 5:2:1 | ||||||
|---|---|---|---|---|---|---|---|---|
| EC50 | EC75 | EC90 | EC95 | EC50 | EC75 | EC90 | EC95 | |
| J89GFP | 0.76 ± 0.16 (++) | 0.57 ± 0.1 (+++) | 0.42 ± 0.07 (+++) | 0.35 ± 0.05 (+++) | 0.47 ± 0.10 (+++) | 0.35 ± 0.07 (+++) | 0.26 ± 0.05 (++++) | 0.22 ± 0.04 (++++) |
| THP89GFP | 4.39 ± 2.02 (−) | 1.92 ± 0.22 (−) | 0.69 ± 0.11 (+++) | 0.46 ± 0.1 (+++) | 1.52 ± 0.62 (−) | 0.84 ± 0.24 (++) | 0.52 ± 0.09 (+++) | 0.4 ± 0.06 (+++) |
CI calculated at the EC50, EC75, EC90 and EC95. CI < 0.9 indicates synergism; 0.9 < CI < 1.1 indicates additive effects, and CI > 1.1 indicates antagonism (−). Synergy level: ± indicates additive effects; 0.85 < CI < 0.9 + (slight synergism); 0.7 < CI < 0.85 ++ (moderate synergism); 0.3 < CI < 0.7 +++ (synergism); CI < 0.1 < 0.3 ++++ (potent synergism). Compound range concentration: BRY (12.5–50 nM), PNB (5–40 nM) and RMD (2.5–20 nM). Each experiment was performed in duplicate. Data are represented as the mean ± SD of at least three independent experiments. Abbreviations: CI, combination index; EC50, 50% effective concentration; EC75, 75% effective concentration; EC90, 90% effective concentration; EC95, 95% effective concentration; SD, standard deviation; BRY, bryostatin-1; PNB, panobinostat; RMD, romidepsin.
Reduction of effective concentration values when bryostatin-1 and HDACIs were used in double or triple combinations
Once demonstrated the synergistic effect of BRY/HDACIs combinations, we analysed their implications in terms of effective concentration. Based on the in vitro results, the CalcuSyn software permits to determine the dose-reduction-index of each drug32,33. Therefore, the effective concentration of each single drug when combined with others can be predicted. We first analysed the reactivation effect of the two-drug combinations containing BRY+PNB, BRY+RMD and PNB+RMD. All the evaluated combinations achieved a statistically significant reduction in the EC75 values, as compared with the EC75 values for each individual drug, in both J89GFP (Fig. 2) and THP89GFP (Table 4) cell lines. For example, BRY alone reactivated HIV expression in J89GFP cells with an average EC75 of 2,020 nM (Fig. 2), meanwhile the combination of BRY and PNB (ratio 2.5:2) or RMD (ratio 2.5:1) reactivated latent HIV with an average EC75 of 24.81 or 19.53 nM, respectively. Therefore, to achieve the same level of HIV reactivation, 80- and 100-fold less concentration of BRY in combination with PNB and RMD respectively was needed, when comparing the effect of BRY as a single drug. The EC75 of PNB and RMD were also reduced in these combinations, showing an average of 4-fold decrease in the concentrations used. In addition, the EC75 of each drug in the triple combination BRY:PNB:RMD (at 5:2:1 ratio), reached very similar EC75 average values in both cell lines: 23.57 nM, 9.45 nM and 4.72 nM (J89GFP) and 20.68 nM, 8.27 nM and 4.14 nM (THP89GFP) for BRY, PNB and RMD, respectively (Fig. 2 and Table 4).
Figure 2. Dose-effect EC75 values of drugs alone or in combinations in J89GFP cells.
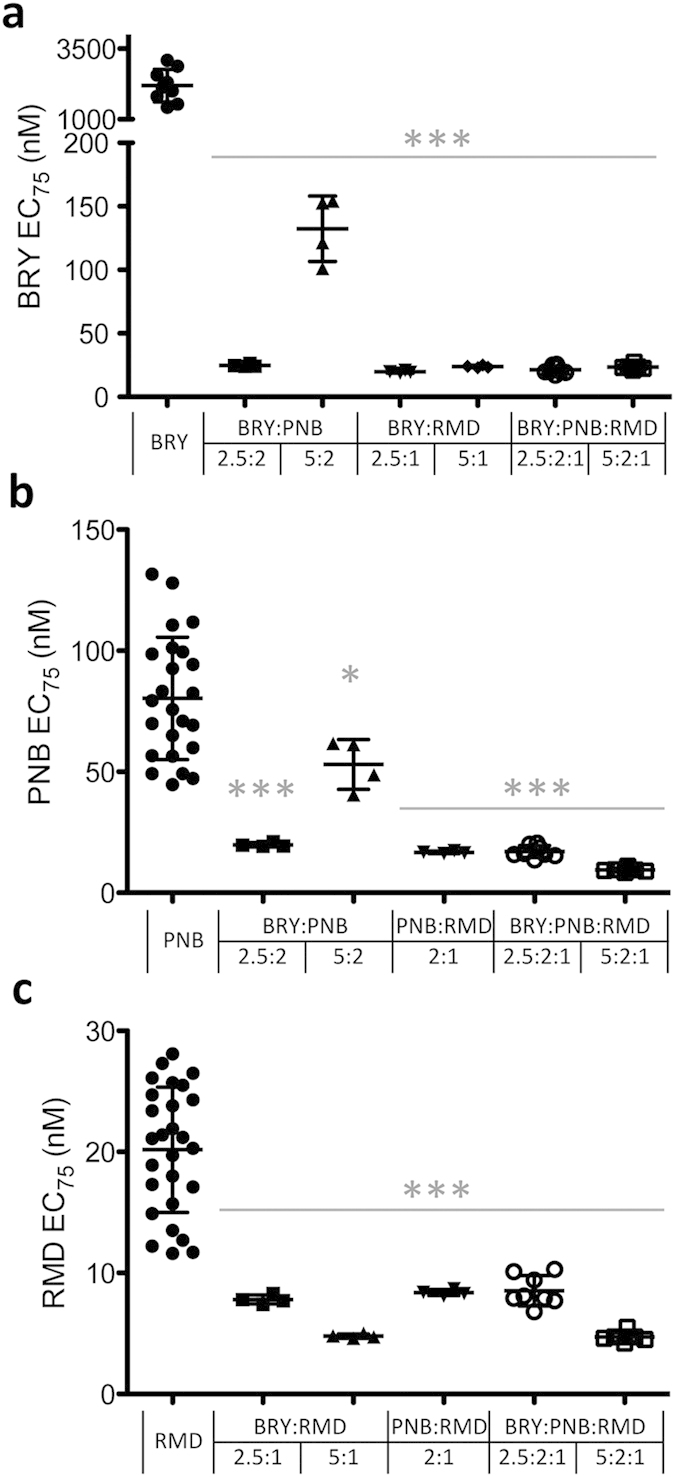
The 75% effective reactivation concentration (EC75) values (nM ± SD) in latently infected J89GFP cells of BRY (a), PNB (b) and RMD (c) drugs alone and in combination at the indicated ratios 24 hours post-treatment are shown. Each symbol represents results from of at least three independent experiments and the mean ± SD is shown. Statistics were performed between the calculated EC75 values of the drug at single and combined treatments. ***p < 0.001; *p < 0.05.
Table 4. Dose-effect EC75 values of drugs alone or in combinations in THP89GFP cells.
| Individual Drug | Double combination |
Triple combination |
||||||
|---|---|---|---|---|---|---|---|---|
| BRY:PNB (2.5:2) | BRY:PNB (5:2) | PNB:RMD (2:1) | BRY:RMD (2.5:1) | BRY:RMD (5:1) | BRY:PNB:RMD (2.5:2:1) | (5:2:1) | ||
| BRY (nM) | 31.4 ± 9.8 | 21.1 ± 3.78* | 26.67 ± 10.32 | 15.49 ± 3.96** | 21.31 ± 3.49 | 37.18 ± 17.21 | 20.68 ± 3.95* | |
| PNB (nM) | 28.87 ± 10.41 | 16.88 ± 3.03* | 10.67 ± 4.13*** | 23.81 ± 7.98 | 29.74 ± 13.75 | 8.27 ± 1.58*** | ||
| RMD (nM) | 19.07 ± 6.9 | 11.9 ± 3.99* | 6.2 ± 1.59** | 4.26 ± 0.7*** | 14.88 ± 6.89 | 4.14 ± 0.79*** | ||
The 75% effective reactivation concentration (EC75) values (nM ± SD) in latently infected THP89GFP cells of BRY, PNB, and RMD drugs alone and in combination at the indicated ratios 24 h post-treatment are shown. Data are represented as the mean ± SD of at least three independent experiments. Statistics were performed between the calculated EC75 values of the drug at single and combined treatments. ***p < 0.001; **p < 0.005; *p < 0.05.
Effect of BRY, PNB and/or RMD combinations on primary human T cells phenotype
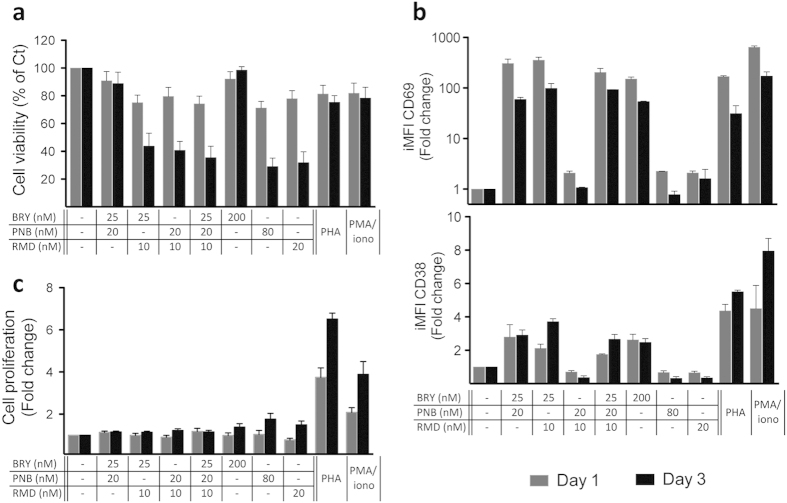
BRY has been previously reported to be potentially associated with uncontrolled T cell activation at concentrations required for efficient HIV reactivation. In this study, we described that BRY, PNB and/or RMD combinations showed a strong synergism in the reversion of latency in J89GFP cells (Tables 2 and 3) and that the EC values of each drug diminished significantly in the combinations in comparison with single drug treatment (Fig. 2). We analysed if the lower EC75 doses achieved in these LRA combinations could improve the effect of single drug treatment at their EC75 in terms of viability, activation and proliferation profile in primary T cells. We selected the combination ratio 2.5:2:1 (BRY:PNB:RMD), since it achieves lower BRY EC75 values, approximately 25 nM in J89GFP cell line (Fig. 2). Isolated primary human CD4 T cells were treated with the double or triple combinations (BRY 25 nM, PNB 20 nM and/or RMD 10 nM) or with the single drugs at their calculated EC75 values (PNB 80 nM, RMD 20 nM) or with the maximal non-toxic concentration (BRY 200 nM, ten-fold below of its EC75 dose). PHA or PMA/ionomycin, which are stimuli that uniformly reactivate latent HIV in almost all the systems previously tested were used as controls24,34. Either one or three days after CD4 T cell treatment, we determined the cell viability and the activation profile by flow cytometry. Cell proliferation was measured by ELISA by BrdU incorporation. Cell death was analysed by 7AAD incorporation by flow cytometry (Fig. 3a). A good cell viability was observed one day post-treatment, whereas it was compromised at day three in BRY+RMD, PNB+RMD and BRY+PNB+RMD combinations, probably due to the effect of RMD, plausible since the highest concentration of BRY or the combination BRY+PNB presented values over 80% of cell viability.
Figure 3. Primary human cells phenotype after treatment with selected drug combinations.
Purified CD4 T cells from healthy subjects were treated with the indicated concentrations of BRY, PNB, RMD or with PHA or PMA/ionomycin for 1 (grey bars) and 3 (black bars) days. (a) Cell viability was determined using 7AAD reagent and analysed by flow cytometry and expressed as percentage. (b) The surface expression of the activation markers CD38 and CD69 in viable CD4 T cells were analysed by flow cytometry and expressed as iMFI. (c) Cell proliferation was determined as BrdU incorporation measured by ELISA. Results are normalized to control vehicle-treated CD4 T cells from the same donors and represent the mean + SEM of three independent experiments.
The effects on T cell activation after BRY, PNB and/or RMD exposure were analysed by measuring the iMFI of CD38 and CD69 activation markers in living cells (Fig. 3b). As expected, CD69 expression was markedly increased and sustained over three days, in CD4 T cells treated with BRY, either alone or in combination with PNB and/or RMD. It is well known that BRY activates NF-κB pathway and that CD69 gene presents three NF-κB binding sites in its promoter region35. On the other hand, CD38 expression was slightly increased in CD4 T cells treated with BRY, either alone or in combination with PNB and/or RMD, but presented levels between 2 and 4 times lower compared to the conventional PHA or PMA/ionomycin treatments.
Representative flow cytometry profiles of CD4 T cell viability and activation markers expression are shown in Supplementary Fig. S6 online.
Finally, cell proliferation was measured on CD4 T cells. Figure 3c shows that the treatment with the LRAs at the maximum concentrations tested produced a 1.5 to 2-fold increase in cell proliferation after three days of treatment, values consistently lower than those obtained after PHA or PMA/ionomycin treatment. Furthermore, the double or triple BRY, PNB and/or RMD combinations diminished the effect of single drugs, presenting proliferation values of non-treated cells.
Discussion
The persistence of HIV-1 involves numerous overlapping cellular pathways, which are interesting from the pharmacological point of view. Thus, targeting multiple steps within the virus latency mechanisms will be important to optimize the reactivation effect and hence permitting the elimination of HIV reservoirs trace.
In a recent study exploring a panel of HDACIs for HIV reactivation, were an in vitro latency assay was used, PNB displayed superior potency to multiple other HDACIs tested including givinostat, belinostat and vorinostat28. Moreover, has already been described33 the great ability of RMD to reverse HIV latency in vitro and ex vivo in resting and memory CD4 T cells from HIV-infected patients on suppressive cART, in comparison with vorinostat and other HDACIs currently in clinical development. BRY has also be remarked as the best effective single agent between a battery of LRAs that disrupt the latent reservoir24. Furthermore, the synergy between prostratin25 or BRY26 and classical HDACIs has been previously described. Nevertheless, the treatment with HDACIs has been shown to mobilize the latent reservoir but could have unintended negative impacts on the effector functions of CTL, thus impairing elimination of infected cells36,37. In addition, the administration of PNB and BRY to patients has been questioned due to significant toxicity levels observed in cell lines and resting CD4 T cells38. Finally, low concentrations of PKC agonists regulate a different set of genes compared to high concentrations of these compounds, which may have clinical implications in terms of potential side effects mediated by the activation of the NF-κB canonical pathway (Muñoz et al., manuscript in preparation). Therefore, a combinatorial strategy can lead to a reduction in the concentration of LRAs used in vivo, resulting in a reduction of adverse effects, limiting the local injuries, the toxicity and the inflammation.
As mentioned above, PNB, RMD and BRY presented a good reactivation activity both in vitro and ex vivo and are currently used in HIV eradication clinical trials23,39,40. To attack HIV latency through HDAC inhibition and NF-κB activation either at epigenetic and transcriptional levels respectively, we studied the possible synergism between these compounds. Such study could bring new insights about the possibility of reactivating virus transcription either with high efficiency and the lowest concentration possible in order to reduce their potential toxicities. For this purpose, we have employed J89GFP and THP89GFP cell lines to explore the impact of PNB and/or RMD in combination with BRY. Although J89GFP and THP89GFP cell lines are not frequently used to assess LRAs41,42,43,44,45,46,47, their exclusive characteristics make from them an experimentally tractable and relevant model to study post-integration HIV latency and reactivation30. Nevertheless, to confirm the results obtained in these cell lines, we have performed a reactivation experiment with BRY, PNB, RMD and the drug combinations selected for primary cells treatment in another two different well- established latency-models, such as ACH-2 and J1.1 cell lines. The obtained data are in concordance to those exposed in this article and are shown in Supplementary Figs S7 and S8 online, which represent cell toxicity (measured by MTT assay) and HIV reactivation (Ag p24 in cell supernatants measured by ELISA), respectively.
All of the tested combinations containing BRY and HDACIs resulted in a more efficient reactivation profile in both the J89GFP and THP89GFP cell lines compared to the effects of the individual drugs. Indeed, the obtained CI values demonstrated synergism between BRY and HDACIs in both HIV latently infected cell lines, whereas slight or no synergism was observed between PNB and RMD. Since both drugs target HDAC, their use in combination could be inducing a competition between them, resulting in an inefficient anti-latency mechanism. On the other hand, the statistically significant reduction in the effective dose values of drugs in combination, as compared to each individual drug, suggested that this kind of combinatorial approach could achieve a strong mobilization of latent reservoirs while diminish side effects.
CD4 T cells are one of the main subsets of cells in which HIV latency is established, and the activation state of this cell type is pivotal for active HIV production. Therefore, the level of phenotype changes in primary human CD4 T cells was assessed after BRY, PNB and/or RMD treatment. We did comparisons between the EC75 doses achieved in drug combinations and the EC75 of single drugs. High cell viability was observed at day one post-treatment, whereas it was compromised at day three in BRY+RMD, PNB+RMD and BRY+PNB+RMD combinations. Since PNB induces prolonged histone hyper acetylation48, pulses instead of continuous treatment or intermittent dosing schedules should be evaluated. In terms of cell activation, we found that none of the combinations augmented the activation effect of the single drugs at their EC75 values, and that double or triple BRY, PNB and/or RMD combinations presented similar proliferation values to non-treated cells, diminishing the effect of single drugs at their EC75 value.
Although administered within the context of cART, the infection of bystander cells remains a concern in a “shock and kill” therapeutic approach. Both HDACIs and BRY may have a negative impact in this aspect, due to their effect on CTL impairment or T cell activation, respectively. On the contrary, these compounds also have a positive impact on HIV inhibition: among other HDACIs, PNB and RMD enable the decrease of HIV release from macrophages via the degradation of intracellular HIV through the canonical autophagy pathway49 and BRY is able to inhibit acute HIV infection by diminishing surface expression of receptors and co-receptors or in a receptor independent manner16.
We are aware that the main limitation of this study is the use of an in vitro cell latency model. The rarity of resting CD4 T cells latently infected makes this type of study challenging. Moreover, it has been remarked the discordance between the effects of non-stimulating LRAs in in vitro models of HIV latency and their effects in ex vivo in resting CD4 T cells from infected individuals on cART. This data indicates that these models do not fully capture all the mechanisms governing HIV latency in vivo. In fact, based on the screening depicted in this article, further efforts should be carried out in ex vivo models in order to find the most appropriate concentrations of these combinations, particularly before any further in vivo studies. On the other hand, the assays presented herein facilitated that is to our knowledge the first in vitro quantitative synergistic evaluation of candidate LRAs administered in combination in both lymphocyte and monocyte/macrophage HIV latently infected lineages.
Methods
Reagents
PE-conjugated anti-human CD69 and FITC-conjugated anti-human CD38 were obtained from BD Biosciences Pharmigen (San Diego, CA, USA). BRY was obtained from Sigma-Aldrich (St. Louis, MO, USA), PNB and RMD were obtained from Selleck Chemicals (Houston, TX). Drugs were dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO, USA) to prepare stock solutions. DMSO concentration in cell cultures was lower than 0.001%. TNF-α was obtained from R&D Systems (Minneapolis, Minn.). PhytohemaGglutinin (PHA), phorbol 12-myristate 13-acetate (PMA) and ionomycin were purchased from Sigma-Aldrich.
Cell lines and culture
J89GFP and THP89GFP cell lines (kindly donated by Dr. David N Levy, NYU, USA) were maintained according to the protocol described in30. ACH-2 and J1.1 cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH from Dr. Thomas Folks45,50,51. Buffy coats from healthy subjects were obtained from the Madrid Transfusion Center. CD4 T cells were purified by negative immunomagnetic separation (CD4+ T Cell Isolation Kit; Miltenyi Biotech, Friedrich, Germany) from peripheral blood mononuclear cells (PBMCs) isolated from heparinised venous blood by fycoll-paque density gradient and maintained in complete RPMI supplemented with 30 U/ml IL2 (Murex Biotech, England, UK)52.
Cell viability assays
The concentration range of each compound assayed in this study is in agreement with previously published results26,28. The toxicity of compounds was measured by MTT (Sigma-Aldrich) assay according to the manufacturer’s instructions in J89GFP and THP89GFP cell lines. 0.001% DMSO treated cells were included in each experiment as vehicle control (Ct); DMSO (10%) was used as positive control of cytotoxicity. Cell death in J89GFP, THP89GFP and primary human CD4 T cells was also determined by 7-aminoactinomicyn-D (7AAD) intercalation and analysed in a Gallios flow-cytometer (Beckman-Coulter, CA, USA). Results were expressed as row data or normalized to control vehicle-treated cells as depicted in each figure.
Analysis of cell activation profile
Cells were stained for 30 minutes at 4°C with the corresponding conjugated antibodies in FACS staining buffer (phosphate-buffered saline (PBS) with 2% FBS), and surface activation markers expression was analysed in live cells in a Gallios flow-cytometer (Beckman-Coulter). At least 50,000 CD4 T cells were collected for each sample and analysed with Kaluza software (Beckman-Coulter). Results were normalized to control vehicle-treated CD4 T cells from the same donors.
Latent HIV-1 reactivation
EGFP-fluorescence pattern measured by flow cytometry was used to determine viral reactivation in J89GFP and THP89GFP cell lines. Whereas Agp24 release measured by enzyme immunosorbent assay (ELISA, INNOTEST® HIV-Antigen mAb, Innogenetics, Belgium) was used to determine viral reactivation in ACH-2 and J1.1 cells. J89GFP and THP89GFP cells were stimulated with the indicated compounds and stained with 7AAD to assess cell viability. At least 30,000 cells were analysed by flow cytometry. The integrated mean fluorescence intensity (iMFI, percentage of EGFP expressing cells *MFI) of live cells was used as a measure of HIV reactivation and standardized with the reactivation obtained after TNF-α treatment. The 50%, 75%, 90% and 95% effective concentrations (EC50, EC75, EC90 and EC95, respectively) were determined, and synergism analysis was performed using the CalcuSyn software (Biosoft, Cambridge, UK), based on the median effect principle33. The combination index (CI) of each drug combination was plotted as a function of the fractional inhibition by computer simulation; the fractional inhibition values ranged from 0.10–0.95. CI values between 0.1–0.9 indicate a synergistic effect; whereas, values between 0.9–1.1 represent an additive effect, and >1.1 represents antagonism. Each experiment was performed in triplicate.
Cell proliferation assays
CD4 T cell proliferation was assayed by using the bromodeoxyuridin (BrdU) Cell Proliferation Kit (Chemicon, Millipore, MA, USA) according to the manufacturer’s instructions. PHA (2 μg/ml) or PMA (2.5 ng/ml) plus ionomycin (250 ng/ml), which are stimuli that uniformly reactivate latent HIV in almost all the systems previously tested24,34 were used as controls. Cell proliferation assays were performed in triplicate.
Statistics
Statistical analysis was performed using SpSS software version 15.0 for Windows. Differences between two groups (control versus different dosages of compounds or BRY-treated versus combined HDACIs and BRY treatment) were assessed by using a paired t-test. (*p < 0.05; **p < 0.005; ***p < 0.001).
Additional Information
How to cite this article: Martínez-Bonet, M. et al. Synergistic Activation of Latent HIV-1 Expression by Novel Histone Deacetylase Inhibitors and Bryostatin-1. Sci. Rep. 5, 16445; doi: 10.1038/srep16445 (2015).
Supplementary Material
Acknowledgments
We thank the Center of Transfusion of Madrid for the buffy coats and the Spanish HIV HGM BioBank, which is supported by ISCIII and CoRISpe and is funded by the RD12/0017/00XX project as part of the plan Nacional R+D+I and cofounded by the ISCIII-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER). We thank Dr. Laura Diaz, of the Flow Cytometry Unit (Instituto de Investigación Sanitaria Gregorio Marañón, IiSGM, Madrid, Spain) for her technical assistance. Funding: This work has been (partially) funded by the RD12/0017/0037, project as part of the project as part of the Acción Estratégica en Salud, Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica 2008–2011 and cofinanced by Instituto de Salud Carlos III (Subdirección General de Evaluación) and Fondo Europeo de Desarrollo Regional (FEDER), RETIC PT13/0010/0028, Fondo de Investigación Sanitaria (FIS) (PI13/02016), Comunidad de Madrid (grant number S-2010/BMD-2332], PENTA and CYTED 214RT0482. CIBER-BBN is an initiative funded by the VI National R&D&i Plan 2008–2011, Iniciativa Ingenio 2010, the Consolider Program, and CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. MMB is supported by “Red de Investigación en SIDA” (RIS).
Footnotes
Author Contributions M.Á.M.F. conceived the study. M.M.B. and M.Á.M.F. participated in the design. M.M.B., M.I.C. and M.J.S. carried out the experimental work and analysed the data. M.M.B., M.I.C. and M.Á.M.F. have participated in writing, revision and discussion of the article. E.M. and S.M. participated in the discussion. M.Á.M.F. took the final decision. All the authors read and approved the article.
References
- Battistini A. & Sgarbanti M. HIV-1 latency: an update of molecular mechanisms and therapeutic strategies. Viruses 6, 1715–1758, 10.3390/v6041715 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D. et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- Wong J. K. et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- Chun T. W. et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J., Gazzard B. & Douek D. C. Where does HIV live? N Engl J Med 350, 1872–1880, 10.1056/NEJMra032395 (2004). [DOI] [PubMed] [Google Scholar]
- Carter C. C. et al. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nature medicine 16, 446–451, 10.1038/nm.2109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J. & Bray S. HAART-persistent HIV-1 latent reservoirs: their origin, mechanisms of stability and potential strategies for eradication. Current HIV research 4, 199–208 (2006). [DOI] [PubMed] [Google Scholar]
- Margolis D. M. Histone deacetylase inhibitors and HIV latency. Curr Opin HIV AIDS 6, 25–29, 10.1097/COH.0b013e328341242d (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirakawa K., Chavez L., Hakre S., Calvanese V. & Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol 21, 277–285, 10.1016/j.tim.2013.02.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S. et al. Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. Journal of virology 85, 6060–6064, 10.1128/JVI.02033-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon G., Zerbato J., Mellors J. W. & Sluis-Cremer N. Disulfiram reactivates latent HIV-1 expression through depletion of the phosphatase and tensin homolog. Aids 27, F7–F11, 10.1097/QAD.0b013e3283570620 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep 2, 807–816, 10.1016/j.celrep.2012.09.008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow D., Gardner J., Darnell G. A., Suhrbier A. & Harrich D. HIV type 1 inhibition by protein kinase C modulatory compounds. AIDS research and human retroviruses 22, 854–864, 10.1089/aid.2006.22.854 (2006). [DOI] [PubMed] [Google Scholar]
- Kulkosky J. et al. Prostratin: activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 98, 3006–3015 (2001). [DOI] [PubMed] [Google Scholar]
- Hamer D. H. et al. Rational design of drugs that induce human immunodeficiency virus replication. Journal of virology 77, 10227–10236 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehla R. et al. Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PloS one 5, e11160, 10.1371/journal.pone.0011160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Real G. et al. Statins inhibit HIV-1 infection by down-regulating Rho activity. The Journal of experimental medicine 200, 541–547, 10.1084/jem.20040061 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin N. M. et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487, 482–485, 10.1038/nature11286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. A., Tolstrup M., Winckelmann A., Ostergaard L. & Sogaard O. S. Eliminating the latent HIV reservoir by reactivation strategies: advancing to clinical trials. Hum Vaccin Immunother 9, 790–799, 10.4161/hv.23202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Zhang Y., Zhou X. & Zhu H. Histonedeacetylase inhibitor Oxamflatin increase HIV-1 transcription by inducing histone modification in latently infected cells. Mol Biol Rep 38, 5071–5078, 10.1007/s11033-010-0653-6 (2011). [DOI] [PubMed] [Google Scholar]
- Victoriano A. F. et al. Novel histone deacetylase inhibitor NCH-51 activates latent HIV-1 gene expression. FEBS Lett 585, 1103–1111, 10.1016/j.febslet.2011.03.017 (2011). [DOI] [PubMed] [Google Scholar]
- Furumai R. et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res 62, 4916–4921 (2002). [PubMed] [Google Scholar]
- Zonder J. A. et al. A phase II trial of bryostatin 1 in the treatment of metastatic colorectal cancer. Clin Cancer Res 7, 38–42 (2001). [PubMed] [Google Scholar]
- Bullen C. K., Laird G. M., Durand C. M., Siliciano J. D. & Siliciano R. F. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nature medicine 20, 425–429, 10.1038/nm.3489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuse S. et al. Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection. PLoS One 4, e6093, 10.1371/journal.pone.0006093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M. et al. Bryostatin-1 synergizes with histone deacetylase inhibitors to reactivate HIV-1 from latency. Current HIV research 8, 418–429 (2010). [DOI] [PubMed] [Google Scholar]
- Chen L. F., Mu Y. & Greene W. C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J 21, 6539–6548 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. A. et al. Comparison of HDAC inhibitors in clinical development: effect on HIV production in latently infected cells and T-cell activation. Hum Vaccin Immunother 9, 993–1001, 10.4161/hv.23800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- International A. S. S. W. G. o. H. I. V. C. et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 12, 607–614, 10.1038/nri3262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsch O., Benveniste E. N., Shaw G. M. & Levy D. N. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. Journal of virology 76, 8776–8786 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. C. Assessment of synergistic and antagonistic effects of chemotherapeutic agents in vitro. Contributions to gynecology and obstetrics 19, 91–107 (1994). [PubMed] [Google Scholar]
- Chou T. C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological reviews 58, 621–681, 10.1124/pr.58.3.10 (2006). [DOI] [PubMed] [Google Scholar]
- Chou T. C. & Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22, 27–55 (1984). [DOI] [PubMed] [Google Scholar]
- Spina C. A. et al. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog 9, e1003834, 10.1371/journal.ppat.1003834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cabrera M. et al. Transcriptional regulation of the gene encoding the human C-type lectin leukocyte receptor AIM/CD69 and functional characterization of its tumor necrosis factor-alpha-responsive elements. J Biol Chem 270, 21545–21551 (1995). [DOI] [PubMed] [Google Scholar]
- Shan L. et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36, 491–501, 10.1016/j.immuni.2012.01.014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. B. et al. Histone deacetylase inhibitors impair the elimination of HIV-infected cells by cytotoxic T-lymphocytes. PLoS Pathog 10, e1004287, 10.1371/journal.ppat.1004287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman F., Ellenberg P., Churchill M. & Lewin S. R. HDAC inhibitors in HIV. Immunol Cell Biol 90, 47–54, 10.1038/icb.2011.95 (2012). [DOI] [PubMed] [Google Scholar]
- Wei D. G. et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog 10, e1004071, 10.1371/journal.ppat.1004071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C., Bouchat S. & Marcello A. HIV-1 transcription and latency: an update. Retrovirology 10, 67, 10.1186/1742-4690-10-67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butera S. T., Perez V. L., Wu B. Y., Nabel G. J. & Folks T. M. Oscillation of the human immunodeficiency virus surface receptor is regulated by the state of viral activation in a CD4+ cell model of chronic infection. Journal of virology 65, 4645–4653 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duh E. J., Maury W. J., Folks T. M., Fauci A. S. & Rabson A. B. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proceedings of the National Academy of Sciences of the United States of America 86, 5974–5978 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks T. M., Justement J., Kinter A., Dinarello C. A. & Fauci A. S. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science 238, 800–802 (1987). [DOI] [PubMed] [Google Scholar]
- Folks T. M. et al. Characterization of a promonocyte clone chronically infected with HIV and inducible by 13-phorbol-12-myristate acetate. J Immunol 140, 1117–1122 (1988). [PubMed] [Google Scholar]
- Perez V. L. et al. An HIV-1-infected T cell clone defective in IL-2 production and Ca2+ mobilization after CD3 stimulation. J Immunol 147, 3145–3148 (1991). [PubMed] [Google Scholar]
- Jordan A., Bisgrove D. & Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22, 1868–1877, 10.1093/emboj/cdg188 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A., Defechereux P. & Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 20, 1726–1738, 10.1093/emboj/20.7.1726 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince H. M., Bishton M. J. & Johnstone R. W. Panobinostat (LBH589): a potent pan-deacetylase inhibitor with promising activity against hematologic and solid tumors. Future oncology 5, 601–612, 10.2217/fon.09.36 (2009). [DOI] [PubMed] [Google Scholar]
- Campbell G. R., Bruckman R. S., Chu Y. L. & Spector S. A. Autophagy Induction by Histone Deacetylase Inhibitors Inhibits HIV Type 1. J Biol Chem, 10.1074/jbc.M114.605428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse K. A. et al. Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. Journal of immunology 142, 431–438 (1989). [PubMed] [Google Scholar]
- Folks T. M. et al. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proceedings of the National Academy of Sciences of the United States of America 86, 2365–2368 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Merino I. et al. The Spanish HIV BioBank: a model of cooperative HIV research. Retrovirology 6, 27, 10.1186/1742-4690-6-27 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.