Abstract
Objective
Numerous studies find elevated depressive symptoms among individuals with type 2 diabetes, yet the mechanisms remain unclear. We examined whether genetic loci previously associated with depressive symptoms predict depressive symptoms among overweight/obese individuals with type 2 diabetes or change in depressive symptoms during behavioral weight loss.
Methods
The Illumina CARe iSelect (IBC) chip and Cardiometabochip were characterized in 2,118 overweight or obese participants with type 2 diabetes from Look AHEAD (Action for Health in Diabetes), a randomized trial to determine the effects of intensive lifestyle intervention (ILI) and Diabetes Support and Education (DSE) on cardiovascular morbidity and mortality. Primary analyses focused on baseline Beck Depression Inventory (BDI) scores and depressive symptom change at one year.
Results
Of eight single nucleotide polymorphisms (SNPs) in six loci, three a priori SNPs in two loci (Chr5: rs60271; LBR: rs2230419, rs1011319) were associated with baseline BDI scores, but in the opposite direction of prior research. In joint analysis of 90,003 IBC and Cardiometabochip SNPs, rs1543654 in the region of KCNE1 predicted change in BDI scores at year 1 in DSE (beta= −1.05, SE=0.21, p=6.9 × 10−7) at the level of chip-wide significance, while also showing a nominal association with baseline BDI (beta=0.35, SE=0.16, p=0.026). Adjustment for antidepressant medication and/or limiting analyses to Non-Hispanic White individuals did not meaningfully alter results.
Conclusions
Previously reported genetic associations with depressive symptoms did not replicate in this cohort of overweight/obese individuals with type 2 diabetes. We identified KCNE1 as a potential novel locus associated with depressive symptoms.
Keywords: genetics, depression, diabetes, weight loss, obesity
Introduction
Numerous studies have documented a disproportionately high prevalence of depression among patients with type 2 diabetes (1–3). Elevated depressive symptoms predict difficulty with adherence to diabetes regimens (4), diabetes complications (5, 6) and mortality (7, 8). Several mechanisms have been suggested to account for the greater prevalence of depression, including poor health behaviors, the stress of managing a chronic disease, altered autonomic and neuroendocrine function, systemic inflammation and cerebrovascular mechanisms (9, 10). Little attention has been paid to the potential for a genetic role.
Diabetes and cardiac disease frequently co-occur, and depressive symptoms are elevated in both. Our team has examined genetic predictors of depressive symptoms in patients with established cardiac disease (11) focusing on candidate genes coding for key elements within select biological pathways thought to contribute to both depression and cardiovascular disease: inflammation, platelet aggregation, endothelial function and omega-3 and –6 fatty acid metabolism (12–14). Following correction for multiple testing, one single nucleotide polymorphism (SNP), rs216873 in intron 38 of the vonWillebrand factor (VWF) gene was found to be significantly associated with depressive symptoms. Several other SNPs relevant to endothelial function and platelet aggregation showed suggestive associations with depressive symptoms, including additional markers within VWF and markers within the vascular cellular adhesion molecule 1 (VCAM1), calcium channel, voltage-dependent, L type, alpha 1C subunit (CACNA1C) and 5-hydroxytryptamine (serotonin) receptor 2A (HTR2A) genes.
A recent genome-wide association study (GWAS) confirmed the role of CACNA1C and the calcium channel pathway in major depression (15). Furthermore, a GWAS examining depressive symptoms identified novel candidate loci, including one in a gene desert region on chromosome 5 and one in the lamin B receptor gene (LBR) (16).
Intensive lifestyle intervention, involving calorie and physical activity goals designed to produce weight loss, may also impact depressive symptoms. Physical activity is well known to produces a moderate reduction in depressive symptoms that is of similar magnitude to behavioral and pharmacologic treatments for depression (17). Additional research suggests that behavioral weight loss, combining diet and physical activity, reduces depressive symptoms (18).
The Action for Health in Diabetes (Look AHEAD) study is a multi-center trial that randomly assigned participants with type 2 diabetes who were overweight or obese to an Intensive Lifestyle Intervention (ILI), with the goal of producing ≥7% weight loss through calorie restriction and physical activity, or to Diabetes Support and Education (DSE) which provided diabetes education but no weight loss treatment per se. After the first year, compared with DSE, participants in the ILI arm lost significantly more weight and showed greater improvement in fitness, waist circumference and indices of diabetes control including metabolic syndrome, diabetes medication use, hemoglobin A1c and fasting glucose (19). Depressive symptoms, as measured by the Beck Depression Inventory (BDI) (20), were not elevated on average at baseline in Look AHEAD (mean=5.5, SD=5.0, range 0–35); nonetheless, participants randomized to ILI exhibited a larger improvement in depressive symptoms at year 1 relative to DSE (ILI= −1.4±4.7, DSE= −0.4±4.5; p<0.001) (21).
The objective of the present study was to examine whether genetic loci previously associated with depressive symptoms would predict BDI scores at baseline and/or change in BDI scores at one year after randomization to ILI or DSE. We further examined whether any SNPs represented on either the Cardiometabochip (22), a genotyping platform of over 200,000 SNPs designed to capture genetic regions associated with cardiovascular and/or diabetes risk traits, or the IBC chip (23), a gene-centric chip including roughly 50,000 SNPs in biologic candidates for cardiovascular disease risk traits, predicted baseline BDI scores or change in BDI scores with weight loss.
Material and Methods
Study cohort
The Look AHEAD study enrolled 5,145 ethnically-diverse overweight/obese subjects with type 2 diabetes (and aged 45 to 76 years) from 16 clinical centers. Of these, 3,905 contributed genetic data on the IBC chip that passed genotyping quality control procedures, while 4,047 did so for the Metabochip. Overall, 3,676 subjects provided genotypes on both the IBC and Metabochip platforms. However, only 8 out of 16 study sites collected consent for genetic analyses sufficiently broad to permit analyses of depressive symptoms. This left 2,120 individuals whose genotypes could be analyzed in the present study. Two of these individuals lacked phenotypic data related to depression, leaving 2,118 subjects with genotyping data from the IBC chip and Metabochip as our final analytic sample.
The design and methods of the Look AHEAD trial have been reported elsewhere (24), as have the baseline characteristics of the randomized cohort (25). Briefly, both the ILI and DSE groups were provided one session of education on diabetes and cardiovascular risk factors. In addition, ILI participants received an intensive lifestyle program, combining diet modification and increased physical activity, designed to produce a loss ≥7% of initial weight and maintain this weight loss. The ILI included one individual and three group meetings per month for six months followed by one individual and two group meetings per month through one year. ILI sessions focused on behavioral weight loss strategies, such as self-monitoring, goal setting and stimulus control, as well as gradually increasing physical activity (primarily walking) to ≥175 minutes per week. The DSE group received the option of attending three sessions per year on nutrition, physical activity and social support with no explicit weight loss goals. The assessments reported herein occurred between June 2001 and December 2005. The Look AHEAD trial, including genetic analyses, was approved by local Institutional Review Boards.
Anthropometric Measures
Participants wore light clothing or a hospital gown and removed their shoes. Weight was measured to the nearest 0.1 kilogram in duplicate at baseline and year 1 using a digital scale. Height was measured in centimeters at baseline using a standard wall-mounted stadiometer. Where discrepant, averages of the duplicate measures were used.
Depressive symptoms
Symptoms of depression were assessed at baseline and year 1 using the Beck Depression Inventory (BDI) (26), a 21-item questionnaire that assesses mood over the previous 2 weeks. This instrument is an effective screening tool for major depression in diabetic patients (27). Total scores range from 0–63, with higher values indicating greater symptoms of depression. In the present study, however, item # 19, which assesses recent weight loss, was excluded from analysis because participants were overweight/obese, were required to be weight stable at entry and half were randomized to weight loss intervention. Thus, our use of the inventory yielded scores of 0–60. Continuous BDI scores were used as the primary outcome.
Genotyping
Genotyping was carried out using the IBC chip (23) and Metabochip (22). On both platforms, we excluded study participants with failed genotyping, gender inconsistency, or familial relatedness (kinship coefficient > 0.025). SNPs with genotyping call rate <95% in any ethnic group were also excluded. After quality control procedures, the mean genotyping success rate per SNP was >99.9%. SNPs derived from the prior literature on depressive symptoms in cardiac patients (11) or genome-wide association studies of depressive symptoms (16) were selected a priori. Where possible, SNPs previously associated with depressive symptoms, but not directly represented on either the IBC chip or the Metabochip, were replaced by proxies (r2 ≥0.97) using phased genotype data from the 1000 Genomes Project and the SNP Annotation and Proxy Search tool (28) based on individuals of European ancestry (CEU) and Yoruba people of Ibadan (YRI). We also examined all SNPs represented on the IBC chip or Metabochip for association with baseline or year 1 change in depressive symptoms.
Observed genotype frequencies were compared with those expected under Hardy Weinberg Equilibrium (HWE) using stratified χ2 tests within the two largest racial/ethnic groups (non-Hispanic White and African-American). As the sample was selected for overweight and diabetes, we did not exclude SNPs a priori based on deviation from HWE on a chip-wide basis. However, we reviewed individual SNP results to ensure SNPs showing significant associations did not deviate from HWE. All markers highlighted in the analyses herein showed no deviation from HWE in either racial stratum at the p<0.05 significance level.
Statistical Analysis
For candidate gene analyses, nominal p-values are reported, as all 8 SNPs under consideration have shown prior associations with depressive symptoms, and our study represents an attempt at replication. For novel marker discovery using chip-wide analyses, we calculated the effective number of independent hypothesis tests among the 90,003 distinct SNPs on the autosomal chromosomes across both genotyping platforms that were relatively common (minor allele frequency (MAF) >0.05). After accounting for linkage disequilibrium (LD) as pooled across ethnicities, we estimated the effective number of uncorrelated SNPs to be 63,444 (29), resulting in a chip-wide significance threshold of p=8.085 × 10−7. We used a false discovery rate (FDR) approach to guide our reporting of suggestive (FDR q<10%) associations, operationalized via a rank ordering of the genetic markers according to their q-values. FDR controls the expected proportion of false negative results among those deemed significant. Q-values are marker-specific quantities that recalibrate the rank ordering of p-values by the probability that they represent a false discovery; they were calculated using the Q-value package (30).
After excluding SNPs in LD (r2>0.30), EIGENSTRAT was used to compute principal components (PCs) from available SNPs to use as covariates to control for population stratification (31). All four primary racial/ethnic groups were adequately captured by the first three PCs: Non-Hispanic White, African American, Hispanic, and Asian.
BDI scores exhibited considerable skewness at both baseline and follow-up, due to the significant number of subjects reporting minimal symptoms of depression. Since regression coefficients are easier to interpret in the original scale, we avoided transfoming the data prior to analysis. Instead, we modeled baseline and year 1 BDI measurements jointly, as bivariate normal variables with an unstructured covariance matrix. However, we subsequently corrected the model-based standard errors using Generalized Estimating Equation (GEE) methodology (32), as implemented in Splus 8.2 (33).
We considered a second outcome in which we adjusted BDI scores for antidepressant medication use. Following the methods used by Hek and colleagues to adjust for antidepressant use (16), previously employed for correcting blood pressure outcomes for antihypertensive medication use (34), we assumed that the BDI score of a person using antidepressants is a right-censored value, i.e., lower than the untreated value would be. Our nonparametric imputation procedure consisted of starting with the highest treated BDI score and replacing it with the mean BDI score of all untreated persons of the same gender that reported the same or higher BDI score. We proceeded in this fashion until all treated values had been replaced by imputed values of the corresponding untreated BDI scores. Regression of the imputed on the observed scores showed that the medication benefit for subjects taking antidepressants could be well characterized over the 0–25 range of treated BDI scores by a linear regression relationship in both men (Intercept=3.82, Slope=0.97) and women (Intercept=4.84, Slope=0.87), with R2 values in excess of 99%. Findings suggested an approximately constant antidepressant medication benefit of 3.82 BDI points for men and a maximum benefit of 4.84 BDI points for women that decreased with depression severity.
Three-way interaction models of individual SNP markers (0, 1 or 2 copies of the minor allele; additive model) with measurement time (baseline vs. year 1) and study arm (ILI vs. DSE) were estimated using GEE procedures. Three distinct types of SNP effects (beta coefficients) are presented in the paper, which can be interpreted as the effect of one additional copy of the corresponding minor allele on a) depression levels within each of the two treatment arms (ILI and DSE); b) depression levels combined across treatment arms; and c) ILI vs. DSE differences in depression levels. When evaluated on baseline BDI scores, type (c) coefficients serve as a check of randomization imbalances across genotypic groups at baseline; when applied to year 1 change in BDI scores, they allow us to test SNP * time * treatment interactions. Longitudinal regression models adjusted all of these genetic effects for age at interview, gender, genetic ancestry (PC1, PC2, PC3), and clinic site. In addition, due to marked differences in allele frequency across major racial and ethnic groups in Look AHEAD for a priori SNPs (Supplemental Table 2), we conducted analyses limited to participants reporting Non-Hispanic White descent to guard against results solely attributable to residual population stratification not captured by PCA adjustment.
Results
Descriptive statistics
Participant characteristics of the genetic sub-cohort of Look AHEAD used in these analyses are shown in Table 1, with the characteristics for the Non-Hispanic White subset presented in Supplemental Table 1. In both subsets, individuals were evenly distributed between the ILI and DSE intervention arms and had comparable age, gender and ethnicity across arms. No baseline differences in weight, clinical characteristics or depressive symptoms across ILI and DSE were observed. Similar to the parent study (19), participants in the genetic substudies that were assigned to ILI lost more weight at year 1 than those assigned to DSE, and showed larger improvements in depressive symptoms (Table 1; Supplemental Table 1). Characteristics for markers chosen for the candidate gene study are presented in Supplemental Table 2.
Table 1.
Demographic Characteristics
| Characteristic | Total (N= 2,118) | DSE (N=1,044) | ILI (N=1,074) | p-value* |
|---|---|---|---|---|
| Women; n (%) | 1189 (56.1) | 589 (56.4) | 600 (55.9) | 0.7981 |
| Age, years; mean (SD) | 59.1 (6.9) | 59.1 (7.0) | 59 (6.9) | 0.6243 |
| Ethnicity; n (%) | ||||
| Non-Hispanic African American | 215 (10.2) | 109 (10.4) | 106 (9.9) | |
| American Indian/Alaskan Native | 8 (0.4) | 5 (0.5) | 3 (0.3) | |
| Asian/Pacific Islander | 11 (0.5) | 3 (0.3) | 8 (0.7) | 0.4438 |
| Non-Hispanic White | 1614 (76.2) | 790 (75.7) | 824 (76.7) | |
| Hispanic/Latino | 230 (10.9) | 113 (10.8) | 117 (10.9) | |
| Other (multiple) | 40 (1.9) | 24 (2.3) | 16 (1.5) | |
| Baseline measurements | ||||
| Diabetes Duration, years; median (IQR) | 5 (2 to 10) | 5 (2 to 10) | 5 (2 to 10) | 0.5205 |
| HbA1c, %; mean (SD) | 7.2 (1.16) | 7.3 (1.19) | 7.2 (1.14) | 0.6882 |
| Insulin use; n (%) | 313 (14.8) | 162 (15.5) | 151 (14.1) | 0.3446 |
| BMI, kg/m2; mean (SD) | 36.3 (6.1) | 36.3 (6.0) | 36.3 (6.1) | 0.8976 |
| Smoking; n (%) | ||||
| Year 0 | 80 (3.8) | 42 (4) | 38 (3.5) | 0.5524 |
| Year 1 | 71 (3.5) | 37 (3.8) | 34 (3.3) | 0.5957 |
| Anti-depressant use; n (%) | ||||
| Year 0 | 396 (18.7) | 189 (18.1) | 207 (19.3) | 0.4898 |
| Year 1 | 396 (19.7) | 201 (20.4) | 195 (19.1) | 0.5223 |
| Weight, kg; mean (SD) | ||||
| Year 0 | 102.5 (19.2) | 102.3 (18.8) | 102.7 (19.5) | 0.6139 |
| Year 1 | 97.2 (19.4) | 101.2 (18.8) | 93.3 (19.2) | <0.0001 |
| Absolute change from baseline | −5.28 (7.89) | −1.05 (5.34) | −9.36 (7.81) | <0.0001 |
| BDI; mean (SD) | ||||
| Year 0 | 5.5 (4.9) | 5.5 (4.6) | 5.5 (5.2) | 0.9321 |
| Year 1 | 4.7 (5.2) | 5.2 (5.0) | 4.2 (5.2) | <0.0001 |
| Absolute change from baseline | −0.78 (4.72) | −0.29 (4.52) | −1.26 (4.85) | <0.0001 |
P-values obtained using: independent samples t test results for quantitative variables with reported mean (SD); Wilcoxon rank sum test results for quantitative variables with reported median (IQR); and chi-square test results for categorical variables with reported n (%).
Candidate SNPs from prior GWAS of depressive symptoms
Two of the five loci with the strongest associations with depressive symptoms in GWAS meta-analysis (16) could be captured by proxies on either the Cardiometabochip or the IBC chip (Chr5 rs161645: proxy rs60271, r2=1.0 in CEU, LD data not available in YRI; LBR rs4653635: proxy rs2230419, r2=1.0 in CEU, r2=0.77 in YRI; LBR rs4653635: proxy rs1011319, r2=1.0 in YRI; r2=0.89 in CEU). Associations of these loci with BDI scores before and after adjustment for antidepressant medication are presented in Table 2.
Table 2.
Association of LBR and RAB9BP1 SNPs with BDI scores at baseline and year 1 in the full sample and the Non-Hispanic White subsample.
| SNP | Chr | Position (Build 137) | Nearest Gene | Alleles* | MAF | N | Time | Effect | Unadjusted BDI | Adjusted BDI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | p-value | Beta | SE | p-value | ||||||||||
| OVERALL SAMPLE | rs1011319 | 1 | 225597846 | LBR | G/A | 19.69 | 2118 | Baseline | Average | 0.49 | 0.19 | 0.0109 | 0.60 | 0.21 | 0.0042 |
| Year 1 | ILI | −0.52 | 0.28 | 0.0628 | −0.57 | 0.29 | 0.0476 | ||||||||
| DSE | 0.01 | 0.26 | 0.9647 | −0.00 | 0.27 | 0.9920 | |||||||||
| Average | −0.25 | 0.19 | 0.1814 | −0.28 | 0.20 | 0.1459 | |||||||||
| ILI-DSE Diff | −0.53 | 0.38 | 0.1627 | −0.56 | 0.39 | 0.1497 | |||||||||
| rs2230419 | 1 | 225607144 | LBR | A/G | 20.02 | 2118 | Baseline | Average | 0.45 | 0.19 | 0.0185 | 0.57 | 0.21 | 0.0071 | |
| Year 1 | ILI | −0.49 | 0.28 | 0.0811 | −0.53 | 0.29 | 0.0702 | ||||||||
| DSE | 0.00 | 0.26 | 0.9923 | −0.02 | 0.27 | 0.9468 | |||||||||
| Average | −0.24 | 0.19 | 0.1987 | −0.27 | 0.20 | 0.1673 | |||||||||
| ILI-DSE Diff | −0.49 | 0.38 | 0.1943 | −0.51 | 0.39 | 0.1967 | |||||||||
| rs60271 | 5 | 104078233 | RAB9BP1 | C/A | 27.79 | 2118 | Baseline | Average | −0.32 | 0.17 | 0.0643 | −0.39 | 0.19 | 0.0350 | |
| Year 1 | ILI | 0.44 | 0.25 | 0.0760 | 0.38 | 0.26 | 0.1450 | ||||||||
| DSE | 0.34 | 0.25 | 0.1636 | 0.25 | 0.23 | 0.2788 | |||||||||
| Average | 0.39 | 0.17 | 0.0251 | 0.31 | 0.17 | 0.0707 | |||||||||
| ILI-DSE Diff | 0.10 | 0.35 | 0.7834 | 0.12 | 0.35 | 0.7197 | |||||||||
| NON-HISPANIC WHITES SAMPLE | rs1011319 | 1 | 225597846 | LBR | G/A | 16.33 | 1614 | Baseline | Average | 0.63 | 0.22 | 0.0039 | 0.74 | 0.24 | 0.0021 |
| Year 1 | ILI | −0.63 | 0.33 | 0.0592 | −0.64 | 0.34 | 0.0567 | ||||||||
| DSE | 0.06 | 0.31 | 0.8413 | −0.02 | 0.32 | 0.9513 | |||||||||
| Average | −0.28 | 0.23 | 0.2078 | −0.33 | 0.23 | 0.1547 | |||||||||
| ILI-DSE Diff | −0.69 | 0.45 | 0.1264 | −0.63 | 0.47 | 0.1807 | |||||||||
| rs2230419 | 1 | 225607144 | LBR | A/G | 16.33 | 1614 | Baseline | Average | 0.63 | 0.22 | 0.0039 | 0.74 | 0.24 | 0.0021 | |
| Year 1 | ILI | −0.63 | 0.33 | 0.0592 | −0.64 | 0.34 | 0.0567 | ||||||||
| DSE | 0.06 | 0.31 | 0.8413 | −0.02 | 0.32 | 0.9513 | |||||||||
| Average | −0.28 | 0.23 | 0.2078 | −0.33 | 0.23 | 0.1547 | |||||||||
| ILI-DSE Diff | −0.69 | 0.45 | 0.1264 | −0.63 | 0.47 | 0.1807 | |||||||||
| rs60271 | 5 | 104078233 | RAB9BP1 | C/A | 32.09 | 1614 | Baseline | Average | −0.33 | 0.18 | 0.0644 | −0.42 | 0.20 | 0.0317 | |
| Year 1 | ILI | 0.30 | 0.27 | 0.2613 | 0.24 | 0.28 | 0.3946 | ||||||||
| DSE | 0.40 | 0.23 | 0.0735 | 0.42 | 0.24 | 0.0869 | |||||||||
| Average | 0.35 | 0.17 | 0.0440 | 0.33 | 0.18 | 0.0771 | |||||||||
| ILI-DSE Diff | −0.10 | 0.35 | 0.7649 | −0.18 | 0.37 | 0.6269 | |||||||||
Major/minor alleles
Three-way interaction models of individual SNP markers (0, 1 or 2 copies of the minor allele; additive model) with measurement time (baseline vs. year 1) and study arm (ILI vs. DSE) were estimated using Generalized Estimating Equation procedures. Beta coefficients reflect the effect of one additional copy of the corresponding minor allele on a) depression levels across ILI and DSE at baseline; b) year 1 depression levels in ILI; c) year 1 depression levels in DSE; d) year 1 depression levels combined across treatment arms; and e) ILI vs. DSE differences in depression levels, or SNP * time * treatment interactions. Longitudinal regression models adjusted all of these genetic effects for age at interview, gender, genetic ancestry, and clinic site.
The minor allele “A” at Chr5 rs60271 (MAF = 0.28) was associated with lower baseline depressive symptoms, reaching nominal significance for BDI scores adjusted for antidepressant use (full cohort: beta=−0.39, SE=0.19, p=0.035; Non-Hispanic Whites: beta=−0.42, SE=0.20, p=0.032). Results were slightly deflated for models prior to adjustment for antidepressant use (full cohort: beta=−0.32, SE=0.17, p=0.064; Non-Hispanic Whites: beta=−0.33, SE=0.18, p=0.064). In either case, however, the direction of association was opposite from the prior report. Minor alleles of the index SNP, rs161645, conferred an increase in depressive symptoms (16) but here we find that minor alleles at the proxy, rs60271 - which is perfectly correlated with the index SNP in the largest racial or ethnic subgroup of Look AHEAD, Non-Hispanic Whites - show a negative association. The minor allele at rs60271 was also associated with year 1 increases in depressive symptoms as averaged across treatment arms. In this case, statistical significance was attained for models without adjustment for antidepressant use (full cohort: beta=0.39, SE=0.17, p=0.025; Non-Hispanic White subsample: beta=0.35, SE=0.17, p=0.044), with slightly deflated results with medication adjustment (full cohort: beta=0.31, SE=0.17, p=0.071; Non-Hispanic Whites: beta=0.33, SE=0.18, p=0.077).
SNP rs2230419 in the LBR region was also associated with baseline depressive symptoms, but, again, in the opposite direction of prior research. Previously, minor alleles in this region were associated with lesser depressive symptoms across studies. However, we found each copy of the minor “G” allele (MAF = 0.20) to be associated with higher depressive symptoms on the BDI scale at baseline in both the full cohort (BDI: beta=0.45, SE=0.19, p=0.018) as well as Non-Hispanic Whites (beta=0.63, SE=0.22, p=0.004) prior to adjustment for antidepressant use. Results were also significant in models after adjustment for antidepressant use (full cohort: beta=0.57, SE=0.21, p=0.007; Non-Hispanic Whites: beta=0.74, SE=0.24, p=0.002). Consistent effects were seen for SNP rs1011319, which was in high linkage disequilibrium with rs2230419, but a better proxy among individuals with African ancestry (r2=0.89 in CEU; r2=1.0 in YRI).
Candidate SNPs with prior association with depressive symptoms among cardiac patients
Five out of seven SNPs previously reported to be associated with depressive symptoms in cardiac patients (11) were either directly represented (HTR2A rs3125; VWF rs216873) or could be captured by proxy SNPs on one of the two genotyping platforms (VCAM1 rs3917010: proxy rs3176861, r2=1.0 in CEU, LD data not available in YRI; CACNA1C rs2239106: proxy rs2239110, r2=1.0 in CEU, r2=0.664 in YRI; VWF rs216856: proxy rs216865, r2=1.0 in CEU; r2 = 1.0 in YRI). None of these SNPs were associated with depressive symptoms at baseline or year 1 follow-up, within or in interaction with treatment arm, with or without adjustment for antidepressant medications, in the full genetic cohort or the genetic cohort limited to Non-Hispanic White individuals (data not shown).
Chipwide analyses across the IBC and Metabochip platforms
Baseline outcomes
No SNPs achieved chip-wide significant associations with depressive symptoms at baseline. Marker prioritization using a false-discovery approach also failed to identify any promising associations with baseline BDI scores, whether or not adjusted for antidepressant medication, in the full genetic cohort and the Non-Hispanic White subset of the genetic cohort (all FDR q-values >0.10). The strongest associations (FDR q-values >0.30) are presented in Supplemental Table 3.
One-year change
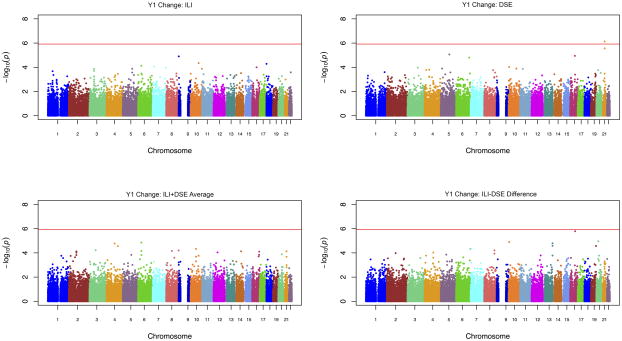
Manhattan plots for change in depressive symptoms in ILI and DSE separately, as averaged across ILI and DSE arms, and as ILI vs. DSE differences (SNP x treatment arm interactions), for the full genetic cohort are presented in Figure 1.
Figure 1.
Manhattan plots of combined IBC and Cardiometabochip results for year 1 depressive symptoms without antidepressant adjustment in ILI, DSE, averaged across intervention arms and difference across intervention arm.
A SNP in the region of KCNE1, rs1543654, was seen to be significantly associated with change in depressive symptoms from baseline to year 1 in DSE alone, prior to antidepressant medication adjustment. The C allele (MAF=0.35) was associated with fewer depressive symptoms at year 1 in DSE (Full cohort: beta= −1.05, SE=0.21, p=6.9 × 10−7; Table 2). No effect was observed in ILI (beta= −0.12, SE=0.21, p=0.565), resulting in a nominally significant SNP x treatment arm interaction (beta= −0.92, SE=0.30, p=0.002). The minor allele at rs1543654 was also nominally associated with increased depressive symptoms at baseline (beta=0.35, SE=0.16, p=0.026). Similar, although slightly attenuated, results were observed for antidepressant-adjusted BDI scores in the full genetic cohort, and for BDI scores within and without medication adjustment in the Non-Hispanic White subsample (Table 2).
No other SNPs were associated with change in depressive symptoms at year 1 in ILI, DSE or in interaction with treatment arm at the level of chip-wide significance.
Discussion
One of the most vexing issues in identifying genetic associations with depressive symptoms is replication. In this paper, we attempted to replicate results from two previous papers on the genetics of depressive symptoms. The first paper employed a candidate gene strategy targeting key pathways thought to contribute to both depressive symptoms and risk for cardiovascular disease and identified one locus in VWF reaching experiment-wide significance and several other promising loci as associated with depressive symptoms among 977 cardiac patients of French-Canadian descent (11). The second paper employed a more agnostic GWAS approach in 51,258 middle-aged to older adults in the general population and identified one SNP reaching genome-wide significance upon replication and several additional promising leads (16). Of the eight SNPs in six loci that we were able to represent using either the IBC or Cardiometabochip in this sample of individuals who are overweight or obese and have type 2 diabetes, we found three nominal associations in two loci (Chr5, rs60271, LBR rs2230419, rs1011319) with baseline depressive symptoms, but in a direction inconsistent with prior research.
This report contributes to an expanding literature finding difficulty identifying and replicating genetic associations with major depression and depressive symptoms (35). Although a number of explanations have been proposed, one of the most compelling is that depression is a heterogeneous disorder with subgroups of individuals having depression attributable to distinct causes, be they genetic, environmental or a combination of both. In this study, we note that the mean of depressive symptoms was low (Mean=5.5; SD=4.9), and variation in depressive symptoms may have reflected a number of causes, including the psychological or physiological impact of having diabetes, long-standing temperament as well as subsets of individuals with liability to major depressive disorder. It is further possible that depressive symptoms may be genetically heterogeneous, with genetic vulnerability related to one set of genes in one individual and a second set of genes in a second. This heterogeneity may be resolved by continuing to increase sample sizes of GWAS to the extent that a signal can be detected despite the noise. Alternatively, intermediate phenotypes, such as neuroimaging, may identify heterogeneity. If the depression phenotype is heterogeneous genetically, it remains plausible that genetic discoveries in very large sample sizes may retain clinical relevance due to their specificity to a subgroup.
It is also plausible that the lack of replication reflects meaningful differences across studies. One prominent difference is the diverse racial and ethnic composition of Look AHEAD compared to the homogeneity of the samples used in the prior GWAS (European-descent) and candidate gene studies (French-Canadian descent) of depressive symptoms. Differences in allele frequency and patterns of co-inheritance between marker and causal variants (linkage disequilibrium) are likely to contribute to difficulty with replication particularly when causal variants are unknown. Here, we noted that the allele frequency of some of the a priori SNPs differed markedly across the major racial and ethnic populations in Look AHEAD (Supplemental Table 2) suggesting the potential of differences in association within the prominent racial and ethnic groups within Look AHEAD. This led us to confirm associations in the Non-Hispanic White subset to ensure that our lack of replication was not solely attributable to differences in racial and ethnic composition. Although our results were largely consistent across the full sample and the Non-Hispanic White subset, it remains plausible that associations observed in prior studies may not generalize to individuals of African, Hispanic or Native American descent.
Additionally, the prior GWAS of depressive symptoms focused primarily on the Center for Epidemiological Studies – Depression Scale (36) to index depressive symptoms, whereas we collected depressive symptoms based on the Beck Depression Inventory. Prior research nonetheless indicates strong correlation between the two measures (e.g., r=0.86 (37)). Another key difference was inclusion/exclusion criteria, with Look AHEAD notably being comprised of overweight individuals with type 2 diabetes. As depressive symptoms were, on average, low, it is plausible that individuals could score in this range on the basis of elevated responses to one or two items (e.g., pain or fatigue).
In chip-wide analyses, one of the first conducted in relation to BDI scores, we identified a novel region associated with change in depressive symptoms from baseline to year 1 in the control arm (DSE). The SNP reaching experiment-wide significance is located within 20 kb of KCNE1, or potassium voltage-gated channel, Isk-related family, member 1, a primary regulator of cellular electrophysiology and function. For example, variation in this gene has previously been associated with cardiac disease, including long QT interval syndrome (38) and atrial fibrillation (39). KCNE1 also regulates neuronal potassium channels and resting membrane electrical potential (40), although no prior associations with depressive symptoms have been reported to our knowledge. It is of note that calcium channel genes (CACNA1C; CACANB2), also primary regulators of voltage-based neuronal activity, were recently associated with risk across multiple psychiatric phenotypes, including schizophrenia, bipolar disorder, autism spectrum disorder, attention deficit-hyperactivity disorder and major depressive disorder (15), although association with major depressive disorder was not seen in the most recent GWAS (35). Nonetheless, given that this is the first report of association with depressive symptoms, and the association with baseline BDI scores was relatively small, it is important to consider that the result may reflect a false positive and seek replication prior to placing too much emphasis on its potential role.
Although this is the largest genetic association study of depressive symptoms among overweight or obese individuals with type 2 diabetes, it is important to note limitations. First, we leveraged existing genotype data available in Look AHEAD, including the IBC chip and Cardiometabochip. These platforms provide complementary approaches with the IBC chip providing candidate gene coverage and the Cardiometabochip covering GWAS loci related to cardiovascular or type 2 diabetes and their risk factors; however, these chips do not provide genome-wide coverage of common variation. As such, we could not test all previously reported associations. Furthermore, although SNPs from several candidate genes often considered in the depressive symptom literature, such as those within serotonin (e.g., SLC6A4 (SERT), HTR1A, HTR2A, TPH1, TPH2), dopamine (e.g., DBH, DRD1, DRD2, DRD4 and COMT) and monoamine oxydase (MAOA, MAOB) pathways, were represented on the IBC chip, no data were available on insertion/deletion or copy number variants.
Overall, this study attempted to replicate prior SNP associations with depressive symptoms in a novel cohort of individuals who were overweight or obese and had type 2 diabetes. We further examined whether any common SNPs represented on the IBC chip or Cardiometabochip were associated with depressive symptoms at baseline or year 1 follow-up after randomization to behavioral weight loss intervention or a minimal contact control condition. No direct replications were found. In chip-wide analyses, a novel locus in close proximity to KCNE1, a key regulator of potassium channels, was associated with change in depressive symptoms over one year, a finding that awaits replication.
Supplementary Material
Supplemental Figure 1. Regional Manhattan plots for the association of KCNE1 with BDI scores without medication adjustment.
Supplemental Table 1. Participant characteristics for Non-Hispanic Whites only sample.
Supplemental Table 2. SNP Characteristics for a priori SNPs in the overall sample and in the three largest racial/ethnic groups, Non-Hispanic White, Non-Hispanic Black, and Hispanic individuals.
Supplemental Table 3. Top hits for baseline depressive symptoms in the full cohort and the Non-Hispanic White subsample for BDI scores without medication adjustment (FDR q-value < 0.30). Note: that no adjusted baseline results met the FDR criteria.)
Table 3.
Association of KCNE1 top SNPs with BDI scores at baseline and year 1 in the full sample and the Non-Hispanic White subsample.
| SNP* | Chr | Position (Build 137) | Nearest Gene | Alleles** | MAF | N | Time | Effect | Unadjusted BDI | Adjusted BDI | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | p-value | Beta | SE | p-value | ||||||||||
| OVERALL SAMPLE | rs1543654 | 21 | 35799050 | KCNE1 | T/C | 35.03 | 2118 | Baseline | Average | 0.35 | 0.16 | 0.0257 | 0.33 | 0.17 | 0.0578 |
| Year 1 | ILI | −0.12 | 0.21 | 0.5487 | −0.19 | 0.22 | 0.3863 | ||||||||
| DSE | −1.05 | 0.21 | 6.9E-07 | −1.02 | 0.22 | 1.9E-06 | |||||||||
| Average | −0.59 | 0.15 | 0.0001 | −0.61 | 0.15 | 0.0001 | |||||||||
| ILI-DSE Diff | 0.92 | 0.3 | 0.0019 | 0.83 | 0.31 | 0.0069 | |||||||||
| rs9975215 | 21 | 35814953 | KCNE1 | G/C | 31.42 | 2118 | Baseline | Average | 0.22 | 0.16 | 0.1808 | 0.22 | 0.18 | 0.2164 | |
| Year 1 | ILI | 0.06 | 0.21 | 0.7923 | 0.08 | 0.23 | 0.7313 | ||||||||
| DSE | −1.05 | 0.22 | 1.5E-06 | −1.07 | 0.23 | 2.1E-06 | |||||||||
| Average | −0.50 | 0.15 | 0.0012 | −0.50 | 0.16 | 0.0019 | |||||||||
| ILI-DSE Diff | 1.11 | 0.31 | 0.0003 | 1.15 | 0.32 | 0.0003 | |||||||||
| NON-HISPANIC WHITE SAMPLE | rs1543654 | 21 | 35799050 | KCNE1 | T/C | 37.42 | 1614 | Baseline | Average | 0.33 | 0.17 | 0.0467 | 0.34 | 0.18 | 0.068 |
| Year 1 | ILI | 0.13 | 0.22 | 0.5468 | 0.09 | 0.23 | 0.7119 | ||||||||
| DSE | −1.10 | 0.23 | 1.1E-06 | −1.08 | 0.24 | 5.3E-06 | |||||||||
| Average | −0.48 | 0.16 | 0.0022 | −0.49 | 0.17 | 0.0028 | |||||||||
| ILI-DSE Diff | 1.23 | 0.32 | 0.0001 | 1.16 | 0.33 | 0.0004 | |||||||||
| rs9975215 | 21 | 35814953 | KCNE1 | G/C | 30.89 | 1614 | Baseline | Average | 0.26 | 0.18 | 0.1485 | 0.28 | 0.2 | 0.152 | |
| Year 1 | ILI | 0.26 | 0.23 | 0.2676 | 0.30 | 0.25 | 0.2205 | ||||||||
| DSE | −1.26 | 0.24 | 1.7E-07 | −1.29 | 0.26 | 4.5E-07 | |||||||||
| Average | −0.50 | 0.17 | 0.0027 | −0.49 | 0.18 | 0.0057 | |||||||||
| ILI-DSE Diff | 1.52 | 0.34 | 5.7E-06 | 1.59 | 0.36 | 7.6E-06 | |||||||||
LD between the pair: D′=0.87, R2 =0.80 based on the full sample and D′=0.97, R2 =0.84 based on the NHW sample.
Major/minor alleles
Three-way interaction models of individual SNP markers (0, 1 or 2 copies of the minor allele; additive model) with measurement time (baseline vs. year 1) and study arm (ILI vs. DSE) were estimated using Generalized Estimating Equation procedures. Beta coefficients reflect the effect of one additional copy of the corresponding minor allele on a) depression levels across ILI and DSE at baseline; b) year 1 depression levels in ILI; c) year 1 depression levels in DSE; d) year 1 depression levels combined across treatment arms; and e) ILI vs. DSE differences in depression levels, or SNP * time * treatment interactions. Longitudinal regression models adjusted all of these genetic effects for age at interview, gender, genetic ancestry, and clinic site.
Acknowledgments
Funding sources: Please see appendix
Funding and Support
This ancillary study is supported by DK056992-14S1. The Look AHEAD study is supported by the Department of Health and Human Services through the following cooperative agreements from the National Institutes of Health: DK57136, DK57149, DK56990, DK57177, DK57171, DK57151, DK57182, DK57131, DK57002, DK57078, DK57154, DK57178, DK57219, DK57008, DK57135, and DK56992. The following federal agencies have contributed support: National Institute of Diabetes and Digestive and Kidney Diseases; National Heart, Lung, and Blood Institute; National Institute of Nursing Research; National Center on Minority Health and Health Disparities; NIH Office of Research on Women’s Health; and the Centers for Disease Control and Prevention. This research was supported in part by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. The Indian Health Service (I.H.S.) provided personnel, medical oversight, and use of facilities. The opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the I.H.S. or other funding sources.
Additional support was received from The Johns Hopkins Medical Institutions Bayview General Clinical Research Center (M01RR02719); the Massachusetts General Hospital Mallinckrodt General Clinical Research Center (M01RR01066); the University of Colorado Health Sciences Center General Clinical Research Center (M01RR00051) and Clinical Nutrition Research Unit (P30 DK48520); the University of Tennessee at Memphis General Clinical Research Center (M01RR0021140); the University of Pittsburgh General Clinical Research Center (M01RR000056 44) and NIH grant (DK 046204); the VA Puget Sound Health Care System Medical Research Service, Department of Veterans Affairs; and the Frederic C. Bartter General Clinical Research Center (M01RR01346).
The following organizations have committed to make major contributions to Look AHEAD: Federal Express; Health Management Resources; Johnson & Johnson, LifeScan Inc.; Optifast-Novartis Nutrition; Roche Pharmaceuticals; Ross Product Division of Abbott Laboratories; Slim-Fast Foods Company; and Unilever.
Abbreviations
- Look AHEAD
The Action in Health and Diabetes trial
- BMI
body mass index
- GWAS
genome-wide association study
- MAF
minor allele frequency
- SNP
single nucleotide polymorphism
- SE
standard error
- LD
linkage disequilibrium
- FDR
false discovery rate
- Chr
chromosome
- ILI
Look AHEAD intensive lifestyle intervention treatment arm
- DSE
Look AHEAD diabetes support and education treatment arm
- BDI
Beck Depression Inventory
- HWE
Hardy-Weinberg Equilibrium
- CEU
Hapmap samples from U.S. residents with northern and western European ancestry from the Centre d’Etude du Polymorphisme Humain
- YRI
Hapmap samples from Yoruba people of Ibadan
- VWF
vonWillebrand factor gene
- VCAM1
vascular cellular adhesion molecule 1 gene
- CACNA1C
calcium channel, voltage-dependent, L type, alpha 1C subunit gene
- HTR2A
5-hydroxytryptamine (serotonin) receptor 2A gene
- LBR
lamin B receptor gene
- RAB9BP1
RAB9B, member RAS oncogene family pseudogene 1
- KCNE1
potassium voltage-gated channel, Isk-related family, member 1
Appendix: Look AHEAD Research Group at Year 1
Clinical Sites
The Johns Hopkins Medical Institutions
Frederick L. Brancati, MD, MHS1; Jeff Honas, MS2; Lawrence Cheskin, MD3; Jeanne M. Clark, MD, MPH3; Kerry Stewart, EdD3; Richard Rubin, PhD3; Jeanne Charleston, RN; Kathy Horak, RD
Pennington Biomedical Research Center
George A. Bray, MD1; Kristi Rau2; Allison Strate, RN2; Brandi Armand, LPN2; Frank L. Greenway, MD3; Donna H. Ryan, MD3; Donald Williamson, PhD3; Amy Bachand; Michelle Begnaud; Betsy Berhard; Elizabeth Caderette; Barbara Cerniauskas; David Creel; Diane Crow; Helen Guay; Nancy Kora; Kelly LaFleur; Kim Landry; Missy Lingle; Jennifer Perault; Mandy Shipp, RD; Marisa Smith; Elizabeth Tucker
The University of Alabama at Birmingham
Cora E. Lewis, MD, MSPH1; Sheikilya Thomas MPH2; Monika Safford, MD3; Vicki DiLillo, PhD; Charlotte Bragg, MS, RD, LD; Amy Dobelstein; Stacey Gilbert, MPH; Stephen Glasser, MD; Sara Hannum, MA; Anne Hubbell, MS; Jennifer Jones, MA; DeLavallade Lee; Ruth Luketic, MA, MBA, MPH; Karen Marshall; L. Christie Oden; Janet Raines, MS;
Cathy Roche, RN, BSN; Janet Truman; Nita Webb, MA; Audrey Wrenn, MAEd
Harvard Center
Massachusetts General Hospital
David M. Nathan, MD1; Heather Turgeon, RN, BS, CDE2; Kristina Schumann, BA2; Enrico Cagliero, MD3; Linda Delahanty, MS, RD3; Kathryn Hayward, MD3; Ellen Anderson, MS, RD3; Laurie Bissett, MS, RD; Richard Ginsburg, PhD; Valerie Goldman, MS, RD; Virginia Harlan, MSW; Charles McKitrick, RN, BSN, CDE; Alan McNamara, BS; Theresa Michel, DPT, DSc CCS; Alexi Poulos, BA; Barbara Steiner, EdM; Joclyn Tosch, BA
Joslin Diabetes Center
Edward S. Horton, MD1; Sharon D. Jackson, MS, RD, CDE2; Osama Hamdy, MD, PhD3; A. Enrique Caballero, MD3; Sarah Bain, BS;
Elizabeth Bovaird, BSN, RN; Ann Goebel-Fabbri, PhD; Lori Lambert, MS, RD;
Sarah Ledbury, MEd, RD; Maureen Malloy, BS; Kerry Ovalle, MS, RCEP, CDE
Beth Israel Deaconess Medical Center
George Blackburn, MD, PhD1; Christos Mantzoros, MD, DSc3; Kristinia Day, RD; Ann McNamara, RN
University of Colorado Health Sciences Center
James O. Hill, PhD1; Marsha Miller, MS, RD2; JoAnn Phillipp, MS2; Robert Schwartz, MD3; Brent Van Dorsten, PhD3; Judith Regensteiner, PhD3; Salma Benchekroun MS; Ligia Coelho, BS;
Paulette Cohrs, RN, BSN; Elizabeth Daeninck, MS, RD; Amy Fields, MPH; Susan Green; April Hamilton, BS, CCRC; Jere Hamilton, BA; Eugene Leshchinskiy; Michael McDermott, MD; Lindsey Munkwitz, BS; Loretta Rome, TRS; Kristin Wallace, MPH; Terra Worley, BA
Baylor College of Medicine
John P. Foreyt, PhD1; Rebecca S. Reeves, DrPH, RD2; Henry Pownall, PhD3; Ashok Balasubramanyam, MBBS3; Peter Jones, MD3; Michele Burrington, RD; Chu-Huang Chen, MD, PhD3; Allyson Clark, RD; Molly Gee, MEd, RD; Sharon Griggs; Michelle Hamilton; Veronica Holley; Jayne Joseph, RD; Patricia Pace, RD: Julieta Palencia, RN; Olga Satterwhite, RD;
Jennifer Schmidt; Devin Volding, LMSW; Carolyn White
University of California at Los Angeles School of Medicine
Mohammed F. Saad, MD1; Siran Ghazarian, MD2; Ken C. Chiu, MD3; Medhat Botrous; Michelle Chan, BS; Kati Konersman, MA, RD, CDE; Magpuri Perpetua, RD
The University of Tennessee Health Science Center
University of Tennessee East
Karen C. Johnson, MD, MPH1; Carolyn Gresham, RN2; Stephanie Connelly, MD, MPH3; Amy Brewer, RD, MS; Mace Coday, PhD; Lisa Jones, RN; Lynne Lichtermann, RN, BSN; Shirley Vosburg, RD, MPH; and J. Lee Taylor, MEd, MBA
University of Tennessee Downtown
Abbas E. Kitabchi, PhD, MD1; Helen Lambeth, RN, BSN2; Debra Clark, LPN; Andrea Crisler, MT; Gracie Cunningham; Donna Green, RN; Debra Force, MS, RD, LDN; Robert Kores, PhD; Renate Rosenthal PhD; Elizabeth Smith, MS, RD, LDN; and Maria Sun, MS, RD, LDN; and Judith Soberman, MD3
University of Minnesota
Robert W. Jeffery, PhD1; Carolyn Thorson, CCRP2; John P. Bantle, MD3; J. Bruce Redmon, MD3; Richard S. Crow, MD3; Scott Crow, MD3; Susan K Raatz, PhD, RD3; Kerrin Brelje, MPH, RD; Carolyne Campbell;
Jeanne Carls, MEd; Tara Carmean-Mihm, BA; Emily Finch, MA; Anna Fox, MA;
Elizabeth Hoelscher, MPH, RD, CHES; La Donna James; Vicki A. Maddy, BS, RD; Therese Ockenden, RN; Birgitta I. Rice, MS, RPh CHES; Tricia Skarphol, BS; Ann D. Tucker, BA; Mary Susan Voeller, BA; Cara Walcheck, BS, RD
St. Luke’s Roosevelt Hospital Center
Xavier Pi-Sunyer, MD1; Jennifer Patricio, MS2; Stanley Heshka, PhD3; Carmen Pal, MD3; Lynn Allen, MD; Diane Hirsch, RNC, MS, CDE; Mary Anne Holowaty, MS, CN
University of Pennsylvania
Thomas A. Wadden, PhD1; Barbara J. Maschak-Carey, MSN, CDE2; Stanley Schwartz, MD3; Gary D. Foster, PhD3; Robert I. Berkowitz, MD3; Henry Glick, PhD3; Shiriki K. Kumanyika, PhD, RD, MPH3; Johanna Brock; Helen Chomentowski; Vicki Clark; Canice Crerand, PhD; Renee Davenport; Andrea Diamond, MS, RD; Anthony Fabricatore, PhD; Louise Hesson, MSN; Stephanie Krauthamer-Ewing, MPH; Robert Kuehnel, PhD; Patricia Lipschutz, MSN; Monica Mullen, MS, RD; Leslie Womble, PhD, MS; Nayyar Iqbal, MD
University of Pittsburgh
David E. Kelley, MD1; Jacqueline Wesche-Thobaben, RN, BSN, CDE2; Lewis Kuller, MD, DrPH3; Andrea Kriska, PhD3; Janet Bonk, RN, MPH; Rebecca Danchenko, BS; Daniel Edmundowicz, MD3; Mary L. Klem, PhD, MLIS3; Monica E. Yamamoto, DrPH, RD, FADA 3; Barb Elnyczky, MA; George A. Grove, MS; Pat Harper, MS, RD, LDN; Janet Krulia, RN, BSN, CDE; Juliet Mancino, MS, RD, CDE, LDN; Anne Mathews, MS, RD, LDN; Tracey Y. Murray, BS; Joan R. Ritchea; Jennifer Rush, MPH; Karen Vujevich, RN-BC, MSN, CRNP; Donna Wolf, MS
The Miriam Hospital/Brown Medical School
Rena R. Wing, PhD1; Renee Bright, MS2; Vincent Pera, MD3; John Jakicic, PhD3; Deborah Tate, PhD3; Amy Gorin, PhD3; Kara Gallagher, PhD3; Amy Bach, PhD; Barbara Bancroft, RN, MS; Anna Bertorelli, MBA, RD; Richard Carey, BS; Tatum Charron, BS; Heather Chenot, MS; Kimberley Chula-Maguire, MS; Pamela Coward, MS, RD; Lisa Cronkite, BS; Julie Currin, MD; Maureen Daly, RN; Caitlin Egan, MS; Erica Ferguson, BS, RD; Linda Foss, MPH; Jennifer Gauvin, BS; Don Kieffer, PhD; Lauren Lessard, BS; Deborah Maier, MS; JP Massaro, BS; Tammy Monk, MS; Rob Nicholson, PhD; Erin Patterson, BS; Suzanne Phelan, PhD; Hollie Raynor, PhD, RD; Douglas Raynor, PhD; Natalie Robinson, MS, RD; Deborah Robles; Jane Tavares, BS
The University of Texas Health Science Center at San Antonio
Steven M. Haffner, MD1; Maria G. Montez, RN, MSHP, CDE2; Carlos Lorenzo, MD3
University of Washington/VA Puget Sound Health Care System
Steven E. Kahn, MB, ChB1; Brenda Montgomery, RN, MS, CDE2; Robert Knopp, MD3; Edward Lipkin, MD3; Matthew L. Maciejewski, PhD3; Dace Trence, MD3; Terry Barrett, BS; Joli Bartell, BA; Diane Greenberg, PhD; Anne Murillo, BS; Betty Ann Richmond, MEd; April Thomas, MPH, RD
Southwestern American Indian Center, Phoenix, Arizona and Shiprock, New Mexico
William C. Knowler, MD, DrPH1; Paula Bolin, RN, MC2; Tina Killean, BS2; Cathy Manus, LPN3; Jonathan Krakoff, MD3; Jeffrey M. Curtis, MD, MPH3; Justin Glass, MD3; Sara Michaels, MD3; Peter H. Bennett, MB, FRCP3; Tina Morgan3; Shandiin Begay, MPH; Bernadita Fallis RN, RHIT, CCS; Jeanette Hermes, MS, RD; Diane F. Hollowbreast; Ruby Johnson; Maria Meacham, BSN, RN, CDE; Julie Nelson, RD; Carol Percy, RN; Patricia Poorthunder; Sandra Sangster; Nancy Scurlock, MSN, ANP-C, CDE; Leigh A. Shovestull, RD, CDE; Janelia Smiley; Katie Toledo, MS, LPC; Christina Tomchee, BA; Darryl Tonemah PhD
University of Southern California
Anne Peters, MD1; Valerie Ruelas, MSW, LCSW2; Siran Ghazarian Sengardi, MD2; Kathryn Graves, MPH, RD, CDE;
Kati Konersman, MA, RD, CDE; Sara Serafin-Dokhan
Coordinating Center
Wake Forest University
Mark A. Espeland, PhD1; Judy L. Bahnson, BA2; Lynne Wagenknecht, DrPH3; David Reboussin, PhD3; W. Jack Rejeski, PhD3; Alain Bertoni, MD, MPH3; Wei Lang, PhD3; Gary Miller, PhD3; David Lefkowitz, MD3; Patrick S. Reynolds, MD3; Paul Ribisl, PhD3; Mara Vitolins, DrPH3; Michael Booth, MBA2; Kathy M. Dotson, BA2; Amelia Hodges, BS2; Carrie C. Williams, MA2; Jerry M. Barnes, MA; Patricia A. Feeney, MS; Jason Griffin, BS; Lea Harvin, BS; William Herman, MD, MPH; Patricia Hogan, MS; Sarah Jaramillo, MS; Mark King, BS; Kathy Lane, BS; Rebecca Neiberg, MS; Andrea Ruggiero, MS; Christian Speas, BS; Michael P. Walkup, MS; Karen Wall; Michelle Ward; Delia S. West, PhD; Terri Windham
Central Resources Centers
DXA Reading Center, University of California at San Francisco
Michael Nevitt, PhD1; Susan Ewing, MS; Cynthia Hayashi; Jason Maeda, MPH; Lisa Palermo, MS, MA; Michaela Rahorst; Ann Schwartz, PhD; John Shepherd, PhD
Central Laboratory, Northwest Lipid Research Laboratories
Santica M. Marcovina, PhD, ScD1; Greg Strylewicz, MS
ECG Reading Center, EPICARE, Wake Forest University School of Medicine
RonaldJ. Prineas, MD, PhD1; Teresa Alexander; Lisa Billings; Charles Campbell, AAS, BS; Sharon Hall; Susan Hensley; Yabing Li, MD; Zhu-Ming Zhang, MD
Diet Assessment Center, University of South Carolina, Arnold School of Public Health, Center for Research in Nutrition and Health Disparities
Elizabeth J Mayer-Davis, PhD1; Robert Moran, PhD
Hall-Foushee Communications, Inc
Richard Foushee, PhD; Nancy J. Hall, MA
Federal Sponsors
National Institute of Diabetes and Digestive and Kidney Diseases
Barbara Harrison, MS; Van S. Hubbard, MD PhD; Susan Z. Yanovski, MD
National Heart, Lung, and Blood Institute
Lawton S. Cooper, MD, MPH; Jeffrey Cutler, MD, MPH; Eva Obarzanek, PhD, MPH, RD
Centers for Disease Control and Prevention
Edward W. Gregg, PhD; David F. Williamson, PhD; Ping Zhang, PhD
Footnotes
Principal Investigator
Program Coordinator
Co-Investigator
Conflicts of interest: None
All other Look AHEAD staffs are listed alphabetically by site.
Contributor Information
Jeanne M. McCaffery, Weight Control and Diabetes Research Center, The Miriam Hospital and Warren Alpert School of Medicine at Brown University, Providence, RI, USA
George D. Papandonatos, Center for Statistical Sciences, Brown University, Providence, RI, USA
Lucy F. Faulconbridge, Department of Psychiatry, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA
Bahar Erar, Center for Statistical Sciences, Brown University, Providence, RI, USA
Inga Peter, Department of Genetics & Genomic Sciences, Mount Sinai School of Medicine, New York, NY, USA
Lynne E. Wagenknecht, Look AHEAD Coordinating Center, Division of Public Health Sciences, Wake Forest School of Medicine, Medical Center Blvd., Winston-Salem, NC, USA
Nicholas M. Pajewski, Department of Biostatistical Sciences, Division of Public Health Sciences, Wake Forest School of Medicine, Winston-Salem, NC, USA
Andrea Anderson, Look AHEAD Coordinating Center, Division of Public Health Sciences, Wake Forest School of Medicine, Medical Center Blvd., Winston-Salem, NC, USA
Thomas A. Wadden, Department of Psychiatry, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA
Rena R. Wing, Weight Control and Diabetes Research Center, The Miriam Hospital and Warren Alpert School of Medicine at Brown University, Providence, RI, USA
References
- 1.Ali S, Stone MA, Peters JL, Davies MJ, Khunti K. The prevalence of co-morbid depression in adults with Type 2 diabetes: a systematic review and meta-analysis. Diabet Med. 2006;23(11):1165–73. doi: 10.1111/j.1464-5491.2006.01943.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderson RJ, Freedland KE, Clouse RE, Lustman PJ. The prevalence of comorbid depression in adults with diabetes: a meta-analysis. Diabetes Care. 2001;24(6):1069–78. doi: 10.2337/diacare.24.6.1069. [DOI] [PubMed] [Google Scholar]
- 3.Nouwen A, Winkley K, Twisk J, Lloyd CE, Peyrot M, Ismail K, Pouwer F. Type 2 diabetes mellitus as a risk factor for the onset of depression: a systematic review and meta-analysis. Diabetologia. 2010;53(12):2480–6. doi: 10.1007/s00125-010-1874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez JS, Peyrot M, McCarl LA, Collins EM, Serpa L, Mimiaga MJ, Safren SA. Depression and diabetes treatment nonadherence: a meta-analysis. Diabetes Care. 2008;31(12):2398–403. doi: 10.2337/dc08-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot M, Anderson R, Freedland KE, Clouse RE, Lustman PJ. Association of depression and diabetes complications: a meta-analysis. Psychosom Med. 2001;63(4):619–30. doi: 10.1097/00006842-200107000-00015. [DOI] [PubMed] [Google Scholar]
- 6.Lustman PJ, Anderson RJ, Freedland KE, de Groot M, Carney RM, Clouse RE. Depression and poor glycemic control: a meta-analytic review of the literature. Diabetes Care. 2000;23(7):934–42. doi: 10.2337/diacare.23.7.934. [DOI] [PubMed] [Google Scholar]
- 7.Bruce DG, Davis WA, Starkstein SE, Davis TM. A prospective study of depression and mortality in patients with type 2 diabetes: the Fremantle Diabetes Study. Diabetologia. 2005;48(12):2532–9. doi: 10.1007/s00125-005-0024-3. [DOI] [PubMed] [Google Scholar]
- 8.Katon WJ, Rutter C, Simon G, Lin EH, Ludman E, Ciechanowski P, Kinder L, Young B, Von Korff M. The association of comorbid depression with mortality in patients with type 2 diabetes. Diabetes Care. 2005;28(11):2668–72. doi: 10.2337/diacare.28.11.2668. [DOI] [PubMed] [Google Scholar]
- 9.Champaneri S, Wand GS, Malhotra SS, Casagrande SS, Golden SH. Biological basis of depression in adults with diabetes. Curr Diab Rep. 10(6):396–405. doi: 10.1007/s11892-010-0148-9. [DOI] [PubMed] [Google Scholar]
- 10.Mezuk B, Eaton WW, Albrecht S, Golden SH. Depression and type 2 diabetes over the lifespan: a meta-analysis. Diabetes Care. 2008;31(12):2383–90. doi: 10.2337/dc08-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCaffery JM, Duan QL, Frasure-Smith N, Barhdadi A, Lesperance F, Theroux P, Rouleau GA, Dube MP. Genetic predictors of depressive symptoms in cardiac patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):381–8. doi: 10.1002/ajmg.b.30824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney RM, Freedland KE, Miller GE, Jaffe AS. Depression as a risk factor for cardiac mortality and morbidity: a review of potential mechanisms. J Psychosom Res. 2002;53(4):897–902. doi: 10.1016/s0022-3999(02)00311-2. [DOI] [PubMed] [Google Scholar]
- 13.Frasure-Smith N, Lesperance F, Julien P. Major depression is associated with lower omega-3 fatty acid levels in patients with recent acute coronary syndromes. Biological psychiatry. 2004;55(9):891–6. doi: 10.1016/j.biopsych.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Lett HS, Blumenthal JA, Babyak MA, Sherwood A, Strauman T, Robins C, Newman MF. Depression as a risk factor for coronary artery disease: evidence, mechanisms, and treatment. Psychosom Med. 2004;66(3):305–15. doi: 10.1097/01.psy.0000126207.43307.c0. [DOI] [PubMed] [Google Scholar]
- 15.Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, Amin N, Bakshis E, Baumert J, Ding J, Liu Y, Marciante K, Meirelles O, Nalls MA, Sun YV, Vogelzangs N, Yu L, Bandinelli S, Benjamin EJ, Bennett DA, Boomsma D, Cannas A, Coker LH, de Geus E, De Jager PL, Diez-Roux AV, Purcell S, Hu FB, Rimm EB, Hunter DJ, Jensen MK, Curhan G, Rice K, Penman AD, Rotter JI, Sotoodehnia N, Emeny R, Eriksson JG, Evans DA, Ferrucci L, Fornage M, Gudnason V, Hofman A, Illig T, Kardia S, Kelly-Hayes M, Koenen K, Kraft P, Kuningas M, Massaro JM, Melzer D, Mulas A, Mulder CL, Murray A, Oostra BA, Palotie A, Penninx B, Petersmann A, Pilling LC, Psaty B, Rawal R, Reiman EM, Schulz A, Shulman JM, Singleton AB, Smith AV, Sutin AR, Uitterlinden AG, Volzke H, Widen E, Yaffe K, Zonderman AB, Cucca F, Harris T, Ladwig KH, Llewellyn DJ, Raikkonen K, Tanaka T, van Duijn CM, Grabe HJ, Launer LJ, Lunetta KL, Mosley TH, Jr, Newman AB, Tiemeier H, Murabito J. A genome-wide association study of depressive symptoms. Biological psychiatry. 2013;73(7):667–78. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooney GM, Dwan K, Greig CA, Lawlor DA, Rimer J, Waugh FR, McMurdo M, Mead GE. Exercise for depression. The Cochrane database of systematic reviews. 2013;9:CD004366. doi: 10.1002/14651858.CD004366.pub6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabricatore AN, Wadden TA, Higginbotham AJ, Faulconbridge LF, Nguyen AM, Heymsfield SB, Faith MS. Intentional weight loss and changes in symptoms of depression: a systematic review and meta-analysis. Int J Obes (Lond) 2011;35(11):1363–76. doi: 10.1038/ijo.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pi-Sunyer X, Blackburn G, Brancati FL, Bray GA, Bright R, Clark JM, Curtis JM, Espeland MA, Foreyt JP, Graves K, Haffner SM, Harrison B, Hill JO, Horton ES, Jakicic J, Jeffery RW, Johnson KC, Kahn S, Kelley DE, Kitabchi AE, Knowler WC, Lewis CE, Maschak-Carey BJ, Montgomery B, Nathan DM, Patricio J, Peters A, Redmon JB, Reeves RS, Ryan DH, Safford M, Van Dorsten B, Wadden TA, Wagenknecht L, Wesche-Thobaben J, Wing RR, Yanovski SZ. Reduction in weight and cardiovascular disease risk factors in individuals with type 2 diabetes: one-year results of the look AHEAD trial. Diabetes Care. 2007;30(6):1374–83. doi: 10.2337/dc07-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck AT, Ward CH, Mendelson M, Mock J, Eraugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–71. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 21.Faulconbridge LF, Wadden TA, Rubin RR, Wing RR, Walkup MP, Fabricatore AN, Coday M, Van Dorsten B, Mount DL, Ewing LJ. One-year changes in symptoms of depression and weight in overweight/obese individuals with type 2 diabetes in the Look AHEAD study. Obesity (Silver Spring) 2012;20(4):783–93. doi: 10.1038/oby.2011.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J, Frayling TM, Heid IM, Jackson AU, Johnson T, Kilpelainen TO, Lindgren CM, Morris AP, Prokopenko I, Randall JC, Saxena R, Soranzo N, Speliotes EK, Teslovich TM, Wheeler E, Maguire J, Parkin M, Potter S, Rayner NW, Robertson N, Stirrups K, Winckler W, Sanna S, Mulas A, Nagaraja R, Cucca F, Barroso I, Deloukas P, Loos RJ, Kathiresan S, Munroe PB, Newton-Cheh C, Pfeufer A, Samani NJ, Schunkert H, Hirschhorn JN, Altshuler D, McCarthy MI, Abecasis GR, Boehnke M. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 2008;3(10):e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan DH, Espeland MA, Foster GD, Haffner SM, Hubbard VS, Johnson KC, Kahn SE, Knowler WC, Yanovski SZ. Look AHEAD (Action for Health in Diabetes): design and methods for a clinical trial of weight loss for the prevention of cardiovascular disease in type 2 diabetes. Control Clin Trials. 2003;24(5):610–28. doi: 10.1016/s0197-2456(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 25.Bray G, Gregg E, Haffner S, Pi-Sunyer XF, WagenKnecht LE, Walkup M, Wing R. Baseline characteristics of the randomised cohort from the Look AHEAD (Action for Health in Diabetes) study. Diab Vasc Dis Res. 2006;3(3):202–15. doi: 10.3132/dvdr.2006.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck AT, Steer RA. Manual for the Beck Depression Inventory. San Antonio: Psychological Corporation; 1987. [Google Scholar]
- 27.Lustman PJ, Clouse RE, Griffith LS, Carney RM, Freedland KE. Screening for depression in diabetes using the Beck Depression Inventory. Psychosom Med. 1997;59(1):24–31. doi: 10.1097/00006842-199701000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–7. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 30.Dabney A, Storey JD. Q-value estimation for false discovery rate control. R package version 1.30.0. 2012 Available from: http://www.bioconductor.org/packages/release/bioc/html/qvalue.html.
- 31.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Diggle P, Heagery P, Liang K-Y, Zeger S. Analysis of Longitudinal Data. Oxford, U.K: Oxford University Press; 2013. [Google Scholar]
- 33.TIBCO Software I. TIBCO Spotfire SPLUS 8.2 for Solaris/Linux User’s Guide. Palo Alto, CA: TIBCO Software, Inc; 2010. [Google Scholar]
- 34.Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36(4):477–83. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 35.Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM, Breen G, Byrne EM, Blackwood DH, Boomsma DI, Cichon S, Heath AC, Holsboer F, Lucae S, Madden PA, Martin NG, McGuffin P, Muglia P, Noethen MM, Penninx BP, Pergadia ML, Potash JB, Rietschel M, Lin D, Muller-Myhsok B, Shi J, Steinberg S, Grabe HJ, Lichtenstein P, Magnusson P, Perlis RH, Preisig M, Smoller JW, Stefansson K, Uher R, Kutalik Z, Tansey KE, Teumer A, Viktorin A, Barnes MR, Bettecken T, Binder EB, Breuer R, Castro VM, Churchill SE, Coryell WH, Craddock N, Craig IW, Czamara D, De Geus EJ, Degenhardt F, Farmer AE, Fava M, Frank J, Gainer VS, Gallagher PJ, Gordon SD, Goryachev S, Gross M, Guipponi M, Henders AK, Herms S, Hickie IB, Hoefels S, Hoogendijk W, Hottenga JJ, Iosifescu DV, Ising M, Jones I, Jones L, Jung-Ying T, Knowles JA, Kohane IS, Kohli MA, Korszun A, Landen M, Lawson WB, Lewis G, Macintyre D, Maier W, Mattheisen M, McGrath PJ, McIntosh A, McLean A, Middeldorp CM, Middleton L, Montgomery GM, Murphy SN, Nauck M, Nolen WA, Nyholt DR, O’Donovan M, Oskarsson H, Pedersen N, Scheftner WA, Schulz A, Schulze TG, Shyn SI, Sigurdsson E, Slager SL, Smit JH, Stefansson H, Steffens M, Thorgeirsson T, Tozzi F, Treutlein J, Uhr M, van den Oord EJ, Van Grootheest G, Volzke H, Weilburg JB, Willemsen G, Zitman FG, Neale B, Daly M, Levinson DF, Sullivan PF. A mega-analysis of genome-wide association studies for major depressive disorder. Molecular psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radloff LS. The CES-D scale: a self-report depression scale for research in the general population. Applied Psychological Measurement. 1977;1:385–401. [Google Scholar]
- 37.Shean G, Baldwin G. Sensitivity and specificity of depression questionnaires in a college-age sample. The Journal of genetic psychology. 2008;169(3):281–8. doi: 10.3200/GNTP.169.3.281-292. [DOI] [PubMed] [Google Scholar]
- 38.Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA, Sotoodehnia N, Smith NL, Rotter JI, Kors JA, Witteman JC, Hofman A, Heckbert SR, O’Donnell CJ, Uitterlinden AG, Psaty BM, Lumley T, Larson MG, Stricker BH. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nature genetics. 2009;41(4):399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang C, Li X, Xu Y, Chen Q, Wu Y, Wang W, Li W, Qiu M. KCNE1 rs1805127 polymorphism increases the risk of atrial fibrillation: a meta-analysis of 10 studies. PloS one. 2013;8(7):e68690. doi: 10.1371/journal.pone.0068690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42(6):927–37. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Regional Manhattan plots for the association of KCNE1 with BDI scores without medication adjustment.
Supplemental Table 1. Participant characteristics for Non-Hispanic Whites only sample.
Supplemental Table 2. SNP Characteristics for a priori SNPs in the overall sample and in the three largest racial/ethnic groups, Non-Hispanic White, Non-Hispanic Black, and Hispanic individuals.
Supplemental Table 3. Top hits for baseline depressive symptoms in the full cohort and the Non-Hispanic White subsample for BDI scores without medication adjustment (FDR q-value < 0.30). Note: that no adjusted baseline results met the FDR criteria.)