Abstract
Objective
The relevance of the microbe-gut-brain axis to psychopathology is of interest in anorexia nervosa (AN), as the intestinal microbiota plays a critical role in metabolic function and weight regulation.
Methods
We characterized the composition and diversity of the intestinal microbiota in AN, using stool samples collected at inpatient admission (T1) (n=16) and discharge (T2) (n=10). At T1, participants completed the Beck Depression and Anxiety Inventories and the Eating Disorder Examination-Questionnaire. Patients with AN were compared to healthy individuals who participated in a previous study (healthy comparison group; HCG). Genomic DNA was isolated from stool samples, and bacterial composition was characterized by 454 pyrosequencing of the 16S rRNA gene. Sequencing results were processed by the Quantitative Insights Into Microbial Ecology pipeline. We compared T1 vs. T2 samples, samples from both points were compared to HCG (n=12), and associations between psychopathology and T1 samples were explored.
Results
In patients with AN, significant changes emerged between T1 and T2 in taxa abundance and beta (between-sample) diversity. Patients with AN had significantly lower alpha (within-sample) diversity than HCG at both T1 (p=0.0001) and T2 (p=0.016), and differences in taxa abundance were found between AN patients and HCG. Levels of depression, anxiety, and eating disorder psychopathology at T1 were associated with composition and diversity of the intestinal microbiota.
Conclusions
We provide evidence of intestinal dysbiosis in AN and an association between mood and the enteric microbiota in this patient population. Future directions include mechanistic investigations of the microbe-gut-brain axis in animal models and association of microbial measures with metabolic changes and recovery indices.
Keywords: eating disorders, anorexia nervosa, intestinal microbiota, gut-brain axis, depression, anxiety
Introduction
The robust and documented role of the intestinal microbiota in metabolic function and weight regulation provides a strong rationale for exploring the role of this complex microbial community in the emergence, maintenance, and recovery from anorexia nervosa (AN) (1). AN is a severe, life-threatening mental illness (2) associated with dangerously low body weight and biochemical, metabolic, immunologic, and sensory abnormalities (3–8), as well as mortality rates among the highest for any psychiatric disorder (9). Despite the significant morbidity and mortality associated with AN (9–12), and decades of research, the evidence base for its treatment is weak—especially during the initial renourishment phase (13, 14). Current models are unable to account for how individuals with AN can achieve and defend such low body weights.
The composition of the human microbiota, which includes the diverse microbial communities living in and on the human body, as well as the genetic material of these microorganisms (microbiome) and their interactions with the surrounding environment, has become a burgeoning area of study. The composition of these microbial communities can vary with age, sex, environment, geography, diet, and disease, but we understand little about the nature of these variations or their impact on human development, physiology, immunity, and nutrition (15). Seminal work by the Human Microbiome Project (HMP) has characterized the microbiome in a cohort of healthy individuals (16), whereas other investigators have focused on how deviations from the norm could contribute to diseases such as inflammatory bowel diseases (IBD) (17), asthma (18–23), and obesity (24–30).
A growing body of evidence from both animal models and human studies shows communication between the intestinal microbiota and the brain (i.e., the so-called “gut-brain axis”) (31). This phenomenon has not been studied in individuals with AN, and the specific mechanism(s) through which enteric microbes affect brain function remains unclear. However, individuals with AN often present with comorbid anxiety and depression—up to 80 percent will experience major depression at some point in their lifetime (32), while up to 75 percent will suffer from some form of anxiety disorder, including social phobia, specific phobia, and generalized anxiety disorder (33–35).
The intestinal microbiota plays a demonstrable role in weight gain/loss (24–30) and energy extraction from the diet (29, 30, 36) in human and animal models. Given that AN is marked primarily by extreme weight dysregulation (37), exploring the role of the intestinal microbiota in AN is a logical and inevitable next step. Consistent evidence implicates this enteric microbial community in obesity and metabolic outcomes, though the degree of that contribution is controversial (27–29, 38, 39). Findings suggest that the composition of the intestinal microbiota differs between obese and lean individuals (27, 28), and that obese individuals may extract more energy from a given diet than their lean counterparts (29), but very little is known about the gut microbiome in individuals with AN.
Intriguing published and preliminary findings suggest a role for the intestinal microbiota in AN. A recent culture-based study of a stool sample from an AN patient at hospital admission identified 11 completely new bacterial species in the Firmicutes (n=7), Bacteroidetes (n=2), and Actinobacteria (n=2) phyla, suggesting distinct characteristics of the gut microbiome in AN (40). Further research is needed to investigate whether these new species are uniquely associated with AN. In addition, a molecular-based study (24) analyzing the intestinal microbiota of nine patients with AN found increased levels of the archaeon Methanobrevibacter smithii (M. smithii). Because M. smithii and other methanogens play an important role in removing excess hydrogen gas from the gut and improving efficiency of microbial fermentation (and associated energy yield), this could demonstrate an adaptive response towards optimizing energy extraction from a very low-calorie diet. Although novel findings were reported, this study analyzed a limited number of microbial groups (two phyla: Bacteroidetes and Firmicutes; one genus: Lactobacillus; and one archaeon: M. smithii). Animal models also suggest that the intestinal microbiota influences satiety mechanisms through interaction with peptide signaling (41) and protective adaptation in a starvation state (42). A more comprehensive characterization of the intestinal microbiota of individuals with acute AN is required, along with exploration of changes in enteric microbes over the course of medically supervised weight restoration.
This study sought to (i) gain insight into the composition and diversity of the intestinal microbiota in a cohort of patients with acute AN; (ii) measure changes in the intestinal microbiota of patients with AN after hospital-based weight restoration; (iii) compare the intestinal microbiota in acutely ill patients with AN to that of a healthy comparison group (HCG); and (iv) examine associations between these microbial measures and depression, anxiety, and eating disorder psychopathology.
Methods and Materials
Ethics Statement
The study was approved by the Biomedical Institutional Review Board (IRB) at the University of North Carolina at Chapel Hill (UNC). All participants provided written consent prior to study participation.
Study Population
Females (n=16) admitted for inpatient treatment at the UNC Center of Excellence for Eating Disorders (CEED) participated in the study. Participants were recruited from consecutive inpatient admissions from December 2012 to May 2013, and inclusion criteria were as follows: (i) age 15–64 years; (ii) meet DSM-IV-TR criteria for AN; (iii) present at <75% of ideal body weight (IBW). Exclusion criteria were based on factors known to influence the composition of the intestinal microbiota: history of gastrointestinal tract surgery (other than appendectomy or cholecystectomy); history of IBD, irritable bowel syndrome (IBS), celiac disease, or any other diagnosis that could explain chronic or recurring bowel symptoms; treatment in the last two months with antibiotics, non-steroidal anti-inflammatory drugs, or steroids; or intentional use of probiotics during the last two months.
Data from HCG (n=12) with no recurring gastrointestinal symptoms were obtained from a previous study (43). This study recruited controls via advertisement from the general population in the same geographical region (central North Carolina) and from UNC outpatient clinics. HCG participants were subject to the same exclusion criteria as patients with AN and were selected for this analysis based on sex (female), age (15–64 years), and body mass index (BMI; 18.5–24.9 kg/m2). They were not screened for psychopathology during recruitment.
Data Collection and Preparation
Body composition and assessments
Weight and height were assessed using a calibrated digital scale and stadiometer. HCG participants were measured once. AN patients were weighed daily as part of standard treatment. Height was measured at admission for all AN patients and again at discharge for those under age 21. Eating disorders diagnosis and psychopathology were established via the Eating Disorder Examination (44) and the Structured Clinical Interview for DSM-IV-TR Axis I Disorders (SCID) (45) conducted by credentialed members of the CEED Assessment Core. AN patients also completed electronic versions of the Beck Anxiety Inventory (BAI) (46), Beck Depression Inventory-II (BDI) (47), and Eating Disorder Examination-Questionnaire (EDE-Q) (48) within 24 hours of admission.
Sample collection, processing, and storage
The first stool sample produced after admission (T1) was collected for all AN patients (n=16), and for a subset of these patients (n=10), an additional sample was collected prior to discharge (T2). Input and output are measured as part of routine treatment, minimizing risk of missing samples, and all samples were collected by nurses trained in collection protocols. Fresh stool samples were collected from HCG in the same manner as AN patients, as previously reported (43). All samples were transferred to the laboratory, where they were mechanically homogenized with a sterile spatula, aliquoted into sterile 2 mL cryovials, and stored in a −80°C freezer for future DNA isolation and nucleotide sequence analysis.
DNA isolation
Bacterial DNA was isolated from collected samples using a phenol/chloroform extraction method combined with physical disruption of bacterial cells and a DNA clean-up kit [Qiagen DNeasy® Blood and Tissue extraction kit (Qiagen, Valencia, CA, USA)], as previously described (43, 49).
454 Pyrosequencing of 16S rRNA Genes
Bacterial community composition in isolated DNA samples was characterized by amplification of the V1-3 (forward, 8f:5′-AGAGTTTGATCMTGGCTCAG-3′; reverse, 518r:5′-ATTACCGCGGCTGCTGG-3′) variable region of the 16S rRNA gene by polymerase chain reaction (PCR), as previously described (43). 16S rRNA PCR products were quantified, pooled, and purified for the sequencing reaction. Sequencing was performed on a 454 Life Sciences Genome Sequencer FLX machine (Roche, Florence, SC, USA) by the Microbiome Core Facility in the UNC School of Medicine.
Analysis of 16S rRNA Sequences using the QIIME Pipeline
16S rRNA sequence data generated by the 454 sequencer were processed by the Quantitative Insights Into Microbial Ecology (QIIME) pipeline (50). Sequences that were less than 200 base pairs (bp) or greater than 1,000 bp in length, contained incorrect primer sequences, or contained more than one ambiguous base were discarded (51). Sequences were clustered into Operational Taxonomic Units (OTUs; similar to species level) based on their sequence similarity at a 97% threshold using BLAST and assigned taxonomy using the Greengenes database (52). Principal coordinates (PCs) were generated using unweighted and weighted UniFrac distances (53–55). The richness of the intestinal microbiota was characterized by the number of observed bacterial species in each sample and the Chao-1 estimator of diversity (56, 57).
Statistical Analysis
Differences in alpha diversity (expressed as number of observed species and Chao-1 estimator), beta diversity (UniFrac distances), and taxa abundance of bacterial groups (at the phylum, class, order, family, and genus levels) were examined in AN patients (n=10) at T1 vs. T2 using 16S rRNA sequence data. Bacterial groups present in at least 25% of all samples at T1 or T2 were included in analyses. As response variables were not normally distributed, non-parametric testing was used. Depending on the symmetry of the distribution of the paired differences, differences were tested at the univariate level using Wilcoxon matched pairs rank test (−2 ≤ skewness ≤ 2) or the sign test (skewness ≤ −2 or ≥ 2). Power analysis was conducted in G*Power 3 to determine the effect size that could be detected with n=10, a two-tailed test, an alpha of 0.05, and power of 80%; under these conditions the Wilcoxon matched pairs rank test can detect a large effect (dz=1.1), as can the sign test (g=0.41). The false discovery rate (FDR) procedure addressed multiple testing (58) and was applied to the number of comparisons per outcome and per taxonomic rank. A global, multivariate hypothesis test developed for high-dimensional small-sample data (i.e., the type of data acquired through high-throughput technology in metabolomics, genomics, and proteomics) was also used to test for differences in alpha diversity, beta diversity, and taxa abundance across all bacterial groups (59). A global test offers additional conceptual advantages to a univariate test, since microbiota can work together or in a pathway and may have greater explanatory power when considered collectively.
Differences in alpha diversity, beta diversity, and taxa abundance of bacterial groups (at the phylum, class, order, family, and genus levels) were compared in AN patients at T1 (n=16) vs. HCG (n=12) and AN patients at T2 (n=10) vs. HCG with two-tailed Wilcoxon-Mann-Whitney tests. The Chi et al. (59) global multivariate test was used for beta diversity and per taxonomic level, and FDR correction was applied as described above.
Associations between T1 psychopathology scores measured as continuous variables [BDI (depression), BAI (anxiety), and EDE-Q (total + subscales for Dietary Restraint, Eating Concern, Shape Concern, and Weight Concern)] and alpha diversity, beta diversity, and taxa abundance of bacterial groups (at the phylum, class, order, family, and genus levels) were examined in AN patients (n=15; one patient did not complete the surveys) with the tau-b correlation coefficient. Bacterial groups present in at least 25% of T1 samples were considered. Univariate analyses used the Wilcoxon-Mann-Whitney test, and the FDR procedure was used to adjust for multiple testing, implemented per outcome and per taxonomic rank. The global multivariate test was implemented for beta diversity and per taxonomic level.
The alpha level used was 0.05, but for FDR correction, a more lenient criterion of 0.1 was used given the exploratory nature and small sample size. All analyses were conducted in SAS 9.3 (Cary, NC, USA).
Results
Demographic and clinical characteristics
Fecal samples were collected at T1 from female patients with AN (n=16). Average age was 28.0 (±11.7) (Mean ± SD) years, and mean BMI at T1 was 16.2 (1.5) kg/m2. A subset of patients (n=10) provided an additional sample at T2, when they had reached a mean BMI of 17.4 (0.9) kg/m2. Female HCG (n=12) who provided samples had a mean age of 29.8 (11.6) years and mean BMI of 21.5 (1.9) kg/m2. Participants were predominately Caucasian (n=14 patients with AN; n=7 HCG), with a small representation of African American participants (n=2 patients with AN; n=1 HCG). Four HCG participants did not provide information on race.
At T1, patients with AN (n=15) had mean BDI and BAI scores of 26.6 (13.4) and 17.7 (11.9), respectively, reflecting moderate depression and anxiety (46, 60). The majority of patients endorsed at least mild levels of depression (80.0%) and anxiety (66.7%). Mean EDE-Q total scores of 3.6 (1.8), and scores on subscales for Dietary Restraint [3.7 (1.9)], Eating Concern [3.4 (1.9)], Shape Concern [3.8 (1.9)], and Weight Concern [3.4 (2.1)] are consistent with other clinical samples of patients with AN (61, 62).
From the 26 fecal samples analyzed from patients with AN, a total of 197,956 16S rRNA sequences with acceptable quality were obtained with an average of 7,613 reads per sample (range: 4,101–9,511). From the 12 fecal samples analyzed from HCG, a total of 122,461 16S rRNA sequences with acceptable quality were obtained with an average of 10,205 reads per sample (range: 5,265–15,596). Using a 97% similarity threshold, we found a total of 1,666 and 2,020 OTUs in the samples analyzed from patients with AN and HCG, respectively.
Changes to the intestinal microbiota during hospital-based weight restoration in patients with AN
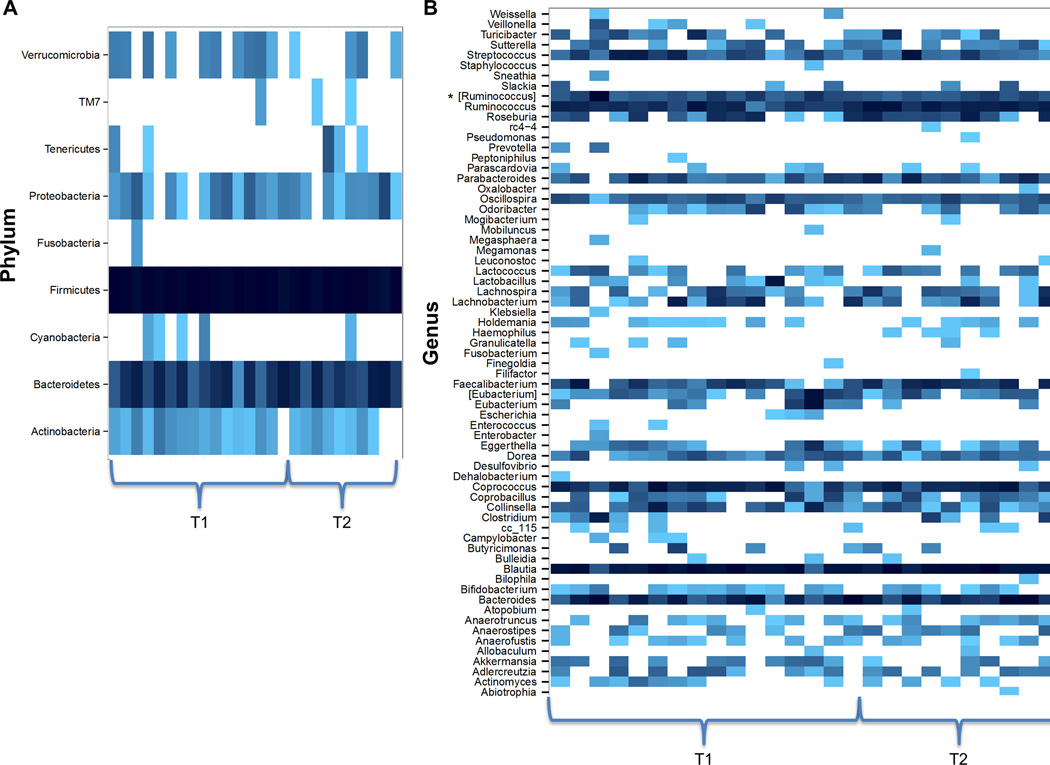
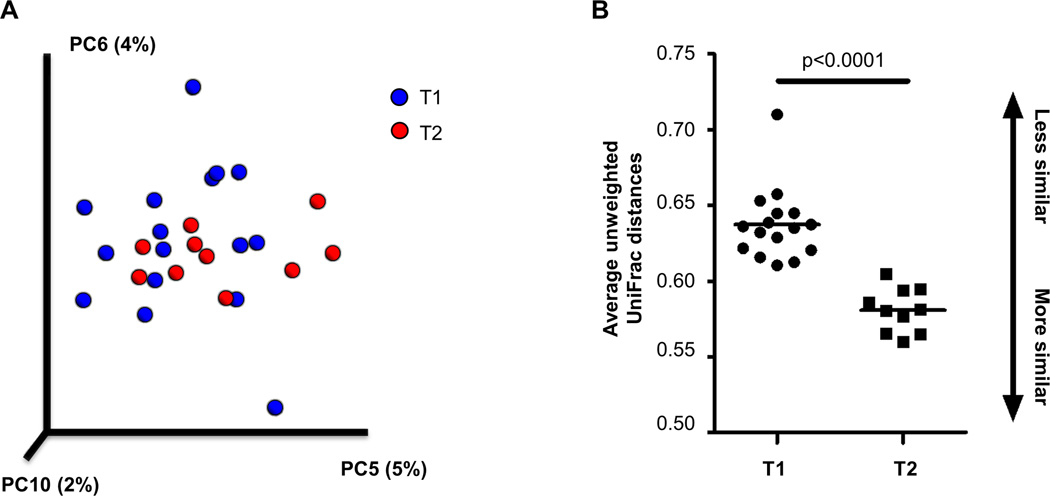
Table 1 presents changes in bacterial composition and diversity over the course of inpatient weight restoration. Global tests indicated significant differences between T1 and T2 in beta (between-sample) diversity (p<0.001) and at the phylum (p=0.042) and genus (p=0.041) taxonomic levels (Figure 1). Based on unweighted UniFrac distances, three principal coordinates (5, 6, 10) were significantly different at hospital admission and discharge and remained significant at an FDR level of 0.1. The average unweighted UniFrac distances were significantly different between groups (p<0.0001), with T2 samples showing greater similarity to each other than T1 samples (Figure 2). The strongest taxonomic changes were seen in the family Ruminococcaceae, with significant changes in specified (Ruminococcus; p=0.002) and unspecified (p=0.004) sub-genera.
Table 1.
Hospital admission (T1) vs. hospital discharge (T2): differences in microbial taxa and diversity measures in females with AN (n=10)
| Taxonomic/diversity level |
Classification | Test statistic |
p | FDR corrected p |
|---|---|---|---|---|
| Global Tests | ||||
| Phylum | 2.66 | 0.042 | ||
| Class | 2.14 | 0.067 | ||
| Order | 1.77 | 0.064 | ||
| Family | 1.70 | 0.064 | ||
| Genus | 1.74 | 0.041 | ||
| Beta diversity | Weighted | 1.50 | 0.22 | |
| Unweighted | 6.07 | 0.0003 | ||
| Univariate Tests | ||||
| Family | Eubacteriaceae | −3.5 | 0.039 | 0.63 |
| Genus | Ruminococcaceae_genus | −26.5 | 0.004 | 0.10 |
| Oscillospira | −22.5 | 0.020 | 0.34 | |
| Ruminococcus | 27.5 | 0.002 | 0.10 | |
| Beta diversity | Unweighted (PC 5) | −23.5 | 0.014 | 0.070 |
| Unweighted (PC 6) | −21.5 | 0.027 | 0.090 | |
| Unweighted (PC 10) | 26.5 | 0.004 | 0.040 | |
FDR = false discovery rate; PC = principal coordinate
Alpha = 0.05; FDR level = 0.1
A global multivariate test (59) was used to test for differences in alpha diversity, beta diversity, and abundance per taxonomic level. Depending on the symmetry of the distribution of the paired differences, differences were tested at the univariate level using Wilcoxon matched pairs rank test (−2 ≤ skewness ≤ 2) or the sign test (skewness ≤ −2 or ≥ 2).
Figure 1.
Heatmaps of samples from patients with anorexia nervosa (AN) at hospital admission (T1; n=16) and discharge (T2; n=10) at the (A) phylum and (B) genus taxonomic levels. Bacterial composition was characterized by 454 pyrosequencing of the 16S rRNA gene, and sequencing results were processed by the Quantitative Insights Into Microbial Ecology (QIIME) pipeline. A global, multivariate hypothesis test was used to test for differences in abundance across all bacterial groups at once, per taxonomic level, and indicated significant differences between T1 and T2 at the phylum (p=0.042) and genus (p=0.041) levels. Bacterial taxa are listed vertically, and samples are grouped horizontally by time point (T1 or T2). Greater abundance is designated by darker shading. * [Ruminococcus] indicates unspecified genera in the Ruminococcaceae family.
Figure 2.
Principal coordinates (PC) plot of samples from patients with anorexia nervosa (AN) and average unweighted UniFrac distances at hospital admission (T1; n=16) and discharge (T2; n=10). Bacterial composition was characterized by 454 pyrosequencing of the 16S rRNA gene, and sequencing results were processed by the Quantitative Insights Into Microbial Ecology (QIIME) pipeline. (A) Based on unweighted UniFrac distances, three principal coordinates (PC5, PC6, PC10) were significantly different at T1 vs. T2. Percentages indicate the amount of variability in the data explained by each PC. Samples from T1 and T2 are designated by royal blue and red dots, respectively. (B) The average unweighted UniFrac distances were significantly different across groups (p<0.0001), with T2 samples showing greater similarity to each other than T1 samples.
Comparison of intestinal microbiota in patients with AN before and after weight restoration in comparison to HCG
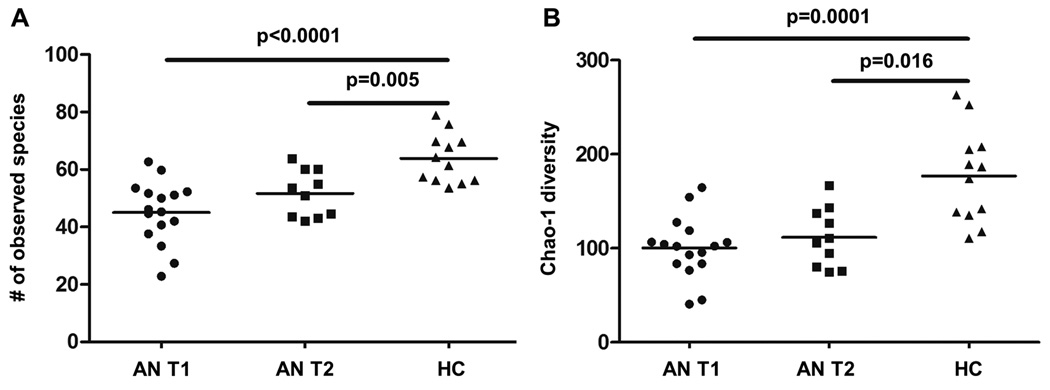
We compared the intestinal microbiota in patients with AN at T1 and T2 to that of age- and sex-matched HCG. At both time points, the alpha (within-sample) diversity remained significantly lower in patients with AN vs. HCG, measured as either the number of observed species or Chao-1 estimator (Figure 3, Tables 2 and 3). However, the bacterial composition of samples from patients with AN at T1 showed greater differences with HCG than samples collected at T2. At T1, patients with AN had greater levels of class Bacilli (p=0.007) and unspecified genera in family Coriobacteriales (p<0.001) and reduced levels of class Clostridia (p=0.007), order Clostridiales (p=0.006), and genera Anaerostipes (p=0.003) and Faecalibacterium (p=0.002) vs. HCG (with all differences remaining significant at an FDR level of 0.1) (Table 2). At T2, the only one of these differences that remained significant was in unspecified genera in family Coriobacteriales (p<0.001), though there were additional differences between patients with AN at T2 and HCG among the family Ruminococcaceae (p=0.002) and genera Parabacteroides (p=0.006) (Table 3).
Figure 3.
Alpha diversity in samples from patients with anorexia nervosa (AN) at hospital admission (T1; n=16) and discharge (T2; n=10) and a healthy comparison group (HCG; n=12). Bacterial composition was characterized by 454 pyrosequencing of the 16S rRNA gene, and sequencing results were processed by the Quantitative Insights Into Microbial Ecology (QIIME) pipeline. Richness was characterized by the (A) number of observed bacterial species in each sample; and (B) Chao-1 estimator of diversity. Differences in alpha (within-sample) diversity were compared in AN T1 vs. AN T2 vs. HCG with two-tailed Wilcoxon-Mann-Whitney tests. At both time points (T1 and T2), the alpha diversity remained significantly lower in patients with AN vs. HCG, measured as either the number of observed species or Chao-1 estimator.
Table 2.
Hospital admission (T1): differences in microbial taxa and diversity measures in females with AN (n=16) vs. healthy comparison group (n=12)
| Taxonomic/diversity level |
Classification | Test statistic |
p | FDR corrected p |
|---|---|---|---|---|
| Class | Bacilli | −2.72 | 0.007 | 0.026 |
| Clostridia | 2.72 | 0.007 | 0.026 | |
| Order | Clostridiales | 2.76 | 0.006 | 0.068 |
| Lactobacillales | −2.25 | 0.024 | 0.15 | |
| Family | Actinomycetaceae | −2.03 | 0.042 | 0.23 |
| Lachnospiraceae | 2.02 | 0.043 | 0.23 | |
| Porphyromonadaceae | −2.60 | 0.009 | 0.14 | |
| Ruminococcaceae | 2.53 | 0.011 | 0.14 | |
| Streptococcaceae | −1.97 | 0.049 | 0.23 | |
| Genus | Anaerostipes | 2.99 | 0.003 | 0.042 |
| Blautia | 2.06 | 0.031 | 0.17 | |
| Coribacteriales_genus | −4.62 | <0.0001 | 0.005 | |
| Faecalibacterium | 3.18 | 0.002 | 0.034 | |
| Lachnospira | 2.16 | 0.030 | 0.17 | |
| Parabacteroides | −2.60 | 0.009 | 0.10 | |
| Ruminococcaceae_genus | 2.30 | 0.022 | 0.16 | |
| Ruminococcus | 2.39 | 0.017 | 0.15 | |
| Alpha diversity | # of observed species | 4.02 | <0.0001 | |
| Chao-1 estimator | 3.83 | 0.0001 | ||
FDR = false discovery rate
Alpha = 0.05; FDR level = 0.1
Differences in alpha diversity, beta diversity, and taxa abundance of bacterial groups were compared with two-tailed Wilcoxon-Mann-Whitney tests. A global multivariate test (59)was used for alpha diversity, beta diversity, and abundance per taxonomic level.
Table 3.
Hospital discharge (T2): differences in microbial taxa and diversity measures in females with AN (n=10) vs. healthy comparison group (n=12)
| Taxonomic/diversity level |
Classification | Test statistic |
p | FDR corrected p |
|---|---|---|---|---|
| Phylum | Bacteroidetes | 2.08 | 0.038 | 0.11 |
| Firmicutes | −2.08 | 0.038 | 0.11 | |
| Class | Bacteroidia | 2.08 | 0.038 | 0.35 |
| Order | Bacteroidales | 2.08 | 0.038 | 0.42 |
| Family | Porphyromonadaceae | 2.74 | 0.006 | 0.15 |
| Genus | Coribacteriales_genus | 4.29 | <0.0001 | 0.004 |
| Parabacteroides | 2.74 | 0.006 | 0.065 | |
| Ruminococcaceae_genus | −3.07 | 0.002 | 0.047 | |
| Alpha diversity | # of observed species | −2.80 | 0.005 | |
| Chao-1 estimator | −2.41 | 0.016 | ||
FDR = false discovery rate
Alpha = 0.05; FDR level = 0.1
Differences in alpha diversity, beta diversity, and taxa abundance of bacterial groups were compared with two-tailed Wilcoxon-Mann-Whitney tests. A global multivariate test (59) was used for alpha diversity, beta diversity, and abundance per taxonomic level.
Association between bacterial composition and diversity and depression, anxiety, and eating disorder psychopathology in patients with AN
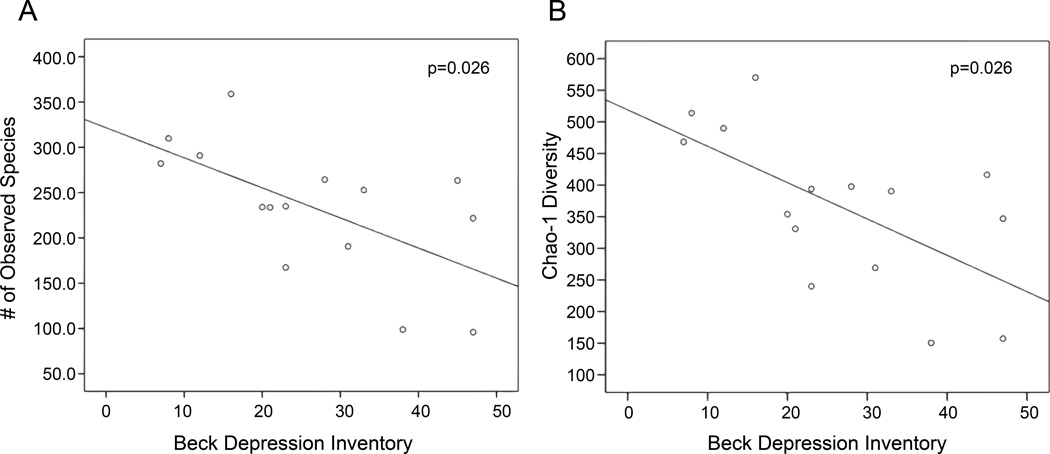
Alpha (within-sample) diversity, measured as bacterial richness or the Chao-1 estimator, was significantly associated with scores on the BDI and EDE-Q. Greater levels of depression were negatively associated with the number of observed bacterial species (p=0.026) and Chao-1 estimator (p=0.026) (Figure 4, Table 4). Lower number of observed species was also associated with greater levels of eating disorder psychopathology, measured as EDE-Q total score (p=0.026) or scores on subscales for Shape Concern (p=0.008) and Weight Concern (p=0.025) (Table 5). All associations remained significant at an FDR level of 0.1.
Figure 4.
Correlation between depression and alpha diversity in samples from patients with AN at hospital admission (T1). Bacterial composition was characterized by 454 pyrosequencing of the 16S rRNA gene, and sequencing results were processed by the Quantitative Insights Into Microbial Ecology (QIIME) pipeline. Richness (vertical axes) was characterized by the (A) number of observed bacterial species in each sample and (B) Chao-1 estimator of diversity. AN patients (n=15; one patient did not complete the surveys) completed the Beck Depression Inventory-II (horizontal axes) within 24 hours of admission. Associations between T1 psychopathology scores and alpha diversity were examined with the tau-b correlation coefficient. Depression was negatively associated with the number of observed bacterial species (p=0.026) and Chao-1 estimator (p=0.026).
Table 4.
Hospital admission (T1): microbial taxa and diversity measures associated with depression and/or anxiety in females with AN (n=15)
| Taxonomic/diversity level |
Classification | Behavioral measure |
Test statistic |
p | FDR corrected p |
|---|---|---|---|---|---|
| Class | Clostridia | BDI | −0.394 | 0.042 | 0.50 |
| Order | Actinomycetales | BDI | 0.406 | 0.040 | 0.40 |
| Clostridiales | BDI | −0.394 | 0.042 | 0.40 | |
| Coriobacteriales | BAI | 0.410 | 0.036 | 0.40 | |
| Family | Actinomycetaceae | BDI | 0.406 | 0.040 | 0.46 |
| Rikenellaceae | BDI | −0.488 | 0.016 | 0.34 | |
| Ruminococcaceae | BDI | −0.587 | 0.002 | 0.16 | |
| BAI | −0.566 | 0.004 | 0.16 | ||
| Genus | Blautia | BDI | −0.433 | 0.026 | 0.47 |
| Faecalibacterium | BDI | −0.386 | 0.047 | 0.47 | |
| Lachnospira | BDI | −0.526 | 0.008 | 0.47 | |
| BAI | −0.421 | 0.037 | 0.47 | ||
| Rikenellaceae_genus | BDI | −0.488 | 0.016 | 0.47 | |
| Roseburia | BDI | −0.406 | 0.040 | 0.47 | |
| BAI | −0.503 | 0.012 | 0.47 | ||
| Ruminococcus | BDI | −0.490 | 0.011 | 0.47 | |
| BAI | −0.527 | 0.007 | 0.47 | ||
| Veillonella | BDI | 0.460 | 0.034 | 0.47 | |
| Alpha diversity | # of observed species | BDI | −0.433 | 0.026 | 0.045 |
| Chao-1 estimator | BDI | −0.433 | 0.026 | 0.090 | |
| Beta diversity | Weighted (PC 2) | BDI | −0.510 | 0.009 | 0.45 |
| BAI | −0.488 | 0.013 | 0.45 | ||
FDR = false discovery rate; PC = principal coordinate; BDI = Beck Depression Inventory-II; BAI = Beck Anxiety Inventory
Alpha = 0.05; FDR level = 0.1
Associations were examined with the tau-b correlation coefficient.
Table 5.
Hospital admission (T1): microbial taxa and diversity measures associated with eating disorder psychopathology in females with AN (n=15)
| Taxonomic/diversity level |
Classification | Behavioral measure |
Test statistic |
p | FDR corrected p |
|---|---|---|---|---|---|
| Order | Actinomycetales | ShapeC | 0.450 | 0.024 | 0.40 |
| WeightC | 0.434 | 0.030 | 0.40 | ||
| Clostridiales | EatingC | −0.452 | 0.020 | 0.40 | |
| EDEQ | −0.448 | 0.020 | 0.40 | ||
| Turicibacterales | EatingC | −0.484 | 0.018 | 0.40 | |
| Family | Actinomycetaceae | ShapeC | 0.450 | 0.024 | 0.34 |
| WeightC | 0.434 | 0.030 | 0.38 | ||
| Clostridiaceae | Restraint | −0.437 | 0.025 | 0.34 | |
| Clostridiales_family | WeightC | −0.453 | 0.022 | 0.34 | |
| Odoribacteraceae | Restraint | 0.450 | 0.024 | 0.34 | |
| Ruminococcaceae | Restraint | −0.554 | 0.005 | 0.16 | |
| EatingC | −0.529 | 0.006 | 0.16 | ||
| ShapeC | −0.534 | 0.006 | 0.16 | ||
| WeightC | −0.579 | 0.003 | 0.16 | ||
| EDEQ | −0.600 | 0.002 | 0.16 | ||
| Turicibacteraceae | EatingC | −0.484 | 0.018 | 0.34 | |
| Genus | Anaerostipes | EatingC | −0.463 | 0.028 | 0.47 |
| ShapeC | −0.491 | 0.021 | 0.47 | ||
| WeightC | −0.508 | 0.017 | 0.47 | ||
| EDEQ | −0.505 | 0.016 | 0.47 | ||
| Bacteroidaceae_genus | EatingC | 0.449 | 0.030 | 0.47 | |
| Clostridiales_genus | WeightC | −0.453 | 0.022 | 0.47 | |
| Clostridium | EatingC | −0.445 | 0.037 | 0.47 | |
| Eubacteriaceae_genus | ShapeC | −0.431 | 0.042 | 0.47 | |
| Faecalibacterium | EatingC | −0.425 | 0.029 | 0.47 | |
| ShapeC | −0.459 | 0.019 | 0.47 | ||
| WeightC | −0.394 | 0.046 | 0.47 | ||
| EDEQ | −0.383 | 0.047 | 0.47 | ||
| Lachnospira | ShapeC | −0.510 | 0.011 | 0.47 | |
| WeightC | −0.495 | 0.015 | 0.47 | ||
| EDEQ | −0.411 | 0.038 | 0.47 | ||
| Ruminococcaceae_genus | EatingC | −0.433 | 0.026 | 0.47 | |
| ShapeC | −0.418 | 0.032 | 0.47 | ||
| WeightC | −0.422 | 0.032 | 0.47 | ||
| TotalC | −0.429 | 0.026 | 0.47 | ||
| Ruminococcus | EDEQ | −0.390 | 0.042 | 0.47 | |
| Turicibacter | EatingC | −0.484 | 0.018 | 0.47 | |
| Veillonella | WeightC | −0.512 | 0.020 | 0.47 | |
| Alpha diversity | # of observed species | ShapeC | −0.515 | 0.008 | 0.045 |
| WeightC | −0.441 | 0.025 | 0.045 | ||
| EDEQ | −0.429 | 0.026 | 0.045 | ||
| Chao-1 estimator | ShapeC | −0.476 | 0.015 | 0.090 | |
| Beta diversity | Weighted (PC 4) | EatingC | −0.394 | 0.042 | 0.83 |
| Unweighted (PC 1) | EatingC | −0.413 | 0.033 | 0.58 | |
| ShapeC | −0.437 | 0.025 | 0.58 | ||
| WeightC | −0.422 | 0.032 | 0.58 | ||
| EDEQ | −0.448 | 0.020 | 0.58 | ||
FDR = false discovery rate; PC = principal coordinate
EDEQ refers to the total score on the Eating Disorder Examination-Questionnaire. The following subscales are also included: Dietary Restraint (Restraint), Eating Concern (EatingC), Shape Concern (ShapeC), Weight Concern (WeightC)
Alpha = 0.05; FDR level = 0.1
Associations were examined with the tau-b correlation coefficient.
Significant associations were also seen between specific bacterial taxa and BDI, BAI, and EDE-Q scores, but none remained significant at an FDR level of 0.1. The strongest (negative) associations were seen with the family Ruminococcaceae (Tables 4 and 5).
Discussion
In examining the composition and diversity of the intestinal microbiota in patients undergoing inpatient treatment for AN, we report (i) changes over the course of hospital-based weight restoration; (ii) significant differences between patients with AN and HCG; and (iii) associations between microbial measures and depression, anxiety, and eating disorder psychopathology. These results extend findings from earlier, smaller studies of patients with AN and provide strong support for future work, including mechanistic studies of gut-brain interaction, to better understand the biological mechanisms at work in the risk and maintenance of AN.
Significant changes in the composition of the intestinal microbiota were seen in patients with AN during renourishment, particularly among genera falling under the family Ruminococcaceae. This family of bacteria has been associated with intestinal disorders marked by inflammation, including IBS and IBD (63, 64).
In comparing the intestinal microbiota of patients with AN to that of HCG, we found that alpha diversity was significantly lower in patients with AN both before and after inpatient weight restoration. Alpha diversity was also significantly associated with depression and eating disorder psychopathology in our patient group, with a lower number of observed bacterial species associated with greater depression and greater weight concern, shape concern, and overall eating disorder psychopathology. These results show intriguing associations and underscore findings across various other disease states, including IBD (17) and arthritis (65), which have shown that a healthier gut is a more diverse one. Moreover, as we found greater differences in bacterial composition between AN and HCG before vs. after hospital-based renourishment, our results may suggest that the intestinal microbiota is trending toward a healthier state during treatment.
Although there has been limited research to date into the role of the intestinal microbiota in AN, some parallels can be drawn to microbial changes associated with malnutrition. Studies have demonstrated that acute malnutrition in children is marked by an intestinal dysbiosis and that the malnutrition phenotype (marked by severe weight loss) can be transmitted via the intestinal microbiota in a gnotobiotic mouse model (66, 67). This microbial dysbiosis may also interact with a nutrient-deficient diet to affect energy metabolism and cause malnourishment to persist (66, 68).
Mounting evidence from animal studies in which the intestinal microbiota have been manipulated through probiotics, antibiotics, or microbial transfer to gnotobiotic mice has shown that behavior is associated with changes in bacterial composition and diversity (69–77). This includes models of depression and anxiety, which are common among patients with AN (34, 35). However, we have little evidence supporting associations between the intestinal microbiota and depression or anxiety disorders in humans (78–80). Our results, particularly those showing that lower bacterial diversity is associated with greater depression and anxiety, are at the forefront of providing evidence for the gut-brain axis in a human population.
Several limitations should be taken into account when considering these results. First, we did not control for diet of either patients with AN or HCG. The composition of the intestinal microbiota is strongly influenced by long-term dietary patterns (81, 82), and short-term dietary changes can also induce dramatic microbial shifts (83). Because patients resided on an inpatient hospital unit, dietary intake was controlled, and all participants consumed a standard diet, with far less variation across individuals than what would be expected from those in a free-living environment. In addition, our sample size limited power to detect differences between patients and controls over the course of renourishment. However, we did see some significant compositional changes during inpatient treatment, as well as significant global changes in composition and diversity using a statistical method that provides greater explanatory power by considering the intestinal microbiota collectively. Third, all of our study participants were female, limiting generalizability of the results to males, who comprise approximately 10 percent of individuals with AN (84). Given that we would be unlikely to recruit a sufficient number of male participants to allow testing for sex differences, we focused recruitment on females in order to maximize sample size. Lastly, we are unable to distinguish between changes to the intestinal microbiota that reflect weight gain vs. recovery from AN, which will be important in future work, as BMI alone is associated with abundance of specific bacteria (85).
Although genetic and neurobiological research underscore that AN is most accurately considered a biologically-based mental illness, the neurobiology of AN remains an enigma, which has hindered the development of novel, safe, and effective treatments. These findings are an important first step in uncovering the role of the intestinal microbiota in AN. Future mechanistic studies examining the impact of specific taxa on behavior and adiposity, including transplantation of the intestinal microbiota of patients with AN into gnotobiotic mice, will allow us to distinguish between microbial markers of renourishment vs. recovery from psychopathology and move us even closer to designing innovative therapies for AN targeting enteric microorganisms. Such studies could identify specific bacterial taxa whose promotion or elimination would improve the efficiency of therapeutic weight restoration, as well as the psychological and physical treatment experience of patients, and lead to pathophysiological-directed therapeutic approaches for the management of AN via probiotic, prebiotic, symbiotic, or antibiotic means.
Acknowledgements
Healthy comparison group data was previously published in the journal of Neurogastroenterology and Motility (2012) (43). Special thanks to Anthony Fodor, PhD and Raad Gharaibeh, PhD for their advice on statistical analysis. The authors would like to thank the UNC Microbiome Core Facility for their contribution to this study.
Source of Funding:
Dr. Bulik is a consultant for Shire Pharmaceuticals. Dr. Carroll is a consultant for Salix Pharmaceuticals, Inc. This study was funded by the Foundation of Hope, the UNC Center of Excellence for Eating Disorders Board of Visitors Fund, and TJ’s Fund for Eating Disorders Research (Academy for Eating Disorders; Bulik: PI) and partially supported by a K01 DK 092330 and a CGIBD pilot feasibility grant P30DK03498 awarded to Dr. Carroll.
Abbreviations
- AN
Anorexia nervosa
- BAI
Beck Anxiety Inventory
- BDI
Beck Depression Inventory-II
- BMI
Body mass index
- BP
Base pairs
- CEED
Center of Excellence for Eating Disorders
- DSM-IV-TR
Diagnostic and Statistical Manual of Mental Disorders, 4th Edition, Text Revision
- EDE-Q
Eating Disorder Examination-Questionnaire
- FDR
False discovery rate
- HCG
Healthy comparison group
- HMP
Human Microbiome Project
- IBD
Inflammatory bowel disease
- IBS
Irritable bowel syndrome
- IBW
Ideal body weight
- IRB
Institutional Review Board
- M
Methanobrevibacter
- OTU
Operational Taxonomic Unit
- PC
Principal coordinate
- PCR
Polymerase chain reaction
- QIIME
Quantitative Insights Into Microbial Ecology
- SCID
Structured Clinical Interview for DSM-IV-TR
- UNC
University of North Carolina at Chapel Hill
Footnotes
Conflicts of Interest: The remaining authors reported no biomedical financial interests or potential conflicts of interest.
References
- 1.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klump KL, Bulik CM, Kaye WH, Treasure J, Tyson E. Academy for eating disorders position paper: eating disorders are serious mental illnesses. The International journal of eating disorders. 2009;42:97–103. doi: 10.1002/eat.20589. [DOI] [PubMed] [Google Scholar]
- 3.Milner MR, McAnarney ER, Klish WJ. Metabolic abnormalities in adolescent patients with anorexia nervosa. J Adolesc Health Care. 1985;6:191–195. doi: 10.1016/s0197-0070(85)80016-4. [DOI] [PubMed] [Google Scholar]
- 4.Mira M, Stewart PM, Vizzard J, Abraham S. Biochemical abnormalities in anorexia nervosa and bulimia. Ann Clin Biochem. 1987;24(Pt 1):29–35. doi: 10.1177/000456328702400104. [DOI] [PubMed] [Google Scholar]
- 5.Mont L, Castro J, Herreros B, Pare C, Azqueta M, Magrina J, Puig J, Toro J, Brugada J. Reversibility of cardiac abnormalities in adolescents with anorexia nervosa after weight recovery. J Am Acad Child Adolesc Psychiatry. 2003;42:808–813. doi: 10.1097/01.CHI.0000046867.56865.EB. [DOI] [PubMed] [Google Scholar]
- 6.Nova E, Gomez-Martinez S, Morande G, Marcos A. Cytokine production by blood mononuclear cells from in-patients with anorexia nervosa. The British journal of nutrition. 2002;88:183–188. doi: 10.1079/BJNBJN2002608. [DOI] [PubMed] [Google Scholar]
- 7.Ulger Z, Gurses D, Ozyurek AR, Arikan C, Levent E, Aydogdu S. Follow-up of cardiac abnormalities in female adolescents with anorexia nervosa after refeeding. Acta Cardiol. 2006;61:43–49. doi: 10.2143/AC.61.1.2005139. [DOI] [PubMed] [Google Scholar]
- 8.Umeki S. Biochemical abnormalities of the serum in anorexia nervosa. J Nerv Ment Dis. 1988;176:503–506. doi: 10.1097/00005053-198808000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan PF. Mortality in anorexia nervosa. The American journal of psychiatry. 1995;152:1073–1074. doi: 10.1176/ajp.152.7.1073. [DOI] [PubMed] [Google Scholar]
- 10.Birmingham CL, Su J, Hlynsky JA, Goldner EM, Gao M. The mortality rate from anorexia nervosa. The International journal of eating disorders. 2005;38:143–146. doi: 10.1002/eat.20164. [DOI] [PubMed] [Google Scholar]
- 11.Hudson JI, Hiripi E, Pope HG, Jr, Kessler RC. The prevalence and correlates of eating disorders in the National Comorbidity Survey Replication. Biol Psychiatry. 2007;61:348–358. doi: 10.1016/j.biopsych.2006.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papadopoulos FC, Ekbom A, Brandt L, Ekselius L. Excess mortality, causes of death and prognostic factors in anorexia nervosa. The British journal of psychiatry : the journal of mental science. 2009;194:10–17. doi: 10.1192/bjp.bp.108.054742. [DOI] [PubMed] [Google Scholar]
- 13.Hart LM, Granillo MT, Jorm AF, Paxton SJ. Unmet need for treatment in the eating disorders: a systematic review of eating disorder specific treatment seeking among community cases. Clinical psychology review. 2011;31:727–735. doi: 10.1016/j.cpr.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Excellence NIfC, editor. NICE. Eating disorders: Core interventions in the treatment and management of anorexia nervosa, bulimia nervosa and related eating disorders. 2004. [PubMed] [Google Scholar]
- 15.HMP. Human Microbiome Project – Core Microbiome Sampling Protocol A. Available from: http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/GetPdf.cgi?id=phd002854.2.
- 16.A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 18.Azad MB, Kozyrskyj AL. Perinatal programming of asthma: the role of gut microbiota. Clinical & developmental immunology. 2012;2012:932072. doi: 10.1155/2012/932072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaser MJ. Equilibria of humans and our indigenous microbiota affecting asthma. Proceedings of the American Thoracic Society. 2012;9:69–71. doi: 10.1513/pats.201108-048MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brar T, Nagaraj S, Mohapatra S. Microbes and asthma: the missing cellular and molecular links. Current opinion in pulmonary medicine. 2012;18:14–22. doi: 10.1097/MCP.0b013e32834dccc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couzin-Frankel J. Bacteria and asthma: untangling the links. Science. 2010;330:1168–1169. doi: 10.1126/science.330.6008.1168. [DOI] [PubMed] [Google Scholar]
- 22.Han MK, Huang YJ, Lipuma JJ, Boushey HA, Boucher RC, Cookson WO, Curtis JL, Erb-Downward J, Lynch SV, Sethi S, Toews GB, Young VB, Wolfgang MC, Huffnagle GB, Martinez FJ. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67:456–463. doi: 10.1136/thoraxjnl-2011-201183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vael C, Vanheirstraeten L, Desager KN, Goossens H. Denaturing gradient gel electrophoresis of neonatal intestinal microbiota in relation to the development of asthma. BMC microbiology. 2011;11:68. doi: 10.1186/1471-2180-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armougom F, Henry M, Vialettes B, Raccah D, Raoult D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PloS one. 2009;4:e7125. doi: 10.1371/journal.pone.0007125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calvani R, Miccheli A, Capuani G, Tomassini Miccheli A, Puccetti C, Delfini M, Iaconelli A, Nanni G, Mingrone G. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes (Lond) 2010;34:1095–1098. doi: 10.1038/ijo.2010.44. [DOI] [PubMed] [Google Scholar]
- 26.Kootte RS, Vrieze A, Holleman F, Dallinga-Thie GM, Zoetendal EG, de Vos WM, Groen AK, Hoekstra JB, Stroes ES, Nieuwdorp M. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes Metab. 2012;14:112–120. doi: 10.1111/j.1463-1326.2011.01483.x. [DOI] [PubMed] [Google Scholar]
- 27.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 29.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cryan JF, O'Mahony SM. The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol Motil. 2011;23:187–192. doi: 10.1111/j.1365-2982.2010.01664.x. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez-Aranda F, Pinheiro AP, Tozzi F, Thornton LM, Fichter MM, Halmi KA, Kaplan AS, Klump KL, Strober M, Woodside DB, Crow S, Mitchell J, Rotondo A, Keel P, Plotnicov KH, Berrettini WH, Kaye WH, Crawford SF, Johnson C, Brandt H, La Via M, Bulik CM. Symptom profile of major depressive disorder in women with eating disorders. The Australian and New Zealand journal of psychiatry. 2007;41:24–31. doi: 10.1080/00048670601057718. [DOI] [PubMed] [Google Scholar]
- 33.Bulik CM, Sullivan PF, Fear J, Pickering A. Predictors of the development of bulimia nervosa in women with anorexia nervosa. J Nerv Ment Dis. 1997;185:704–707. doi: 10.1097/00005053-199711000-00009. [DOI] [PubMed] [Google Scholar]
- 34.Godart NT, Flament MF, Perdereau F, Jeammet P. Comorbidity between eating disorders and anxiety disorders: a review. The International journal of eating disorders. 2002;32:253–270. doi: 10.1002/eat.10096. [DOI] [PubMed] [Google Scholar]
- 35.Kaye WH, Bulik CM, Thornton L, Barbarich N, Masters K. Comorbidity of anxiety disorders with anorexia and bulimia nervosa. The American journal of psychiatry. 2004;161:2215–2221. doi: 10.1176/appi.ajp.161.12.2215. [DOI] [PubMed] [Google Scholar]
- 36.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik CM, Berkman ND, Brownley KA, Sedway JA, Lohr KN. Anorexia nervosa treatment: a systematic review of randomized controlled trials. The International journal of eating disorders. 2007;40:310–320. doi: 10.1002/eat.20367. [DOI] [PubMed] [Google Scholar]
- 38.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, Derrien M, Druesne A, Van Hylckama Vlieg JE, Bloks VW, Groen AK, Heilig HG, Zoetendal EG, Stroes ES, de Vos WM, Hoekstra JB, Nieuwdorp M. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913 e7–916 e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 40.Pfleiderer A, Lagier JC, Armougom F, Robert C, Vialettes B, Raoult D. Culturomics identified 11 new bacterial species from a single anorexia nervosa stool sample. Eur J Clin Microbiol Infect Dis. 2013 doi: 10.1007/s10096-013-1900-2. [DOI] [PubMed] [Google Scholar]
- 41.Tennoune N, Chan P, Breton J, Legrand R, Chabane YN, Akkermann K, Jarv A, Ouelaa W, Takagi K, Ghouzali I, Francois M, Lucas N, Bole-Feysot C, Pestel-Caron M, do Rego JC, Vaudry D, Harro J, De E, Dechelotte P, Fetissov SO. Bacterial ClpB heat-shock protein, an antigen-mimetic of the anorexigenic peptide alpha-MSH, at the origin of eating disorders. Translational psychiatry. 2014;4:e458. doi: 10.1038/tp.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickard JM, Maurice CF, Kinnebrew MA, Abt MC, Schenten D, Golovkina TV, Bogatyrev SR, Ismagilov RF, Pamer EG, Turnbaugh PJ, Chervonsky AV. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–530. e248. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooper Z, Fairburn C. The eating disorders examination: a semi-structured interview for the assessment of the specific psychopathology of eating disorders. The International journal of eating disorders. 1987;6:1–8. [Google Scholar]
- 45.First M, Spitzer R, Gibbon M, Williams J. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition. New York: New York State Psychiatric Institute; 2002. [Google Scholar]
- 46.Steer RA, Rissmiller DJ, Ranieri WF, Beck AT. Structure of the computer-assisted Beck Anxiety Inventory with psychiatric inpatients. Journal of personality assessment. 1993;60:532–542. doi: 10.1207/s15327752jpa6003_10. [DOI] [PubMed] [Google Scholar]
- 47.Steer RA, Ball R, Ranieri WF, Beck AT. Dimensions of the Beck Depression Inventory-II in clinically depressed outpatients. Journal of clinical psychology. 1999;55:117–128. doi: 10.1002/(sici)1097-4679(199901)55:1<117::aid-jclp12>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 48.Luce KH, Crowther JH. The reliability of the Eating Disorder Examination-Self-Report Questionnaire Version (EDE-Q) The International journal of eating disorders. 1999;25:349–351. doi: 10.1002/(sici)1098-108x(199904)25:3<349::aid-eat15>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 49.Carroll IM, Ringel-Kulka T, Keku TO, Chang YH, Packey CD, Sartor RB, Ringel Y. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–G807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonald D, Price MN, Goodrich J, Nawrocki EP, Desantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. Isme J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chao A, Colwell RK, Lin CW, Gotelli NJ. Sufficient sampling for asymptotic minimum species richness estimators. Ecology. 2009;90:1125–1133. doi: 10.1890/07-2147.1. [DOI] [PubMed] [Google Scholar]
- 57.Preheim SP, Perrotta AR, Martin-Platero AM, Gupta A, Alm EJ. Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl Environ Microbiol. 2013;79:6593–6603. doi: 10.1128/AEM.00342-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B. 1995;57:289–300. [Google Scholar]
- 59.Chi YY, Gribbin M, Lamers Y, Gregory JF, 3rd, Muller KE. Global hypothesis testing for high-dimensional repeated measures outcomes. Statistics in medicine. 2012;31:724–742. doi: 10.1002/sim.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beck JG, Stanley MA, Zebb BJ. Characteristics of generalized anxiety disorder in older adults: a descriptive study. Behaviour research and therapy. 1996;34:225–234. doi: 10.1016/0005-7967(95)00064-x. [DOI] [PubMed] [Google Scholar]
- 61.Aardoom JJ, Dingemans AE, Slof Op't Landt MC, Van Furth EF. Norms and discriminative validity of the Eating Disorder Examination Questionnaire (EDE-Q) Eating behaviors. 2012;13:305–309. doi: 10.1016/j.eatbeh.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 62.Welch E, Birgegard A, Parling T, Ghaderi A. Eating disorder examination questionnaire and clinical impairment assessment questionnaire: general population and clinical norms for young adult women in Sweden. Behaviour research and therapy. 2011;49:85–91. doi: 10.1016/j.brat.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Berry D, Reinisch W. Intestinal microbiota: a source of novel biomarkers in inflammatory bowel diseases? Best practice & research Clinical gastroenterology. 2013;27:47–58. doi: 10.1016/j.bpg.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 64.Rajilic-Stojanovic M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 65.Scherz G, Stahl J, Glunder G, Kietzmann M. Effects of carry-over of fluoroquinolones on the susceptibility of commensal Escherichia coli in the intestinal microbiota of poultry. Berliner und Munchener tierarztliche Wochenschrift. 2014;127:478–485. [PubMed] [Google Scholar]
- 66.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr, Ahmed T, Gordon JI. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmed T, Auble D, Berkley JA, Black R, Ahern PP, Hossain M, Hsieh A, Ireen S, Arabi M, Gordon JI. An evolving perspective about the origins of childhood undernutrition and nutritional interventions that includes the gut microbiome. Ann N Y Acad Sci. 2014;1332:22–38. doi: 10.1111/nyas.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–264. e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 71.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Molecular psychiatry. 2013;18:666–673. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 72.Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. e1–e3. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 73.Barden N. Implication of the hypothalamic-pituitary-adrenal axis in the physiopathology of depression. Journal of psychiatry & neuroscience : JPN. 2004;29:185–193. [PMC free article] [PubMed] [Google Scholar]
- 74.Heuser IJ, Schweiger U, Gotthardt U, Schmider J, Lammers CH, Dettling M, Yassouridis A, Holsboer F. Pituitary-adrenal-system regulation and psychopathology during amitriptyline treatment in elderly depressed patients and normal comparison subjects. The American journal of psychiatry. 1996;153:93–99. doi: 10.1176/ajp.153.1.93. [DOI] [PubMed] [Google Scholar]
- 75.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience. 2010;170:1179–1188. doi: 10.1016/j.neuroscience.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 78.Naseribafrouei A, Hestad K, Avershina E, Sekelja M, Linlokken A, Wilson R, Rudi K. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil. 2014;26:1155–1162. doi: 10.1111/nmo.12378. [DOI] [PubMed] [Google Scholar]
- 79.Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends in neurosciences. 2013;36:305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 80.Rees JC. Obsessive-compulsive disorder and gut microbiota dysregulation. Medical hypotheses. 2014;82:163–166. doi: 10.1016/j.mehy.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 81.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoek HW. Incidence, prevalence and mortality of anorexia nervosa and other eating disorders. Current opinion in psychiatry. 2006;19:389–394. doi: 10.1097/01.yco.0000228759.95237.78. [DOI] [PubMed] [Google Scholar]
- 85.Million M, Angelakis E, Maraninchi M, Henry M, Giorgi R, Valero R, Vialettes B, Raoult D. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii and Escherichia coli. Int J Obes (Lond) 2013;37:1460–1466. doi: 10.1038/ijo.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]