Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that can regulate their target gene expressions at the post-transcriptional level. Moreover, they have been reported as either oncomirs or tumor suppressors and possessed therapeutic potential in cancer. In this study, we investigated differential co-expression of miRNAs across four cancer types. We observed that losing positive co-expressions among miRNAs frequently occurs in the studied cancer types. This observation suggests that the disruption of positive co-expressions among miRNAs may be prevalent during tumorigenesis. By systematically collecting these losing positive co-expression among miRNAs in cancer, we constructed a cross-cancer miRNA differential co-expression network. We observed that the influential miRNAs in the proposed network, i.e. hubs or in larger cliques, tended to be involved in more cancer types than other miRNAs. Moreover, we found that miRNAs losing positive co-expression in cancers might make co-contribution to cancer development, and even could be used to predict the cancer types in which miRNAs were involved. Finally, we identified two potential miRNA-regulated onco-modules, mitosis and DNA replication, that are associated with poor survival outcomes in patients across multiple cancers. Collectively, our study suggested that the disruption of miRNA positive co-expression in cancer might make contribution to cancer development. Our findings also form an important basis for identifying miRNAs with potential co-contribution to carcinogenesis.
Introduction
MicroRNAs (miRNAs) are small (~22 nucleotide) noncoding RNAs that regulate gene expression at the post-transcriptional level in eukaryotic cells1,2. They are involved in numerous biological processes3–5, including tumorigenesis6–8, and have therapeutic potential in cancer9,10. In humans, more than 2,500 mature miRNAs have been found and recorded in miRBase release 2011, and the majority of human genes are reported to be potentially regulated by miRNAs12. Accordingly, miRNAs constitute one of the most abundant gene regulators in human cells; they form complicated regulatory networks with target genes in living cells.
In the miRNA regulatory network, co-regulations of miRNAs are reported to cause biological consequences13–16. Zhou et al. showed that co-regulations by miRNA pairs are significant and abundant in the miRNA regulatory network14; Xu et al. constructed a miRNA-miRNA synergistic network to uncover miRNA co-regulating functional modules and their implications in several diseases16. These observed co-regulations of miRNAs may suggest the existence of cooperative behaviors among miRNAs. Moreover, the co-expression among miRNAs is required for forming their co-regulation. Therefore, disruption of miRNA co-expression might be influential in cellular system, and even lead to disease.
To address this knowledge gap, we investigated the differential co-expression among miRNAs across four cancer types (lung, prostate, ovarian, and stomach cancer). We observed that the positive co-expression among miRNAs in normal samples were frequently disrupted in all the examined cancer types. This phenomenon motivated us to build the cross-cancer miRNA differential co-expression network. By analyzing this network, we found that the critical miRNAs in the network might also play pivotal roles in cancer, and identified two miRNA-regulated onco-modules. In summary, through the cross-cancer miRNA differential co-expression network that we built, we uncovered possible influence of disrupting miRNA positive co-expression in cancer and revealed the potential miRNA-regulated modules that play pivotal roles during tumorigenesis.
Results and discussion
Differential co-expression of microRNAs in cancer
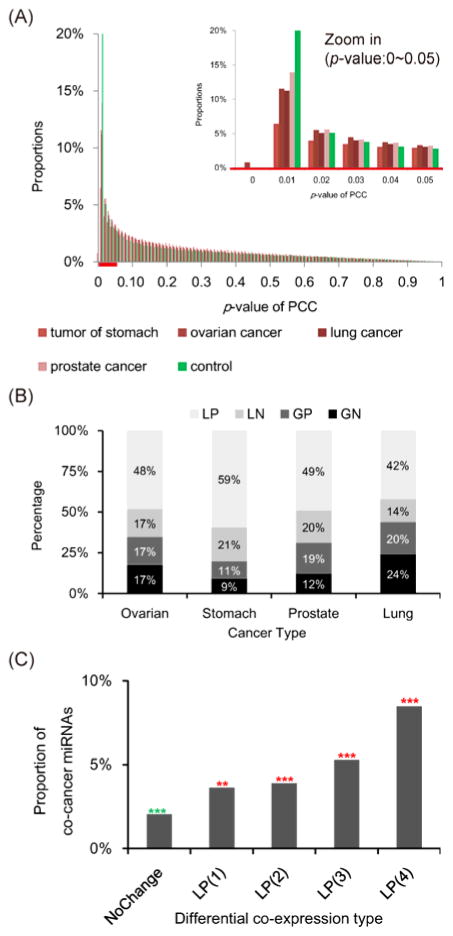
In this study, we investigated miRNA differential co-expression between normal and four cancer types (lung, stomach, ovarian, and prostate cancers). We observed that the proportion of significantly co-expressed miRNA pairs (Pearson correlation coefficient P ≤ 0.01) in normal samples was higher than those in the four types of cancer samples (Fig. 1A). Notably, we did not observe this difference in mRNA co-expression (Fig. S1A) or randomly permutated expression profiles (Fig. S2). This observation implies that the co-expressed miRNA pairs in normal samples might be disturbed in cancer. By comparing the co-expression measurement of miRNA pairs between normal and cancer samples, we observed that miRNAs frequently lost their positive co-expressions in cancer (42~59%, Fig. 1B and Table S1). This consistent loss of positive co-expression was not observed in mRNA co-expression (Fig. S1B). These results suggested that the loss of positive co-expressions among miRNAs might be a common mechanism in cancer.
Fig. 1.
Differential co-expression among miRNAs. (A) The PCC P-value distributions of expression profiles in the control and the four cancer samples. (B) The distributions of miRNA differential co-expression in the four cancer types. Among four types of miRNA differential co- expression, losing positive co-expression was the most frequently observed in each of the four cancer types. The four differential co-expression categories are losing positive (LP), losing negative (LN), gaining positive (GP), and gaining negative (GN) co-expression, respectively. (C) The co-cancer miRNAs were defined as those paired miRNAs reported to be differentially expressed in the same cancer type according to the miRCancer database. The numbers in parentheses are the number of cancer types in which miRNA pairs losing their positive co-expression. The “NoChange” category contains non-differentially co-expressed (PCC P-value > 0.01) miRNA pairs. The significance of each bar was tested by Fisher’s exact test (***: P ≤ 1e-30, **: P ≤ 1e-20, *: P ≤ 1e-10; green asterisk: underrepresented, red asterisk: overrepresented).
To further investigate the association of losing positive miRNA co-expression with cancer, we collected cancer-miRNA relationships from miRCancer database17. The miRCancer database accumulated and curated differentially expressed miRNAs in cancer(s) from literature. We further denoted these miRNAs collected by miRCancer database as cancer-associated miRNAs. We found that the paired miRNAs losing positive co-expressions in more cancer types tended to be reportedly differentially expressed in the same cancer types according to miRCancer database (Fig. 1C). Importantly, the trend was not observed in other three differential co-expression categories, i.e. LN, GP, and GN (Fig. S3A). Of note, only lung, stomach, prostate, and ovarian cancers were considered in Figure 1C and Figure S3A to keep the consistency with the studied cancer types. The similar trend of miRNA differential co-expression was also observed when all the cancer types in miRCancer database were considered (Fig. S3B). In other words, miRNA pairs which lose their positive co-expressions in all the four studied cancer types were more recurrently observed to be involved in the same cancer types in the previous studies than the other miRNA pairs (Fig. 1C; P ≤ 1e-30, Fisher’s exact test). These observations emphasize the association between these miRNA pairs and cancer and even suggest that they may co-contribute to carcinogenesis, and further motivated us to focus on those losing positive co-expressions among miRNAs in all the four cancer types.
Cross-cancer miRNA differential co-expression network
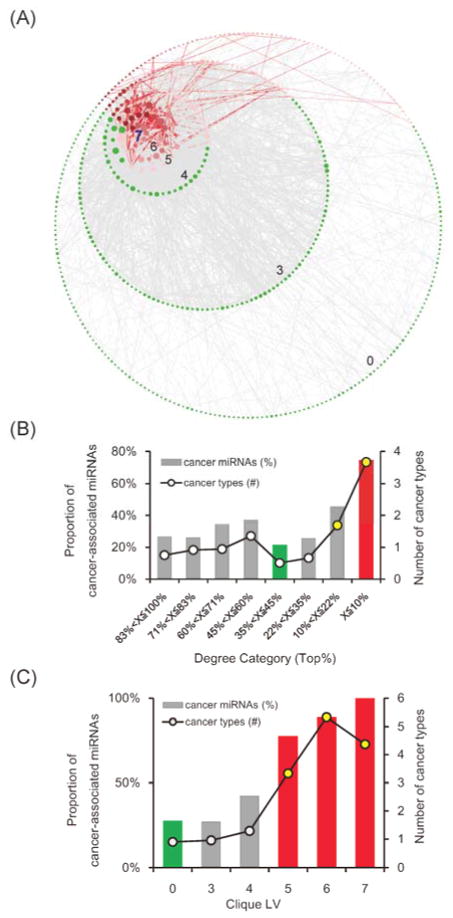
We then collected the 2,036 miRNA pairs that lose positive co-expression in all the four cancer types to construct the cross-cancer miRNA differential co-expression network (Fig. 2A). In this network, 507 nodes were miRNAs; 2,036 edges were positive co-expression among miRNAs in normal samples, but these 2,036 positive co-expression lost in all the four cancer types. We found that this network significantly enriched with known cancer-associated miRNAs (36%, P = 0.002, Fisher’s exact test). The significant overrepresentation of cancer-associated miRNAs in the network might show the relevance of miRNAs losing positive co-expression to cancer. In addition, six miRNA families, mir-8, mir-15, mir-17, mir-33, mir-146, and mir-515, were significantly enriched in the network (Table S2). Among them, mir-17 is a well-known oncomir family18–21. In to mir-17 family, the remaining five miRNA families have been reported to be differentially expressed in cancer(s) in the miRCancer database. The enrichment of these cancer-associated miRNA families might further emphasize the relevance of the proposed network to cancer. However, these enriched cancer-associated families accounted for only 14% (72/507) of the miRNAs in the network, suggesting that only a small proportion of the relevance of the network to cancer might be attributed to the miRNA family feature. More underlying scenarios describing how losing positive co-expression among miRNAs contributed to carcinogenesis remain to be discovered.
Fig. 2.
The cross-cancer miRNA differential co-expression network. (A) The red links are connected by two cancer-associated miRNAs sharing at least one cancer type. Node size represents the degree of miRNAs in the network. The network was organized by the clique levels of miRNAs. The miRNAs in the innermost circle possess the highest clique level, 7, and the clique levels then descend from the inner toward outer circles. (B) The hub miRNAs significantly enriched with the cancer-associated miRNAs and participated in more cancer types than other miRNAs. (C) The clique level of miRNA positively correlates with the proportion of cancer-associated miRNAs and the number of miRNA-involved cancer types. In (B) and (C), the red bars show the significantly overrepresented cancer miRNAs in the corresponding categories of miRNAs (P < 0.001, Fisher’s exact test), and the green bars denote those significantly underrepresented (P < 0.001, Fisher’s exact test). The yellow circles show the significantly higher number of miRNA-participated cancer types compared to miRNAs that form no cliques (P < 0.001, Wilcoxon rank sum test).
The cross-cancer miRNA differential co-expression network is scale-free (Fig. S4). The scale-free feature suggests that this network might be robust against randomly attack to the network structure, such as arbitrarily removing nodes22,23. However, scale-free network might be relatively weak from targeted attack, such as removal of hub nodes22,23. Notably, links in the proposed network were formed in normal samples but disconnected in tumor samples. Disconnection of links could lead removal of nodes and further network structure failure. Accordingly, the scale-free structure of our cross-cancer miRNA differential co-expression network implies that the pivotal nodes (miRNAs), such as hubs or nodes forming larger cliques, in the network might be critical in cancer. The hub proteins and proteins forming larger cliques in the human protein interaction network have been reported to be highly associated with essentiality and cancer24–26. Moreover, their corresponding topological properties, which are degree and clique level (CLV), have been widely used to predict essential genes27,28. Indeed, we found that cancer-associated miRNAs possessed significantly higher degree and CLV than other miRNAs (Table S3). Interestingly, after organizing the network by clique level, the hotspot formed by cancer-associated miRNAs obviously could be observed near the largest maximum cliques, i.e. cliques with a size of 7 (Fig. 2A). This observation might confirm that the cancer-associated miRNAs tend to locate on the hub or form larger cliques in the network, the highly influential locations in the network structure.
We also observed that the hubs, which are miRNAs with top 10% highest degree, were significantly enriched with cancer-associated miRNAs (75%, P < 0.001, Fisher’s exact test, Fig. 2B, bar chart). Moreover, hub miRNAs were involved in more cancer types than non-hubs (Fig. 2B, line chart). According to the miRCancer database, hubs participated in nearly four cancer types on average, but non-hubs participated in less than one (3.67 vs. 0.88, P < 0.001, Wilcoxon rank sum test). Briefly, the hub miRNAs in the proposed network tend to be differentially expressed in multiple cancer types. To note, the hub miRNAs formed relatively larger number of positive co-expressions in normal samples, but these positive co-expressions were disrupted in all four studied cancer types. This result suggests that the degree of miRNAs in the proposed network, i.e. the number of losing positive co-expression in cancers, might be associated with the importance of the given miRNA in cancer development. It further implies that attacking hub miRNAs, through either removing them or disrupting their positive co-expression, in normal sample might influence carcinogenesis.
Like hubs, miRNAs with higher CLV (≥ 5) also tended to enrich with cancer-associated miRNAs (86%, P < 0.001, Fisher’s exact test, Fig. 2C, bar chart), whereas cancer-associated miRNAs were significantly underrepresented in miRNAs that did not form cliques (CLV = 0, 28%, P < 0.001, Fisher’s exact test, Fig. 2C, bar chart). Additionally, miRNAs in larger cliques were involved in more cancer types according to the miRCancer database (Fig. 2C, line chart). On average, miRNAs with CLV ≥ 5 participated in more than four cancer types, while rest of the miRNAs participated in less than one cancer type (4.34 vs. 0.96, P < 0.001, Wilcoxon rank sum test). Briefly, the miRNAs forming relatively larger clique (CLV ≥ 5) tend to be differentially expressed in multiple cancer types, while those independent miRNAs, i.e. forming no cliques, tend to be not differentially expressed in cancer. To note, miRNAs with higher CLV formed larger and denser positively co-expressed modules in normal samples, and these modules were disrupted in all four studied cancer types. This result pinpoints that the miRNA CLV in the proposed network might be associated with their importance in cancer. It further suggests that the disruption of larger miRNA positively co-expressed cliques might have catastrophic effects on cellular systems, and even lead to carcinogenesis. Taken together, our network analyses revealed that the miRNAs that are critical in the network (i.e., hubs and cliques) tend to be differentially expressed in cancer and even multiple cancer types, and therefore they may play pivotal roles in cancer. Furthermore, the relevance of these pivotal miRNAs in the network might be attributed by their connecting losing positive co-expression, which formed the proposed network. Therefore, the above network analyses further implied that the disruption of positive co-expression among miRNAs might influence cancer development.
MicroRNAs losing positive co-expression might be co-involved in carcinogenesis
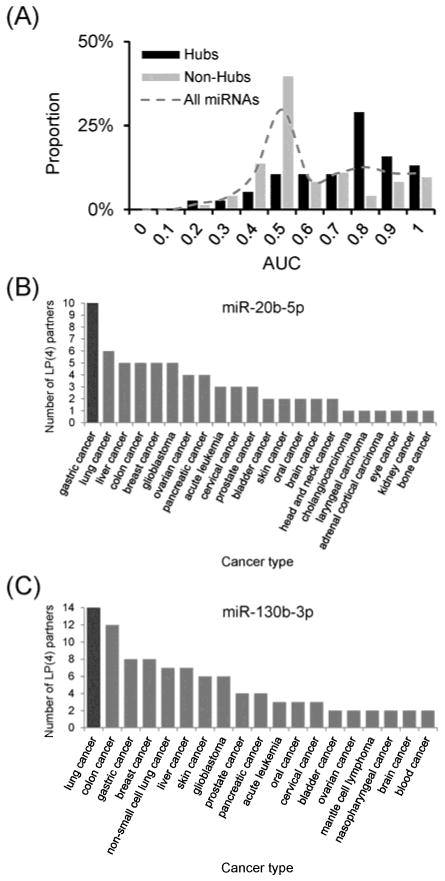
In the above network analysis, we found that network-critical miRNAs might play pivotal roles in cancer. Moreover, the relevance of these network-critical miRNAs in cancer might be attributed by their connecting losing positive co-expression. In addition, we observed that the paired miRNAs losing positive co-expression in more cancer types tended to be reportedly involved in the same cancer types (Fig. 1C). To further investigate the relevance of miRNA losing positive co-expression in cancer, we defined co-cancer probability between two miRNAs via Jaccard index (See Methods in detail). We found that those miRNA pairs losing positive co-expression in all the four studied cancer types had significantly higher co-cancer probability than remaining pairs (P < 0.001, Wilcoxon rank sum test). In other words, two connected miRNAs in our network tend to be involved in the same cancer type. This observation implied that miRNAs losing positive co- expression in cancer may co-contribute to cancer development. To verify this implication, we designed a classifier to predict the cancer types of the studied miRNAs using the cancer types of their connecting partners in the network. Briefly, the concept of this classifier is “A given miRNA tends to participate in those cancer types in which its connecting partners participate recurrently.” Interestingly, this classifier performed well on hub miRNAs (Fig. 3A, black bar, the median of area under curve (AUC) is 0.73) but not for all miRNAs (Fig. 3A, dashed line, the median: 0.55) or non-hubs (Fig. 3A, grey bar, the median: 0.50). This result indicates that the degree could affect the performance of the classifier. Indeed, we observed that the AUC value was significantly and positively correlated with the degree (Spearman’s rank correlation coefficient = 0.40, P < 0.001). That is, more connecting partners could provide more information about cancer involvement to predict the cancer types of studied miRNAs. The result confirmed the concept of this classifier: the studied miRNAs tended to participate in the cancer types that were recurrently observed to be involved by enough number of connecting partners. It further connects to the conclusion from our network analysis: disrupting positive co-expression among miRNAs may influence cancer development.
Fig. 3.
Losing positive co-expression predicts miRNA-regulated cancer and uncovered cancer-associated miRNAs. (A) The AUC distribution of all miRNAs, hub miRNAs, and non-hub miRNAs. (B) and (C) The cancer types in which (B) miR-20b-5p and (C) miR-130b-3p partners in the network are involved. The cancer types are reportedly associated with the (B) miR-130b-3p or (C) miR-20b-5p connecting partners in the network were listed along with the X-axis. The number of interacting partners that were reported to be differentially expressed in the corresponding cancer type is shown on the Y-axis.
For example, miR-20b-5p had the highest degree in the network (62 partners). Among the cancer types involved by miR-20b-5p connecting partners in the network, gastric cancer was the most frequently observed (10 partners, Fig. 3B). According to our classifier, miR-20b-5p might be involved in gastric cancer. As expected, miR-20b-5p has also been reported to be associated with gastric cancer29–32. Another example is miR-130b-3p, which is located in one of the largest cliques in the network. Among the cancer types of miR-130b-3p connecting partners, lung cancer was the most frequently observed (14 partners, Fig. 3C). However, there has been no report in literature of miR-130b-3p in lung cancer. Importantly, our recent study33 confirmed the de-regulation of miR-130b-3p in lung cancer using the reverse transcription polymerase chain reaction (RT-PCR). This result indicated that miR-130b-3p could be involved in lung cancer development. These two examples confirmed that recurrently losing positive co-expression in cancers could be a robust predictor of cancer types involved by miRNAs. Furthermore, these observations suggest that the paired miRNAs forming connections in the network may be mutually implicated in the pathogenesis of the same cancer type.
Clique miRNA-regulated pan-cancer activated functional modules
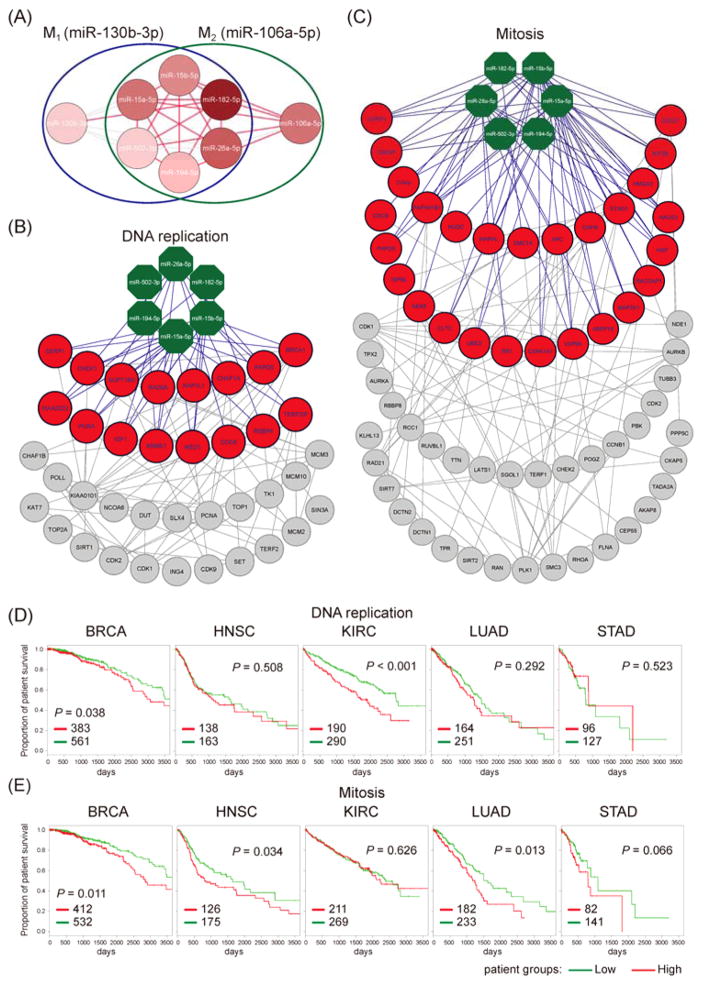
Within the proposed network, there are two maximum cliques measuring as size-7 (K7) (Fig. 4A). We termed these two K7 cliques M1 and M2 (Fig. 4A). According to the above results, we’ve observed that paired miRNAs forming connections in the proposed network may be mutually implicated in the pathogenesis of the same cancer type. Therefore, the miRNAs in these two maximum cliques might have co-contribution to carcinogenesis. In other words, they might form co-expressed modules in normal samples to regulate cancer development either positively or negatively. Notably, all the miRNAs in these two maximum cliques are reported to be differentially expressed in cancer(s) in miRCancer database. Furthermore, their target genes were significantly enriched with cancer-associated genes annotated by the Cancer Gene Census (CGC) (M1: 4.11%, M2: 3.87%, P < 0.001, Fisher’s exact test). More importantly, we found that genes targeted by more M1 or M2 miRNAs had a higher probability of being cancer-associated genes (Fig. S5). This result suggests that the clique miRNAs might co-contribute to carcinogenesis through co-regulating their cancer-associated target genes.
Fig. 4.
The two largest maximum cliques and their regulatory pan-cancer activated functional modules. (A) The two largest maximum cliques (size 7), M1 and M2, in the proposed network. These two cliques share six miRNAs. Accordingly, we tagged them by their exclusive miRNAs. M1 was tagged by miR-130b-3p and M2 by miR-106a-5p. Red nodes denote the cancer-associated miRNAs, and red edges between two miRNAs losing positive co-expression in the four cancer types. Notably, all member miRNAs in these two maximum cliques are cancer-associated miRNAs. The miRNAs associated with more cancer types are represented by darker red. (B) (C) The two identified miRNA-regulated functional modules, (B) DNA replication and (C) Mitosis. In these two modules, green octagons are miRNAs, red circles are their predicted targets, and the other grey circles are genes that possess at least two PPIs with the targets. The purple edges are miRNA regulations, and the grey edges are PPIs between genes. (D) (E) The Kaplan-Meier of 10-year survival curves for two pan-cancer activated functional modules in five used cancer types. Patients were grouped into lowly (green) and highly (red) expressed groups based on the average expression levels of genes in the identified modules. The P-values were derived from the Cox’s regression model with age as an explanatory variable. The number of patients in each group was labeled on the lower left corner of each panel. (D) DNA replication. (E) Mitosis.
To further explore the downstream regulation of K7 miRNAs in cancer, we applied an integrative approach to identify miRNA-regulated functional modules in cancer (Supporting Information S4). Among them, there were two functional modules co-regulated by both M1 and M2 (Fig. 4B and C). The number of functional modules regulated by M1 and/or M2 during the identification process is depicted in Figure S6. According to their enriched functions, we termed these two modules as “DNA replication” and “Mitosis.” Dysregulation of DNA replication and/or mitosis have been known to lead to genome instability and even carcinogenesis34,35. Notably, these two modules enriched with co-expressed protein-protein interactions (PPIs) in all four cancer types, as well as in eleven examined TCGA cancer types (Table S4). These results further suggest that these two modules may be commonly activated in pan-cancer. Accordingly, they could be onco-modules, i.e. their activation could stimulate or promote tumorigenesis. Moreover, we could not similarly identify common functional modules across the four cancer types when only mRNA co-expression data was used (Table S5). In other words, when the influence of miRNA regulation was removed from the differential co-expression analysis, no consistent functional module across four studied cancers was discovered. This observation implied that the activation of these two functional modules might be attributed to the disruption of the co-expressed miRNA modules, which are the two maximum miRNA cliques (M1 and M2) in the proposed network. To substantiate the oncogenic potential of the two proposed modules, we performed survival analysis on them by utilizing gene expression profiles. We calculated the mean expression level of the observed module gene set for each sample (patient). Then, we categorized as “highly expressed” those samples in which patients exhibited a mean expression level that was higher than the mean of the averaged expression level across patients. Accordingly, we can divide patients into two groups: 1) those with highly expressed gene set and 2) those with lower expressions of the gene set in the observed functional module. We found that patients with higher expression levels of the genes in the modules possessed poor survival rates (Fig. 4D and E). However, we did not observe significant difference in survival rate for COAD, LUSC, OV, or UCEC (Fig. S7). Overall, the high expression of the genes in these two modules may be associated with poor survival rates in patients across multiple cancer types. This result verifies that these two identified functional modules may have oncogenic potential, and their activation may promote tumorigenesis. Furthermore, it emphasizes that the disruption of miRNA co-expression might make contribution to cancer development. Additionally, our previous study demonstrated that the co-expression network analysis might be a promising approach to identify potential drugs for cancer treatment, such as drug repositioning36. Similar to the findings in our previous study, the miRNA co-expression results in this study may be useful for the development of the miRNA-based diagnosis or therapy in cancer.
Conclusions
In this study, we investigate miRNA differential co-expression in four cancer types. Our miRNA differential co-expression analysis further suggested that losing positive co-expression among miRNAs might be a common mechanism during cancer development. We then proposed a cross-cancer miRNA differential co-expression network by collecting the disrupted positive co-expression among miRNAs in all the studied cancer types. We found that the influential miRNAs, i.e. hub miRNAs or miRNAs in larger cliques, in the network could play pivotal roles in cancer. Moreover, the results of our network analysis imply that disruption of positive co-expression among miRNAs might potentially influence cancer development. Furthermore, we found that miRNAs losing positive co-expression in cancers might co-contribute to cancer development, and even could be used to predict the cancer types in which miRNAs were involved. Finally, we identified two miRNA-regulated onco-modules that are likely activated in pan-cancer and that are associated with the poor survival rate of patients. Collectively, our study sheds light on the overall effects of disrupting miRNA positive co-expression in cancer, which might make contribution to cancer development. In addition, our findings form an important basis for identifying miRNAs with potential co-contribution to carcinogenesis.
Materials and methods
Expression profiles and clinical data
The miRNA expression profiles were produced by the miRNome project37 and downloaded from the Gene Expression Omnibus (GEO)38,39. The accession number is GSE31568. In this study four types of cancer (lung, prostate, ovarian, and gastric cancer) were used. There were 32, 23, 15, and 13 samples from lung, prostate, ovarian, and gastric cancer, respectively. The expression profiles from 70 healthy individuals were used as a control (normal samples). Furthermore, only miRNAs with expression levels registering higher than 25% of other miRNAs in at least 75% samples were used to perform the analysis (See Table S1 for the detail number of expressed miRNAs in control and cancer samples). In this study, we used this miRNA expression profile to build the cross-cancer miRNA differential co-expression network.
The mRNA expression profiles, which were used to validate the differential co-expression of miRNA and identify miRNA regulatory functional modules for the four cancer types, were also downloaded from the GEO38,39. These accession numbers are GSE32863 (lung cancer)40, GSE17951 (prostate cancer)41,42, GSE18520 (ovarian cancer)43, and GSE13861 (gastric cancer)44. There were 58 tumor samples from lung adenocarcinoma and 58 controls, which were extracted from adjacent non-tumor lung tissues. For prostate cancer, the numbers of tumor and control samples were 109 and 13, respectively. With respect to ovarian cancer samples, 53 samples from high-grade primary tumor specimens and 10 from normal ovarian samples were used. The gastric cancer samples contained 65 tumor samples from primary gastric adenocarcinoma and 19 control samples from surrounding normal tissue.
In this study, we also used mRNA expression profiles and clinical data from The Cancer Genome Atlas (TCGA). For the cross validation of module activity in cancer, mRNA expression profiles of eleven cancer types in TCGA were investigated: breast cancer (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSC), clear cell kidney carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), papillary thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC). We used RNA-Seq V2 data from BRCA, COAD, HNSC, KIRC, LUAD, LUSC, OV, PRAD, THCA, and UCEC, and RNA-Seq data from STAD. The RSEM (RNA-Seq by Expectation Maximization)45 and RPKM (Reads Per Kilobase of transcript per Million mapped reads) values of mRNA were used as gene expression levels for RNA-Seq V2 and RNA-Seq data, respectively.
Among these eleven cancer types, the mRNA expression profiles and clinical data of nine cancer types, i.e. BRCA, COAD, HNSC, KIRC, LUAD, LUSC, OV, STAD, and UCEC, were used to perform the survival analyses. Of note, PRAD and THCA data were excluded from survival analyses because their clinical data contained only 1 and 15 death events, respectively. For other cancer types, the number of death events was more than 20 (BRCA: 114, COAD: 89, HNSC: 158, KIRC: 167, LUAD: 125, LUSC: 140, OV: 303, STAD: 24, and UCEC: 60).
Construction of the cross-cancer miRNA differential co-expression network
The miRNA expression profiles collected from control (normal) and cancer samples were used to calculate the expression correlations between miRNAs. In this study, we utilized the Pearson Correlation Coefficient (PCC) of miRNA expression profiles in control or cancer samples to access the co-expressions among miRNAs. While the distribution of PCC values varies by sample size, the distribution of PCC P-values does not (Supporting Information S5). We utilized the PCC P-value instead of the PCC value to filter out insignificantly correlated miRNA pairs. In our case, we observed that the proportion of significantly correlated (P ≤ 0.01) miRNA pairs in control (normal) samples was higher than that in any of the four examined cancer samples (Fig. 1A). Therefore, we considered miRNA pairs with a PCC P-value ≤ 0.01 as significantly correlated and formed co-expressions.
By collecting co-expressed miRNA pairs, we constructed the miRNA co-expression networks for normal (GN) and cancer samples (GTi) respectively. We applied a difference function df (GN,GTi) to obtain the differentially co-expressed miRNA pairs between normal and cancer samples. This function contains two parts. The first part was the symmetric difference of synergistic interaction sets in normal and cancer networks, as defined below:
where E(GN) and E(GTi) is the set of co-expressions in network G from normal and tumor i, respectively. To decrease uncertainty, we additionally considered the second function, which calculates the differences between PCC z-scores (standard scores) of co-expressed miRNA pairs produced by the first function. The difference of PCC z-scores is defined as below:
where r(a,b,S) is the PCC between miRNA a and b on condition S; S can be normal or one of the examined cancer types. and σ(r)S are the mean PCC and standard deviation of PCC for all paired miRNAs in condition S, respectively. The PCC z-score difference of paired miRNAs a and b between normal and cancer can then be defined as ΔZr(a,b) Zr(a,b). In this study, we set one as the cut-off for ΔZr(a,b).
After applying these two functions, we could obtain the set of miRNA differential co-expressions termed as DGTi in cancer i. Furthermore, we divided interactions in DGTi into the two groups shown below:
where DGL|Ti represents those co-expressions formed in normal samples but lost in cancer i; DGL|Ti contains those co-expressions gained in cancer i but not in normal samples. Finally, we defined DGL(m) or DGG(m) as an assessment of the specificity or generality of the miRNA differential co-expression across cancer types. The bracketed m represents the number of cancer types in which the differential co-expression was observed. In other words, a miRNA differential co-expression with a larger m value was observed in more cancer types and therefore can be more general for cancer development. We further divided miRNA differential co-expressions into four groups by changing signs of PCCs: losing positive (LP) or negative (LN) and gaining positive (GP) or negative (GN). For example, losing positive co-expression of lung cancer were formed by miRNA pairs with positive co-expressions in control samples but losing co-expressions in lung cancer samples. Finally, the cross-cancer miRNA differential co-expression network contained 507 nodes (mRNAs) and 2,036 edges between miRNAs (losing positive co-expression in the four used cancer types). The descriptive statistics of miRNA differential co-expression are listed in Table S1.
In this study, we examined two network properties of miRNA, i.e. degree and clique level, in the cross-cancer miRNA differential co-expression network. The degree is the number of connecting partners of one miRNA in the network. We defined those miRNAs with the top 10% highest degree as hubs in the network. Clique level is the size of the largest clique in which the observed miRNAs participate. Clique is a complete graph structure in network.
Calculation of co-cancer probability and the classifier for predicting potentially involved cancer types of miRNAs
We used the Jaccard index to investigate the level co-involvement between miRNAs in cancer. We termed this index as the co-cancer probability between miRNAs. The co-cancer probability between two miRNAs measured the proportion of shared cancer types and was defined as |Cm1∩Cm2|/|Cm1∪Cm2|. Cm was denoted as the set of cancer types that were reportedly involved with the observed miRNA m in the miRCancer database. Notably, we only analyzed the co-cancer probability between cancer-associated miRNAs, i.e. both |Cm1| and |Cm2| were required to be greater than 1.
For a given miRNA m, we obtained a set of cancer types (Cm) that were reportedly affected by m in the miRCancer database. In addition, we obtained another set of cancer types (Cmp), which were reportedly influenced by m’s partners in the network. By using Cmp as the gold standard, we further divided Cmp into two groups: 1) false: not observed in Cm, and 2) true: observed in Cm and designated as a binary classifier. Moreover, for a given cancer type t in Cmp, the number of m’s partners that reportedly participated in t could be determined, and we denoted this number as nt. Furthermore, utilizing nt as the varied discrimination threshold, we drew the receiver operating characteristic (ROC) curve for the binary classifier. The performance of the classifier in precisely categorizing the Cmp was evaluated by plotting the sensitivity and 1-specificity at all varied discrimination thresholds, followed by calculating the area under the curve (AUC).
Supplementary Material
Acknowledgments
This work was partially supported by National Institutes of Health grants (R01LM011177, R03CA167695, P30CA068485, P50CA095103, and P50CA098131) and Ingram Professorship Funds (to ZZ). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Electronic Supplementary Information (ESI) available: Supporting Information file 1. See DOI: 10.1039/x0xx00000x
References
- 1.Doench JG, Sharp PA. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H, Ingolia NT, Weissman JS, Bartel DP. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Flynt AS, Lai EC. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN, Nam JW. Trends Genet. 2006;22:165–173. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Ling H, Fabbri M, Calin GA. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rooij E, Purcell AL, Levin AA. Circ Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 11.Kozomara A, Griffiths-Jones S. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman RC, Farh KK, Burge CB, Bartel DP. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shalgi R, Lieber D, Oren M, Pilpel Y. PLoS Comput Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Ferguson J, Chang JT, Kluger Y. BMC Genomics. 2007;8:396. doi: 10.1186/1471-2164-8-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin CC, Chen YJ, Chen CY, Oyang YJ, Juan HF, Huang HC. BMC Syst Biol. 2012;6:18. doi: 10.1186/1752-0509-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Li CX, Li YS, Lv JY, Ma Y, Shao TT, Xu LD, Wang YY, Du L, Zhang YP, Jiang W, Li CQ, Xiao Y, Li X. Nucleic Acids Res. 2011;39:825–836. doi: 10.1093/nar/gkq832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie B, Ding Q, Han H, Wu D. Bioinformatics. 2013;29:638–644. doi: 10.1093/bioinformatics/btt014. [DOI] [PubMed] [Google Scholar]
- 18.Tan W, Li Y, Lim SG, Tan TM. World J Gastroenterol. 2014;20:5962–5972. doi: 10.3748/wjg.v20.i20.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Guan X, Sun Y, Mi J, Shu X, Liu F, Li C. Exp Cell Res. 2014;323:1–6. doi: 10.1016/j.yexcr.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 20.Concepcion CP, Bonetti C, Ventura A. Cancer J. 2012;18:262–267. doi: 10.1097/PPO.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendell JT. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 22.Albert R, Jeong H, Barabasi AL. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 23.Holme P, Kim BJ, Yoon CN, Han SK. Phys Rev E. 2002;65:056109. doi: 10.1103/PhysRevE.65.056109. [DOI] [PubMed] [Google Scholar]
- 24.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Greenbaum D, Xin Lu H, Zhu X, Gerstein M. Trends Genet. 2004;20:227–231. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Jonsson PF, Bates PA. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang YC, Lin CC, Chang JY, Mori H, Juan HF, Huang HC. Mol Biosyst. 2009;5:1672–1678. doi: 10.1039/B900611G. [DOI] [PubMed] [Google Scholar]
- 28.Manimaran P, Hegde SR, Mande SC. Mol Biosyst. 2009;5:1936–1942. doi: 10.1039/B905264j. [DOI] [PubMed] [Google Scholar]
- 29.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espinosa-Parrilla Y, Munoz X, Bonet C, Garcia N, Vencesla A, Yiannakouris N, Naccarati A, Sieri S, Panico S, Huerta JM, Barricarte A, Menendez V, Sanchez-Cantalejo E, Dorronsoro M, Brennan P, Duarte-Salles T, BAB-d-MH, Weiderpass E, Lund E, Clavel-Chapelon F, Boutron-Ruault MC, Racine A, Numans ME, Tumino R, Canzian F, Campa D, Sund M, Johansson M, Ohlsson B, Lindkvist B, Overvad K, Tjonneland A, Palli D, Travis RC, Khaw KT, Wareham N, Boeing H, Nesi G, Riboli E, Gonzalez CA, Sala N. Int J Cancer. 2014;135:2065–2076. doi: 10.1002/ijc.28850. [DOI] [PubMed] [Google Scholar]
- 31.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 32.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. Int J Oncol. 2009;34:537–542. [PubMed] [Google Scholar]
- 33.Mitra R, Edmonds MD, Sun J, Zhao M, Yu H, Eischen CM, Zhao Z. RNA. 2014;20:1–13. doi: 10.1261/rna.042754.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 35.Gaillard H, Garcia-Muse T, Aguilera A. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 36.Lin CC, Jiang W, Mitra R, Cheng F, Yu H, Zhao Z. Sci Rep. 2015;5:12063. doi: 10.1038/srep12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller A, Leidinger P, Bauer A, Elsharawy A, Haas J, Backes C, Wendschlag A, Giese N, Tjaden C, Ott K, Werner J, Hackert T, Ruprecht K, Huwer H, Huebers J, Jacobs G, Rosenstiel P, Dommisch H, Schaefer A, Muller-Quernheim J, Wullich B, Keck B, Graf N, Reichrath J, Vogel B, Nebel A, Jager SU, Staehler P, Amarantos I, Boisguerin V, Staehler C, Beier M, Scheffler M, Buchler MW, Wischhusen J, Haeusler SF, Dietl J, Hofmann S, Lenhof HP, Schreiber S, Katus HA, Rottbauer W, Meder B, Hoheisel JD, Franke A, Meese E. Nat Methods. 2011;8:841–843. doi: 10.1038/nmeth.1682. [DOI] [PubMed] [Google Scholar]
- 38.Edgar R, Domrachev M, Lash AE. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, Lam S, Gazdar AF, Laird-Offringa IA. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Xia XQ, Jia Z, Sawyers A, Yao H, Wang-Rodriquez J, Mercola D, McClelland M. Cancer Res. 2010;70:6448–6455. doi: 10.1158/0008-5472.CAN-10-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia Z, Wang Y, Sawyers A, Yao H, Rahmatpanah F, Xia XQ, Xu Q, Pio R, Turan T, Koziol JA, Goodison S, Carpenter P, Wang-Rodriguez J, Simoneau A, Meyskens F, Sutton M, Lernhardt W, Beach T, Monforte J, McClelland M, Mercola D. Cancer Res. 2011;71:2476–2487. doi: 10.1158/0008-5472.CAN-10-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, Ozbun L, Samimi G, Brady J, Randonovich M, Pise-Masison CA, Barrett JC, Wong WH, Welch WR, Berkowitz RS, Birrer MJ. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, Noh SH, Park ES, Chu IS, Hong WK, Ajani JA, Lee JS. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li B, Dewey CN. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.