Summary
Actin polymerization powers membrane deformation during many processes including clathrin-mediated endocytosis (CME). During CME in yeast, actin polymerization is triggered and coordinated by a six-protein WASP/Myosin complex that includes WASP, class I myosins (Myo3 and Myo5), WIP (Vrp1), and two other proteins. We show that a single engineered protein can replace this entire complex while still supporting CME. This engineered protein reveals that the WASP/Myosin complex has four essential activities; (1) recruitment to endocytic sites, (2) anchorage to the plasma membrane, (3) Arp2/3 activation, and (4) transient actin filament binding by the motor domain. The requirement for both membrane and F-actin binding reveals that myosin-mediated coupling between actin filaments and the base of endocytic sites is essential for allowing actin polymerization to drive membrane invagination.
Introduction
Clathrin-mediated endocytosis (CME) is an ancient and highly conserved pathway for internalizing nutrients, recycling plasma membrane components, and regulating signaling receptors. Endocytic sites progress through a series of steps in which early-arriving proteins establish an endocytic site, clathrin and coat proteins form a patch that matures into a bud, and a burst of actin polymerization occurs as the bud invaginates more deeply and then undergoes scission (reviewed in (Boettner et al., 2012)). Evidence from genetic (Kubler and Riezman, 1993), fluorescence imaging (Kaksonen et al., 2005), pharmacological (Aghamohammadzadeh and Ayscough, 2009), and electron microscopy studies (Idrissi et al., 2002; Kukulski et al., 2012) indicates that this burst of actin polymerization drives membrane invagination and scission in yeast CME. Moreover, these studies support a model in which Arp2/3-mediated actin branching is triggered at the base of endocytic invaginations, older actin filaments are pushed inward as new actin is polymerized, and the bud tip is dragged inward with the older filaments by membrane-actin coupling proteins Sla2, Ent1, Ent2, and Pan1, in the endocytic coat (Kaksonen et al., 2003; Skruzny et al., 2012). The burst of actin polymerization is triggered by a group of proteins termed the WASP/Myosin Module. These proteins are recruited to endocytic sites but remain at the cortex as endocytic invaginations and their associated coat proteins are internalized (Jonsdottir and Li, 2004; Kaksonen et al., 2005; Sun et al., 2006). The kernel of this module is a complex of six proteins that can be co-purified from cells (Feliciano and Di Pietro, 2012; Soulard et al., 2002). These proteins include the yeast homologues of N-WASP (Las17), WIP (Vrp1), Class I myosins (Myo3 and Myo5), and Toca-1 (Bzz1), all of which positively regulate actin polymerization, as was well as Bbc1, which negatively regulates actin polymerization (Sun et al., 2006). For ease of discussion, we refer to this complex as the WASP/Myosin complex (Figure 1A).
Figure 1. Simplification of the WASP/Myosin complex.
(A) A diagram of the components of the WASP/Myosin complex (circled with dotted line), the coat protein Sla1 (green) that is hypothesized to recruit Las17, and proteins of the Actin Module (red) that are directly or indirectly recruited by WASP/Myosin complex NPF activity. Black arrows indicate known or theorized physical interactions between complex components while gray lines indicate regulatory relationships.
(B) Single frames from movies of wild-type (WT) cells and cells with various WASP/Myosin complex components deleted, all expressing Sla1-GFP and Abp1-RFP. A kymograph of a representative endocytic patch from each cell type is shown to the right of each image.
(C) Proportion of Sla1-GFP patches that internalize more than 3 pixels (194 nm). At least 10 patches from the mother cell (or two similar cells if fewer than 10 patches could be counted for a cell) were scored for movement and an average was obtained for each cell. At least fifteen cells on three or more separate days of imaging were analyzed. Values represent the mean +/− SEM. *P<0.001 vs MYO5 (myo3Δ).
(D) Growth of WASP/Myosin complex components on YPD plates. Starter cultures were diluted to an O.D600 of 0.02 and plated. Two additional 1:5 dilutions were also plated. Plates were grown at the indicated temperature for 48 hours and imaged. See also Figure S1, Movie S1, Tables S1 and S2.
Studies in mammalian cells have confirmed that the modular arrangement of endocytic components is a conserved feature of CME (Taylor et al., 2011). Like the WASP/Myosin complex in yeast, mammalian proteins including N-WASP, cortactin, and the class I myosin Myosin 1E are recruited to endocytic sites and orchestrate actin polymerization (Merrifield et al., 2005)(Cheng et al., 2012). The requirement for actin polymerization at endocytic sites is particularly acute in metazoan cells when the plasma membrane is under tension, (Boulant et al., 2012; Aghamohammadzadeh and Ayscough, 2009). In both yeast and metazoan cells, actin polymerization extends shallow invaginations into deeply invaginated pits. This phenotype is apparent in electron micrographs of yeast cells (Idrissi et al., 2012; Kukulski et al., 2012) and can also be observed in metazoan cells that lack dynamin (Ferguson et al., 2010).These similarities suggest that the mechanisms by which actin contributes to CME are highly conserved between yeast and metazoan cells. The conservation of this pathway and the genetic tractability of yeast make S. cerevisiae a powerful organism for elucidating the molecular mechanisms by which actin powers CME.
The Arp2/3 complex is activated by nucleation promoting factors (NPFs). Within the WASP/Myosin complex, Las17 can activate the Arp2/3 complex, as can Myo3 or Myo5 in cooperation with Vrp1 (Galletta et al., 2008; Sun et al., 2006). Deletion of the NPF domains of either Las17 or Myo3 and Myo5 is tolerated by cells but simultaneous deletion of the NPF domains from Las17, Myo3, and Myo5 causes endocytic failure and growth defects (Galletta et al., 2008; Sun et al., 2006). These findings demonstrate the importance of NPF activity, but there is also evidence that the complex has other crucial roles in CME besides Arp2/3 activation. For example, complete deletion of Las17, Vrp1, or Myo3 and Myo5 causes profound defects in growth and CME that are not observed when only the NPF activity of these proteins is disrupted (Galletta et al., 2008; Sun et al., 2006), indicating that these proteins play additional roles in CME beyond Arp2/3 activation. The Class I myosins are particularly interesting because Myo5 has been shown to bind and move actin filaments in vitro, and mutations that disrupt its contractile cycle also stop CME (Sun et al., 2006). However, a comprehensive explanation of how the motor domain or the myriad other domains of this complex may contribute to CME remains elusive. Actin polymerization powers membrane remodeling in numerous biological processes including the extension of lamellipodia and filopodia and podosome growth (Suetsugu et al., 2014), so the mechanisms of membrane reshaping during CME will likely apply broadly.
Here, we employed a synthetic biology approach to identify the minimal constellation of WASP/Myosin complex activities that are necessary for CME and sufficient for WASP/Myosin complex function. We found that the six proteins of the complex can be replaced by a single engineered protein with only four activities: a recruitment mechanism, an NPF activity, a nonspecific phospholipid anchor, and the ability to transiently bind actin filaments. These findings revealed that membrane-actin coupling at the base of endocytic sites is required for CME. Unlike membrane-actin coupling mediated by Sla2, Pan1, and Epsins at the tip of endocytic buds, the linkage at the base utilizes a motor that can both bind and release actin filaments. The ubiquitous and highly conserved features of CME suggest that this second membrane-actin linkage is likely conserved across eukaryotic taxa.
Results
The WASP/Myosin complex can be replaced by a single protein consisting of fused Las17 and Myo5
For the purpose of identifying the minimal set of biochemical or biophysical activities required for function of the WASP/myosin complex, we attempted to eliminate as much of the complexity as possible without stopping CME or inhibiting growth. First, we deleted the genes encoding each protein in the complex and determined the effects on growth and CME. We quantified endocytic defects by imaging the coat marker Sla1-GFP and actin binding protein Abp1-RFP in a medial focal plane. In cells with a functional WASP/Myosin complex, Sla1-GFP is recruited to patches at the cortex, Abp1-RFP is recruited subsequently as actin is polymerized, and the fluorescent patch moves away from the cortex (Movie S1). This inward movement is apparent as a terminal hook when the data are represented as a kymograph (Figure 1B). Previous studies established that this inward movement corresponds to the invagination and scission steps of CME (Kaksonen et al., 2003; Kukulski et al., 2012). Consistent with previous reports (Sun et al., 2006), we found that the movement of Sla1-GFP patches was not affected by deletion of only MYO3 so most additional experiments were conducted in the simpler myo3Δ background , except where noted otherwise (Figure 1B and C, Movie S1). We further confirmed that Sla1-GFP patches do not move away from the cortex in myo3Δ cells lacking LAS17, VRP1, or MYO5 (Figure 1B and C, Movie S1) and the lifetimes of both Sla1-GFP and Abp1-mRFP are extended (Figure S1B). Also, cells lacking LAS17, VRP1, or MYO5 were unable to grow at 39°C and had severe growth defects at 30°C (Figure 1D). These findings are consistent with reports showing that LAS17 or VRP1 deletion also affect endocytosis and growth in cells that have MYO3 (Galletta et al., 2008; Sun et al., 2006). Finally, we found that deleting MYO5 in myo3Δ cells caused internal actin structures to form in a large proportion of cells (Figure 1B)(Movie S1), a distinctive phenotype that has not been reported previously. In contrast, Bbc1 and Bzz1 were not required for growth (Figure 1D) or CME (Figure 1C), (Kishimoto et al., 2011).
Vrp1 is a central and essential protein of the WASP/Myosin complex. It binds to Las17 and recruits the class I myosins and possibly other SH3 domain-containing proteins through a proline-rich region (PRR) (Evangelista and Klebl, 2000; Geli et al., 2000; Sun et al., 2006). It also contains an actin monomer-binding WH2 domain that combines with the Arp2/3 complex-binding CA domain of Myo3 and Myo5 to stimulate actin polymerization (Evangelista and Klebl, 2000; Galletta et al., 2008; Geli et al., 2000; Sun et al., 2006). We hypothesized that the essential functions of Vrp1 are (1) to recruit type I myosins to endocytic sites and (2) to stimulate actin polymerization. We tested this hypothesis by creating a synthetic gene that linked MYO5 and LAS17 at the MYO5 locus to produce a Myo5-Las17 fusion protein, which would ensure Myo5 localization in the absence of Vrp1, since Las17 is recruited to endocytic sites independently of Vrp1. We investigated the effects of VRP1 deletion in myo3Δ cells when Myo5 and Las17 were linked. While VRP1 deletion normally prevents all detectable internalization of Sla1-GFP puncta, Sla1-GFP internalization was restored to 78% by the Myo5-Las17 fusion protein (versus 0% for vrp1Δ, myo3Δ cells and 97% for myo3Δ cells) (Figure 1B and C) (Movie S1). We also observed a complete growth rescue at all temperatures (Figure1D, Figure S1A). This result demonstrated that Vrp1 is not required for CME if Las17 and Myo5 are linked.
Based on the rescue by the fusion protein and the single deletion results, we hypothesized that the Myo5-Las17 fusion protein might rescue deletion of the entire WASP/Myosin complex. First, we deleted each component of the WASP/Myosin complex in myo3Δ cells (Sun et al., 2006) with Myo5 linked to Las17. We found that bzz1Δ cells had no additional growth phenotype when Myo5 was linked to Las17, but bbc1Δ cells became temperature sensitive (Figure1D). However, this growth defect did not appear to be caused by insufficient CME because at least 67% of Sla1-GFP patches internalized (Figure1C) and the lifetimes of Sla1-GFP and Abp1-RFP were only slightly longer than in normal cells (FigureS1B). Because Myo5-Las17 may have more NPF activity than Las17 alone from the extra CA domain of Myo5 and because deleting Bbc1 increases NPF activity (Sun et al., 2006), we hypothesized that bbc1Δ, MYO5-LAS17, myo3Δ cells might have excessive actin polymerization at endocytic sites. Consistent with this hypothesis, deleting BZZ1, which encodes an NPF activator, in bbc1Δ, MYO5-LAS17, myo3Δ cells partially rescued the growth defect (Figure S1C) and deletion of VRP1 in bbc1Δ, MYO5-LAS17, myo3Δ cells restored normal growth (Figure1D). Linking Myo5 and Las17 and deleting the additional unlinked copy of LAS17 inhibited growth at 39°C (Figure S1C). This result may indicate that when present, unlinked Las17 is able to compete with the Myo5-Las17 fusion protein at endocytic sites and reduce the number of NPF domains, and deletion of unlinked Las17 thus leads to excessive actin polymerization at endocytic sites. Further evidence supporting this notion is that deletion of both LAS17 and VRP1 in the presence of Myo5-Las17 rescues growth at 39°C (Figure 1D), likely because deletion of VRP1 alleviates excessive NPF activity at endocytic sites in MYO5-LAS17, las17Δ, myo3Δ cells. Overall, these results suggest that endocytic sites need a balance of positive and negative regulators of actin polymerization for normal cell growth.
Finally, we replaced the entire WASP/Myosin complex with only Myo5-Las17. Cells were nevertheless able to grow at all temperatures (Figure 1D) and 71% of Sla1-GFP patches internalized (Figure1C). These findings show that Las17 and Myo5 can provide all essential functions of the WASP/Myosin complex.
The TH2 and SH3 domains can be eliminated from Myo5 if it is artificially recruited to endocytic sites
To further investigate why a the type I myosin is required for CME we focused on the role of each of Myo5’s five domains (Figure 2A) (Grötsch et al., 2010). Previous qualitative analysis of N-terminally-tagged mutants suggested that truncating Myo5 after the TH1 domain, deleting the motor-lever domain, or deleting the TH1 domain all cause defects in cortical recruitment (Grötsch et al., 2010). We expanded upon this analysis by quantitatively analyzing the effects of SH3 and TH2 domain deletions on cortical recruitment. We also chose to append GFP to the Myo5 carboxy terminus because amino-terminal modifications have the potential to interfere with motor domain function while carboxy-terminal modifications are tolerated (Sun et al., 2006). To normalize for brightness variation between slides and fields, we mixed cells expressing each Myo5-GFP variant with reference cells expressing wild-type Myo5-GFP. The reference cells also expressed a tagged kinetochore protein (Mtw1-mCherry) for identification (Figure 2B). We then compared the fluorescence of each Myo5-GFP punctum to a reference Myo5-GFP punctum and quantified the fluorescence intensity (Figure S2D). Each cell also had a functional MYO3 gene so any changes in Myo5-GFP fluorescence intensity were not the result of general perturbations in cell physiology or endocytosis. This approach revealed that deletion of the SH3 domain dramatically reduced cortical recruitment to 35% of the level of the reference Myo5-GFP (Figure 2F). Deletion of the Myo5 SH3 domain also eliminated the ability of myo3Δ cells to grow at 39°C (Figure 2D) and reduced the percent of internalized Sla1-GFP puncta to 43%, versus 96% in cells with wild-type MYO5 (Figure 2C and E). Deleting the Myo5 TH2 domain in MYO3 cells also decreased cortical recruitment to 65% of the level of the reference Myo5-GFP (Figure 2F, Figure S2D), and Sla1-GFP internalization was also reduced to 79% in myo3Δ cells (Figure 2C and E). Myo5 TH2 domain deletion also abolished growth at 39°C in myo3Δ cells (Figure 2D). Finally, deletion of both the TH2 and SH3 domains resulted in undetectable Sla1-GFP internalization in myo3Δ cells (Figure S2E and F) and lengthened Sla1-GFP and Abp1-RFP lifetimes (Figure S2G).
Figure 2. Endocytic roles of the Myo5 TH2 and SH3 domains.
(A) Diagram of the domains in Myo5 and each Myo5 variant analyzed in Figure 2.
(B) Maximum intensity projection of Myo5-GFP and Myo5-Vrp1-GFP patches (Green) imaged over the course of a 120 second movie in the medial focal plane. Each culture also contains cells with a reference Myo5-GFP and Mtw1-mCherry for identification. A maximum intensity Z-projection of Mtw1-mCherry fluorescence is shown to identify the reference cells in each field (red dots). Scale bar is 5 μm.
(C) Kymographs of representative endocytic patches from cells expressing Sla1-GFP and Abp1-RFP and the indicated Myo5 variant.
(D) Growth of Myo5 variants on YPD plates, as described for Figure 1D.
(E) Proportion of Sla1-GFP patches that internalize, as described in Figure 1C. *p<0.001 vs MYO5(myo3Δ); ** p<0.05 between bracketed variants; *** p<0.001 between bracketed variants.
(F) Histogram of normalized fluorescence intensity peaks of the indicated GFP-tagged Myo5 variants. Values represent the mean +/− SEM. * p<0.01 vs Myo5-GFP; **p<0.05 between bracketed variants; ***p<0.001 between bracketed variants. See also Figure S2, Movie S2, Tables S1 and S2.
To determine whether deleting the TH2 and SH3 domains caused growth and CME defects because of recruitment failure, we engineered a synthetic allele of MYO5 with VRP1 appended to its carboxy-terminal end (Figure 2A) and integrated the construct at the MYO5 locus. We confirmed that both Myo5 and Vrp1 components were expressed (Figure S2C), functional (Figure S2A), and recruited to cortical puncta in MYO3 cells (Figure 2B, Figure S2D). Because Vrp1 is independently recruited to endocytic sites (Sun et al., 2006), we hypothesized that linking Myo5 to Vrp1 might rescue recruitment defects of Myo5 mutants that are not efficiently recruited to endocytic sites. Indeed, we found that when Myo5 without the SH3 domain was linked to Vrp1, recruitment was restored from 35% to 70% of the level of the reference Myo5-GFP in MYO3 cells (Figure 2F, Figure S2D). Vrp1 linkage also rescued Sla1-GFP internalization (Figure 2C and E), Abp1-RFP lifetimes (Figure S2B), and cell growth at 39°C in myo3Δ cells (Figure 2D). Vrp1 linkage did not significantly increase the recruitment of Myo5 lacking the TH2 domain in MYO3 cells (Figure 2F, Figure S2D), but Sla1-GFP internalization (Figure 2C and E) and growth at 39°C were rescued in myo3Δ cells (Figure 2D). Vrp1 linkage also rescued endocytosis and lifetimes of Sla1-GFP and Abp1-RFP in myo3Δ cells simultaneously lacking the TH2 and SH3 domains of Myo5 (Figure S2E, F and G). Together these data show that while the TH2 and SH3 domains are required for normal Myo5 function, they become unnecessary when Myo5 is artificially linked to Vrp1.
CME requires actin binding and release by the Myo5 motor domain
Myo5 has a motor domain that can bind, release, and move actin filaments in an ATP-dependent manner (Sun et al., 2006). Next to the motor domain is a lever arm that has two conserved IQ motifs that can bind to calmodulin (Figure 3A) (Grötsch et al., 2010). While a functional type I motor domain is required for CME, the mechanism by which the motor domain contributes to CME has not been established.
Figure 3. Endocytic roles of the Myo5 motor domain.
(A) Diagram of each Myo5 variant analyzed in Figure 3.
(B) Kymographs of representative endocytic patches from cells expressing Sla1-GFP and Abp1-RFP.
(C) Proportion of Sla1-GFP patches that internalize, as described in Figure 1C. *P<0.001 vs MYO5 (myo3Δ).
(D and E) Growth of Myo5 variants on YPD plates, as described for Figure 1D.
(F) Proportion of Sla1-GFP patches that internalize, as described in Figure 1C. *P<0.001 vs MYO5 (myo3Δ).
(G) Growth of Myo5 variants on YPD plates, as described for Figure 1D.
(H) Proportion of Sla1-mCherry patches that internalize, as described in Figure 1C. *P<0.001 vs MYO5-GFP (MYO3). Myo5-GFP fluorescence was not imaged.
(I) Single frame from movies of sla2Δ cells expressing Sla1-GFP and Abp1-RFP with the indicated motor domain variants indicated. Also, kymographs from each cell are shown to the right of each image to show the extent of Abp1-RFP internalization relative to the Myo5-GFP signal. Image scale bar represents 5 μm. Vertical scale bar in kymograph is 1.0 μm, horizontal scale bar in kymograph is 20 seconds. See also Figure S3, Movies S3, Tables S1 and S2.
First, absence of the motor domain caused a profound decrease in Myo5 recruitment to cortical patches in MYO3 cells (Figure S3A). Deletion of the motor domain also increased the lifetime of GFP puncta and caused a subtle halo of GFP recruitment to the cortex making it difficult to quantify the defects in cortical recruitment (Figure S3A). Also, myo5-motorΔ, myo3Δ cells exhibited a near complete cessation of Sla1-GFP patch internalization (Figure 3B and C) and cells became temperature sensitive (Figure 3D).
Linkage of Myo5-motorΔ to Vrp1 in MYO3 cells did appear to rescue the recruitment of cortical GFP puncta to daughter cell buds, but it remained impossible to quantitate recruitment accurately in the mother cell (Figure S3A). Furthermore, Vrp1 linkage did not rescue Sla1-GFP patch internalization (Figure 3B and C), lifetimes of Sla1-GFP and Abp1-RFP (Figure S2C), or growth defects (Figure 3D) when MYO3 was deleted. These data demonstrate the importance of the motor domain for CME, although we cannot rule out the possibility that some of the CME defect is caused by poor Myo5 recruitment.
During the myosin motor power cycle, the motor domain binds and releases actin filaments in an ATP-dependent manner. The power cycle can be blocked in a state of high actin affinity by a rigor-type point mutation in the ATP binding site (G132R), which is predicted to prevent actin filament release (Lechler et al., 2000; Sun et al., 2006). Previous studies have proposed that a major role of the motor domain is to promote the formation of an actin network that is dynamic and can propel itself away from the cell cortex (Sun et al., 2006). Cells lacking the SLA2 gene have a distinct phenotype in which actin filaments are generated at the cortex and flux toward the cell interior as large comet-like tails (Figure 3I, Movie S3) (Kaksonen et al., 2003). This phenotype has become a useful tool for evaluating whether cells can generate a dynamic actin flux by assembling new filaments at endocytic sites (Kaksonen et al., 2003; Okreglak and Drubin, 2007, 2010; Skruzny et al., 2012; Sun et al., 2006). Sun et al. (Sun et al., 2006) found that the G132R point mutation in the motor domain prevented the inward flux of actin at most endocytic sites and suggested that the motor domain may be necessary for generating a normal, dynamic actin network at endocytic sites. We confirmed that the G132R mutation prevents most actin flux in sla2Δ, myo3Δ cells (Figure 3I, Movie S3). However, we also found that complete deletion of the Myo5 motor domain restored normal actin flux behavior in sla2Δ, myo3Δ cells (Figure 3I, Movie S3). This finding demonstrates that the motor domain per se is not necessary for generating a dynamic actin network, but that actin release by the motor domain is crucial.
Consistent with a model wherein F-actin release is a crucial Myo5 activity, we found that myo5-G132R, prevents all detectable movement of endocytic patches away from the cortex in myo3Δ cells (Figure 3F). The G132R mutation also caused a greater growth defect than Myo5-motorΔ mutation (Figure3E), longer lifetimes of Sla1-GFP and Abp1-RFP patches (Figure S3C). These findings indicate that complete loss of actin binding is less detrimental to cells than the inability to release actin. If prolonged actin binding by Myo5 prevents endocytic internalization by preventing actin filaments from moving away from the cortex, the G132R mutation should cause endocytic defects even in the presence of a functional myosin. Indeed, we found that myo5-G132R caused a decrease in the proportion of Sla1-mCherry patches that move away from the cortex in cells with MYO3 while deletion of the motor domain had little effect on CME in the MYO3 background (Figure 3H, Figure S3B). Also, the G132R mutation caused a decrease in growth at 25°C and 39°C that was not observed when the entire motor domain was absent (Figure 3G).
Together, these results suggest that while an actin-binding Myo5 motor domain is required for successful CME, actin release is equally important and is also necessary to allow actin filaments to flux away from the cortex.
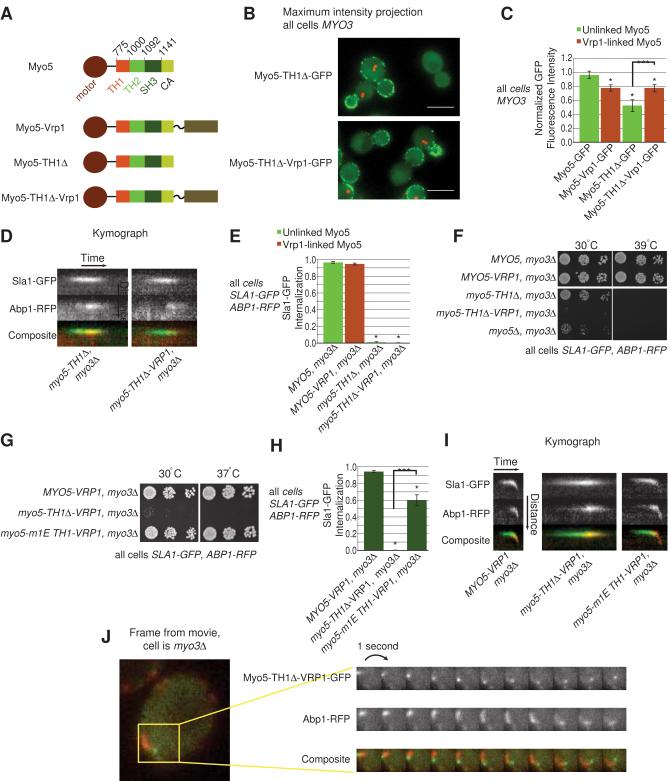
TH1 domain function
The TH1 domain of Myo5 can bind to phosphatidyl insositol phosphates (Fernández-Golbano et al., 2014). Furthermore, Grötsch et al. (Grötsch et al., 2010) identified a transient intramolecular interaction between the Myo5 TH1 and SH3 domain that may permit increased Vrp1 association in the presence of high levels of phosphatidyl inositol (4,5) bisphosphate (PI(4,5)P2) (Fernández-Golbano et al., 2014). However, these studies did not address whether the TH1 domain also makes additional SH3 domain-independent contributions to CME.
We first analyzed how TH1 domain deletion affects the recruitment of Myo5-GFP to cortical patches, as described above. We found that TH1 domain deletion substantially reduces Myo5 recruitment to the cell cortex (Figure 4B and C, Figure S4A). In myo3Δ myo5-TH1Δ cells, Sla1-GFP internalization was not observed (Figure 4D and E), and there was no cell growth at 39°C (Figure 4F). Interestingly, we observed internal Abp1-RFP structures (Movie S2), reminiscent of the internal structures that appeared when both MYO3 and MYO5 were deleted (Movie S1). The difference between these phenotypes and the phenotypes caused by SH3 domain deletion suggests that the TH1 domain has important roles in CME beyond simply modulating SH3 domain availability.
Figure 4. Endocytic roles of the Myo5 TH1 domain.
(A) Diagram of each Myo5 variant analyzed in Figure 4.
(B) Maximum intensity projection of Myo5-TH1Δ-GFP and Myo5-TH1Δ-Vrp1-GFP patches mixed with reference Myo5-GFP cells; all as described for Figure 2B. Scale bar is 5 μm
(C) Histogram of normalized fluorescence intensity peaks of the indicated GFP-tagged Myo5 variants. * p<0.01 vs Myo5-GFP; ***p<0.001 between bracketed variants.
(D) Kymographs of representative endocytic patches from cells expressing Sla1-GFP and Abp1-RFP and the indicated Myo5 variant.
(E) Proportion of Sla1-GFP patches that internalize, as described in Figure 1C. *P<0.001 vs MYO5 (myo3Δ).
(F-G) Growth of Myo5 variants on YPD plates at the indicated temperatures, as described for Figure 1D.
(H) Proportion of Sla1-GFP patches that internalize, as described in Figure 1C. *P<0.001 vs MYO5(myo3Δ); ***P<0.001 for bracketed variants.
(I) Kymographs of representative endocytic patches from cells expressing Sla1-GFP and Abp1-RFP and the Myo5 variant indicated below.
(J) Single frame from a movie of cells expressing Myo5-TH1Δ-Vrp1-GFP and Abp1-RFP. To the right is a montage showing the boxed from the image over several consecutive frames. The co-localization of Myo5-TH1Δ-Vrp1-GFP with the leading edge of Abp1-RFP tails is a feature seen in numerous cells with this genotype. See also Figure S4, Movie S4, Tables S1 and S2.
Because Vrp1 linkage rescued the recruitment of other Myo5 variants, we tested whether the linkage could also rescue recruitment of Myo5 lacking the TH1 domain. In MYO3 cells, the linkage rescued recruitment from 53% to 78% of the level of the reference Myo5-GFP (Figure 4B and C, Figure S4A), but it did not rescue the Sla1-GFP internalization defect (Figure 4D and E) and it exacerbated the growth defect caused by TH1 domain deletion (Figure 4F). The linkage also did not prevent appearance of internal Abp1-RFP structures (Movie S2) when MYO3 was also deleted. These results indicate that the TH1 domain performs a crucial function during CME besides recruiting Myo5 to endocytic sites.
Much speculation about the role of the TH1 domain has focused on the binding to PI(4,5)P2 or other phosphatidyl inositol phosphates, which are required for CME (Sun et al., 2005)(Cooke et al., 1998). To determine whether a specific interaction between the TH1 domain and PI(4,5)P2 is important for endocytosis, we replaced the TH1 domain of Myo5 with a TH1 domain from human Myosin 1E, the orthologue of Myo3 and Myo5 (Cheng et al., 2012; Krendel et al., 2008). Importantly, previous detailed analysis of the TH1 domain of human Myosin 1E revealed that it binds to all anionic phospholipids through nonspecific electrostatic interactions (Feeser et al., 2010). We generated this substitution in the background of Myo5-Vrp1 and myo3Δ to amplify the CME defects caused by TH1 deletion. Replacing the TH1 domain of Myo5 with the Myosin 1E TH1 domain completely rescued growth at 37°C (Figure 4G) and the proportion of Sla1-GFP patches that moved away from the cortex was restored to 60% (Figure 4H and I). The Myosin 1E TH1 domain also eliminated the internal actin structure phenotype (Figure S4B), rescued the Sla1-GFP lifetime and partially rescued the Abp1-RFP lifetime (S4C). These data suggest that endocytosis does not require that the TH1 domain bind to PI(4,5)P2 or other phosphoinositides specifically because it can be replaced with a domain that binds any anionic phospholipid based on charge. Conversely, neither the TH1 domain from mouse Myosin 1C that specifically binds PI(4,5)P2 and PI(3,4,5)P3 (Hokanson and Ostap, 2006) nor a mutant version with low affinity for all phospholipids (K892A) (Hokanson et al., 2006) rescued growth (Figure S4D), suggesting that PI(4,5)P2 binding activity alone is not sufficient to replace Myo5 TH1 function. Although any lack of rescue may be due to improper folding or domain orientation (For western blot, see Figure S4E), the positive growth and endocytosis rescue by the Myosin 1E TH1 domain is strong evidence that the primary role of the TH1 domain is as an electrostatic phospholipid anchor and that this function is conserved between fungi and metazoans.
The appearance of mobile, internal Abp1-RFP patches in cells lacking a TH1 domain is an unusual phenotype (Movie S1, Movie S2). These patches were not associated with the endocytic coat marker Sla1 (Movie S1, Movie S2), but Myo5-TH1Δ-GFP did associate with the leading edge of the mobile internal Abp1-RFP patches (Figure 4J, Movie S4). The appearance of these patches suggests that the TH1 domain couples actin polymerization to endocytic sites or the cortex and deletion of the TH1 domain causes ectopic internal actin polymerization. Although the mechanism for this is activity is not yet clear, this ectopic actin polymerization might contribute to the substantial growth defect that occurs when the TH1 domain is deleted.
A synthetic protein comprised of only a motor domain, lever arm, TH1 domain, PRR and NPF domains can perform the essential WASP/Myosin complex endocytic functions
Las17 is the first component of the WASP/Myosin complex to be recruited to endocytic sites. Three domains have been defined in Las17 including the WASP Homology Domain 1 (WH1) that interacts with Vrp1 (Wong et al., 2010), a proline-rich region (PRR) that contains multiple PXXP motifs and binds to SH3 domains, and the WH2 and CA domains that constitute an NPF (Figure 5A). The PRR region from residue 339 to 361 interacts with Sla1 and contains two PXXP motifs (Feliciano and Di Pietro, 2012). The role of the remaining PXXP motifs of the PRR is less clear, but they might be involved in recruiting additional SH3 domain-containing proteins including Bbc1, Bzz1, Myo3 and Myo5 (Tonikian et al., 2009). Since we have shown that artificially linking Myo5 to Las17 eliminates the need for the remaining WASP/Myosin complex components, we hypothesized that we may also be able to eliminate much of Las17 when it is fused to Myo5. To test this idea, we generated several internally truncated versions of Myo5-Las17, shown in Figure 5A. Las17 truncated up to residue 324 contains the Sla1-interacting PXXP motifs, but those motifs are absent when Las17 is truncated up to residue 426. We found that truncated Las17 fused to Myo5 supported growth when Las17 was truncated up to residue 324, but truncating Las17 up residue 426 prevented growth at 39°C (Figure 5B)
Figure 5. WASP/Myosin engineered protein can be simplified to contain only motor, TH1, PRR and NPF domains.
(A) Diagram of all truncations of the Myo5-Las17 fusion protein analyzed in this Figure.
(B) Growth of Myo5-Las17 fusion protein variants on YPD, as described in Figure 1D.
(C) Kymographs of representative endocytic patches from cells expressing Sla1-GFP and Abp1-RFP and the indicated Myo5-Las17 variant indicated below. In all cells analyzed, the entire WASP/Myosin complex has been deleted and Myo5 has been replaced by the Myo5-Las17 fusion protein or fusion protein variant.
(D) Proportion of Sla1-GFP patches that internalize, as described in Figure 1C. See also Figure S5.
We next asked whether a minimal Myo5 fused to a minimal Las17 could still rescue complete deletion of the WASP/Myosin complex. We generated a new construct consisting of only the motor-lever and TH1 domains from Myo5 and Las17 residues starting at 325 (TH2-CAΔ-WH1-324Δ) (Figure 5A). Like full length Myo5-Las17, TH2-CAΔ-WH1-324Δ supported CME in the absence of the entire WASP/Myosin complex with 69% of Sla1-GFP patches internalizing (Figure 5C and D). In fact, the only difference we noted between cells with Myo5-Las17 and TH2-CAΔ-WH1-324Δ was that the latter cells had extended Sla1-GFP lifetime (45+/− 1 seconds vs 34+/−1 seconds; Figure S5) and an extended delay between the arrival of Sla1-GFP and Abp1-RFP (31+/−1 seconds vs 18+/−1 seconds; Figure S5). This delay seems to occur only in cells lacking the N-terminal half of the PRR up to residue 324 since strains lacking only the WH1 domain do not have this delay (Figure S5). Nevertheless, these data demonstrate that the motor-lever, the TH1, the NPF domain, and the PRR regions provide all of the WASP/Myosin complex activities that are essential for CME.
Discussion
From yeast to humans, the endocytic machinery is characterized by high overall complexity including the highly conserved proteins that couple actin assembly to membrane invagination and scission. Our results demonstrate that the WASP/Myosin complex can be dramatically simplified without major consequences for CME or cell growth, identifying the crucial activities for this complex. We found that six proteins: Las17, Vrp1, Bzz1, Bbc1, Myo3, and Myo5 could all be replaced by a single synthetic protein containing only a motor-lever domain, a TH1 domain, a truncated PRR, and NPF domains. While considerable research has already highlighted the role of the NPF domains in triggering actin polymerization (Galletta et al., 2008; Sun et al., 2006), our results offer insights into the roles of the motor domain, the TH1 domain, and the PRR. Furthermore, our results show that all remaining proteins and domains in the complex do not provide any essential biochemical or biophysical functions because CME still occurs when they are all eliminated.
It was not expected that combining all of the essential activities of the complex into a single protein would fulfill the functions of the complex because in the normal situation, these proteins are recruited to endocytic sites in a regular order and with distinct timing for each protein (Sun et al., 2006). For example, Sun et al. (2006) showed that there is greater than a ~10 second delay between Las17 and Vrp1 recruitment and another ~10 second delay between Vrp1 and class I myosin/Bbc1/actin recruitment. Electron and fluorescence microscopy have further revealed that Las17 and Myo5 are localized differently along the outside of an endocytic invagination; Las17 appears to be mostly recruited to the base and bud neck while Myo5 may be primarily limited to the base (Idrissi et al., 2012; Picco et al., 2015). Surprisingly, a single protein can function in place of all of these proteins. This finding indicates that the differences in recruitment timing and position along the membrane invagination are not essential for CME, although they may fine-tune the performance of the system.
Another striking finding from this study was that Vrp1 is dispensable when Myo5 and Las17 are linked. Vrp1 had been considered one of the key proteins of the WASP/Myosin complex because VRP1 deletion stops virtually all CME (Evangelista and Klebl, 2000; Geli et al., 2000; Grötsch et al., 2010; Idrissi et al., 2012; Sun et al., 2006; Wong et al., 2010). The only other reported mutation that can partially suppress the vrp1Δ growth and CME phenotypes is a mutation in the Sla2-interaction region of clathrin light chain (clc-Δ19-76) (Boettner et al., 2011). In this case, the mechanism of this suppression was proposed to be increased actin binding by the coat protein Sla2, which might partially compensate for a semi-functional WASP/Myosin complex (Boettner et al., 2011). In contrast, our results indicate that Myo5-Las17 linkage preserves the function of the WASP/Myosin complex in the absence of Vrp1. This result strongly suggests that the only essential role of Vrp1 is as a scaffolding protein that brings together Myo5 and Las17.
It was noteworthy that deleting BBC1 in cells with Myo5 linked to Las17 caused temperature sensitivity but no severe Sla1-GFP internalization defect. Both of these mutations are predicted to increase actin polymerization at endocytic sites, suggesting that they may be inhibiting growth by excessive actin polymerization. Indeed, deleting BZZ1 or VRP1, positive regulators of the Arp2/3 complex, reduced or eliminated this temperature sensitivity. Competition for actin monomers between distinct processes was recently shown in fission yeast, wherein Arp2/3 inhibitors caused excessive formin-assembled actin cables (Burke et al., 2014; Suarez et al., 2015). Our data are consistent with the hypothesis that S. cerevisiae cells also require a balance of positive and negative regulators of actin polymerization at endocytic sites.
Because class I myosins are required for CME, developing a mechanistic understanding of the process requires a detailed understanding of the role of each its domains. However, because some domains are also required for class I myosin recruitment to endocytic sites, previous studies have not been able to distinguish whether a particular endocytic defect caused by deletion of a particular domain is the result of recruitment failure or disruption of some other essential biophysical or biochemical activity. In this study, we addressed this problem by artificially recruiting Myo5 mutants to endocytic sites by Vrp1 linkage. In the process we learned that each domain of Myo5 contributes to recruitment, but we found that when Myo5 was artificially recruited to endocytic sites by Vrp1 linkage, we could eliminate the SH3 domains without impacting CME. Vrp1 linkage also rescued the growth phenotypes caused by deleting the TH2 domain, although cortical recruitment of Myo5-TH2Δ-GFP was not rescued and the CME rescue was subtle. Therefore, the mechanisms for this growth rescue are likely more complicated than simply rescuing Myo5 recruitment or CME. Nevertheless, these studies focused our attention on the TH1 and motor domains, which remained essential for CME regardless of whether Myo5 was linked to Vrp1. Therefore, models for how the WASP/Myosin complex contributes to CME must incorporate the activities of the motor-lever and TH1 domains.
Our functional analysis of the Myo5 motor domain showed that actin release, in addition to actin binding, is essential for CME. This finding clarifies several conflicting results from previous reports with important mechanistic implications. Sun et al. (2006) found that a rigor-type mutation (G132R) in the motor domain prevented actin fluxing from the cell cortex in sla2Δ cells. This result strongly suggested that the motor domain is required to produce a dynamic actin cytoskeleton that can propel itself away from the cortex. Subsequently, Idrissi et al. (2012) investigated the motor domain mutant E414V that is predicted to increase the amount of time spent in the low F-actin affinity state (Friedman et al., 1998). Idrissi et al. (2012) found that endocytic sites failed to deeply invaginate, but there was still a large ribosome-free area seen by electron microscopy, which suggests that F-actin could still be propelled away from the cortex. The authors therefore concluded that the F-actin network could still internalize, but the motor domain was required to utilize the network for productive CME. To reconcile these fundamentally different mechanisms, we analyzed the motorΔ mutant that is functionally analogous to the E414V mutant and we analyzed the G132R mutant that blocks actin release. Both mutations prevented CME. Like Sun et al. (2006), we also found that the G132R mutant prevented the flux of F-actin away from the cortex in sla2Δ cells, but complete deletion of the motor domain did not. Furthermore, the G132R mutation was partially dominant over the MYO3 paralogue, while complete deletion of the motor domain was not. These results suggest that actin release is required to produce a dynamic, branched actin network that can be pushed away from the cortex, but transient actin binding by the motor domain is required to utilize that network to power CME.
The TH1 domain has been studied less than other domains. We found that deleting the TH1 domain in myo3Δ cells caused the most pronounced CME defect of all the mutants we analyzed. The TH1 domain contributed to Myo5 recruitment, and linking Myo5-TH1Δ to Vrp1 rescued recruitment. However, the growth defect was actually made worse by the linkage to Vrp1 in myo3Δ cells. Furthermore, myo5-TH1Δ in myo3Δ cells exhibited the frequent appearance of internal comet-like tails of Abp1-RFP that move throughout the cells. These structures resemble the structures that occur when endocytic coat proteins Pan1 and End3 are depleted (Sun et al., 2015), although this study provides evidence of this phenotype associated with a WASP/Myosin complex mutation. This phenotype also occurs in myo5Δ myo3Δ cells which also have no functional TH1 domains. These data establish that the TH1 domain is required for CME and for retaining the endocytic actin assembly machinery at the plasma membrane. Lipid strip binding assays suggested that the Myo5 TH1 domain has the capacity to bind PI(4,5)P2 and other phosphoinositide phosphates (Fernández-Golbano et al., 2014). However, the human orthologue Myosin 1E, binds to all negatively charged phospholipids through an apparently non-specific electrostatic mechanism (Feeser et al., 2010). Because the TH1 domain from Myosin 1E could replace the Myo5 TH1 domain and rescue phenotypes due to loss of the TH1 domain, we conclude that the primary role of the TH1 domain is as an electrostatic phospholipid anchor. Because PI(4,5)P2 is negatively charged and present at endocytic sites, our data do not exclude a role in PI(4,5)P2 binding by the TH1 domain or an intramolecular interaction between the TH1 and SH3 domains. However, our data do suggest that specificity for PI(4,5)P2 versus phosphatidyl serine or other negatively charged phospholipids is not necessary. Finally, these results are consistent with the hypothesis that Myo5 and human Myosin 1E perform a similar role during CME.
We also found that several Las17 domains could be eliminated when Myo5 and Las17 are linked. The WH1 domain is believed to interact with Vrp1 (Wong et al., 2010) and we could completely eliminate it without endocytic consequence. We could also remove about half of the PRR (up to residue 324) without consequence, but when we truncated the PRR up to residue 426, growth was impacted. These findings are predicted by recent results by Feliciano and Di Pietro (Feliciano and Di Pietro, 2012) in which Las17 PRR residues 339 to 361 were found to bind to Sla1, to help recruit Las17, and to allow Sla1 to repress the NPF activity of Las17 (Feliciano and Di Pietro, 2012). An even more dramatically truncated Myo5-Las17 in which Las17 lacked residues up to 324 and Myo5 lacked the TH2 through CA domains could still replace the entire WASP/Myosin complex and support robust CME. In fact, the only difference between the CME phenotype of Myo5-Las17 and the TH2-CAΔ-WH1-324Δ variant of Myo5-Las17 is that the latter had an extended Sla1-GFP lifetime and an extended delay between the appearance of Sla1-GFP and Abp1-RFP. These results suggest that the N-terminal half of the PRR may contribute to the derepression of NPF activity of Las17, although this function is clearly not essential for CME because the overall proportion of Sla1-GFP patches that internalize is the same between the two variants. Overall, these results are consistent with the notion that the Las17 PRR mediates critical WASP/Myosin complex interactions.
Together these results suggest that the WASP/Myosin complex has only four activities that are essential for CME (see Figure 6A). First, the complex requires an initial recruitment mechanism, which is mediated by the PRR of Las17 binding to Sla1 and potentially other coat module proteins. Second, the WH2 and CA domains stimulate actin polymerization by activating the Arp2/3 complex. The current study has also revealed a third required activity, actin binding and release by the motor domain. Finally, the TH1 domain must anchor the complex to the plasma membrane.
Figure 6. WASP/Myosin complex functions as a single protein with only four activities.
(A) Model: The WASP/Myosin complex has four activities that are necessary for CME and sufficient for a functional WASP/Myosin complex.
(B) Theoretical outward-directed movement of individual, growing actin filaments that are tethered to the bud tip but not the base.
(C) Model: The TH1 and motor domains constitute a membrane-actin linkage that tethers F-actin to the base of endocytic sites. While the F-actin at endocytic sites is a branched, cross-linked network, we have shown only two individual filaments for clarity of presentation. See also Movie S5.
The membrane and F-actin binding by the TH1 and motor domains, respectively, suggests that these domains constitute a membrane-actin linkage. This linkage is fundamentally distinct from the membrane-actin linkage at the bud tip that is mediated by Sla2, Pan1 and epsins (Skruzny et al., 2012). First, this linkage likely occurs near the base or neck of endocytic sites because immunogold-labeled EM images (Idrissi et al., 2008) and highly precise fluorescence localization data (Picco et al., 2015) show that Myo5 most frequently occupies that region. Second, the F-actin-binding component of this linkage must have the ability to release actin filaments in addition to bind filaments so the filaments can be pushed inward as they grow (Figure 6B, Movie S5). Class I myosins bind to actin filaments, move the filaments in the pointed end direction, and release the filaments. Therefore, this actin linkage appears to be specifically evolved to catch and release actin filaments. This mechanism offers plausible explanations for how actin filaments may be oriented to produce this observed inward flux in the presence of coupling between F-actin and the bud tip (Figure 6C, Movie S5 bottom panel) and for how individual filaments are prevented from propelling themselves away from endocytic sites, which could occur in the absence of a membrane-actin linkage at the endocytic base (Figure 6B, Movie S5 top panel). Consistent with this hypothesis, deletion of the membrane-binding TH1 domain results in actin polymerization that is uncoupled from endocytic sites (Figure 4I). Similar internal actin structures were not seen when the motor domain was deleted, which could indicate that other Myo5 domains are able to anchor the actin cytoskeleton to endocytic sites in the absence of the motor domain, although not in a fashion that supports robust endocytic internalization. These domains along with other actin-binding domains that have yet to be identified may cooperate with the motor domain to orient actin filaments.
At this point, it is unclear whether Myo5 and Myo3 also provide a force on the actin filaments to add to the force generated by F-actin polymerization. There is precedence for class I myosin motors to either have force generating power strokes (Myosin 1C) (Greenberg et al., 2012), or power strokes that act as a tension sensitive anchors that produce negligible force (Myosin 1B) (Laakso et al., 2008). Biophysical characterization of the Myo5 motor domain will be necessary to elucidate the specific function provided by the motor domain during CME. However, the data presented in this paper and the identification of an essential myosin-based actin-membrane linkage substantially enhances our understanding of this process and will inform future models for how the WASP/Myosin complex contributes to CME.
Experimental Procedures
Plasmids and Strains: Plasmids were used as template DNA for genomic integration as described (Sun et al., 2006) and are listed in Table S1. For MYO5 mutations, plasmid pDD2453 containing MYO5 plus the flanking genomic sequence and the URA3 gene cassette immediately after the MYO5 terminator was used as the template DNA for wild-type MYO5 marked with URA3. All other MYO5 variants and fusion proteins were generated in this plasmid background by site directed mutagenesis using the BsaI restriction enzyme and recognition site in the 5’ end of primers. For genomic integration, a linear fragment from each plasmid was excised, gel purified, and transformed into DDY1102 or DDY1102-derived strains using standard transformation procedures and plated on selective media . Proper genomic integration was confirmed by PCR using primers in the flanking genomic region outside each insert and within the insert (data not shown) and by Western blot analysis for myc- or GFP-tagged variants (Figure S2C, Figure S4E, and data not shown). Strains las17Δ and vrp1Δ genotypes were generated as described (Longtine et al., 1998) using a modified version of pFA6 plasmid with a HygMX6 cassette. Growth of yeast strains on YPD plates was performed essentially as described (Sun et al., 2006). For comparing growth on YPD plates, starter liquid YPD cultures were diluted to OD600 of 0.02, 0.004, and 0.0008 and 2.5 microliters of each dilution were spotted on the plates. Plates were then grown at the indicated temperatures for 48 hours and photographed. Additional gene combinations were generated by crosses to existing strains. The strains used in this study are listed in Table S2. For strains with the sla2Δ mutation, diploids were generated from haploid parent strains, sporulated, and the spore colonies were immediately analyzed to avoid selection for secondary mutations.
Live cell imaging: Strains with Sla1-GFP, Sla1-Cherry, and Abp1-RFP, were imaged in a medial focal plane as described previously (Kaksonen et al., 2005). Sla1-GFP or Sla1-mCherry patches were considered to have internalized when the fluorescence centroid moved more than three pixels (194nm) away from the cortex. All imaging experiments were conducted on at least three separate imaging sessions. At least 150 Sla1-GFP and Abp1-RFP patches were analyzed for each cell type. See the Supplemental Experimental Procedures for further details.
Quantitative fluorescence measurements: For quantitative measurement of Myo5-GFP recruitment, cells were imaged in a medial focal plane for 120 seconds, 1 second per exposure. Fluorescence intensity of cortical patches was measured in mother cells. Background fluorescence was calculated by measuring the local fluorescence intensity before and after the Myo5-GFP patch and a linear regression was used to extrapolate and subtract background fluorescence from each time point over the course of the patch lifetime. Cells expressing each Myo5-GFP variant were mixed with reference cells expressing wild-type Myo5-GFP and Mtw1-mCherry (kinetochore marker) for differentiation. Fluorescence intensity of Myo5-GFP patches were normalized by dividing by the mean peak fluorescence of the reference Myo5-GFP within the same viewing field. Normalized fluorescence intensity measurements were pooled and compared. Exactly 45 patches were measured for each variant and associated reference cells.
Statistical analysis: For each pair of variables, pooled data from all days of analysis were compared by a 2-sided Mann-Whitney test using the program M-stat version 6.0. The null hypothesis was that the two populations were the same. We rejected the null hypothesis if the p value was below the threshold value of 0.05, 0.01, or 0.001, as indicated for each comparison.
Supplementary Material
Acknowledgements
We would like to thank Nathaniel Krefman, Yidi Sun and Kathryn Sieverman for providing plasmids, Jasper Weinberg for providing the Image J plugin used for kymograph analysis, Christa Cortesio and Akemi Kunibe for feedback on the design and analysis, Norman Drinkwater for providing the the statistical package Mstat 6.0, and Eric D. Metzgar for video editing assistance. Funding was provided by NIH Research grants R37 GM42759 and R01 GM50399 and postdoctoral fellowship F32 GM 100709 for E.B.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions:
Conceptualization, E.B.L. D.G.D.; Methodology, E.B.L, J.H., R.T.A.P., D.G.D.; Validation, E.B.L., R.T.A.P., D.G.D.; Formal Analysis, E.B.L., J.H., R.T.A.P., Investigation, E.B.L., R.T.A.P., J.H., R.L., H.M.M.; Resources, D.G.D.; Data curation, E.B.L.; Writing-Original Draft, E.B.L.; Writing-Reviewing and Editing, E.B.L., R.T.A.P., D.G.D.; Visualization, E.B.L.; Supervision, D.G.D.; Project Administration, E.B.L.; Funding Acquisition, D.G.D., E.B.L.
Supplemental Information:
Figures S1, S2, S3, S4, S5
Tables S1, S2
Movie S1, S2, S3, S4, S5
Supplemental experimental procedures
References
- Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat. Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol. Biol. Cell. 2011;22:3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, Chi RJ, Lemmon SK. Lessons from yeast for clathrin-mediated endocytosis. Nat. Cell Biol. 2012;14:2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Kural C, Zeeh J-C, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 2012;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T. a, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr. Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Grassart A, Drubin DG. Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis. Mol. Biol. Cell. 2012;23:2891–2904. doi: 10.1091/mbc.E11-04-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke FT, Parker PJ, Hall MN. MSS4 , a Phosphatidylinositol-4-phosphate 5-Kinase Required for Organization of the Actin Cytoskeleton in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:15787–15793. doi: 10.1074/jbc.273.25.15787. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Klebl B. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeser EA, Ignacio CMG, Krendel M, Ostap EM. Myo1e binds anionic phospholipids with high affinity. Biochemistry. 2010;49:9353–9360. doi: 10.1021/bi1012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano D, Di Pietro SM. SLAC, a complex between Sla1 and Las17, regulates actin polymerization during clathrin-mediated endocytosis. Mol. Biol. Cell. 2012;23:4256–4272. doi: 10.1091/mbc.E11-12-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, Raimondi A, Paradise S, Mesaki K, Ferguson A, Destaing O, Takasaki J, Cremona O, Toole EO, De P. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev. Cell. 2010;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Golbano IM, Idrissi F-Z, Giblin JP, Grosshans BL, Robles V, Grötsch H, Borrás MDM, Geli MI. Crosstalk between PI(4,5)P2 and CK2 Modulates Actin Polymerization during Endocytic Uptake. Dev. Cell. 2014;30:746–758. doi: 10.1016/j.devcel.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Friedman a. L., Geeves M. a., Manstein DJ, Spudich J. a. Kinetic characterization of myosin head fragments with long-lived myosin??ATP states. Biochemistry. 1998;37:9679–9687. doi: 10.1021/bi973143f. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Chuang DY, Cooper J. a. Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Lombardi R, Schmelzl B, Riezman H. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, Lin T, Goldman YE, Shuman H, Ostap EM. PNAS Plus: Myosin IC generates power over a range of loads via a new tension-sensing mechanism. Proc. Natl. Acad. Sci. 2012;109:E2433–E2440. doi: 10.1073/pnas.1207811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grötsch H, Giblin JP, Idrissi F-Z, Fernández-Golbano I-M, Collette JR, Newpher TM, Robles V, Lemmon SK, Geli M-I. Calmodulin dissociation regulates Myo5 recruitment and function at endocytic sites. EMBO J. 2010;29:2899–2914. doi: 10.1038/emboj.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Ostap EM. Myo1c binds tightly and specifically to phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3118–3123. doi: 10.1073/pnas.0505685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol. Biol. Cell. 2006;17:4856–4865. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi F, Wolf BL, Geli MI. Cofilin , But Not Profilin , Is Required for Myosin-I-Induced Actin Polymerization and the Endocytic Uptake in Yeast. Mol. Biol. Cell. 2002;21:4074–4087. doi: 10.1091/mbc.02-04-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi F-Z, Grötsch H, Fernández-Golbano IM, Presciatto-Baschong C, Riezman H, Geli M-I. Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J. Cell Biol. 2008;180:1219–1232. doi: 10.1083/jcb.200708060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi F-Z, Blasco A, Espinal A, Geli MI. Ultrastructural dynamics of proteins involved in endocytic budding. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2587–E2594. doi: 10.1073/pnas.1202789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsdottir GA, Li R. Dynamics of Yeast Myosin I : Evidence for a Possible Role in Scission of Endocytic Vesicles. Curr. Biol. 2004;14:1604–1609. doi: 10.1016/j.cub.2004.08.055. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A Pathway for Association of Receptors, Adaptors, and Actin during Endocytic Internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Sun Y, Buser C, Liu J, Michelot A, Drubin DG. Determinants of endocytic membrane geometry, stability, and scission. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E979–E988. doi: 10.1073/pnas.1113413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Osterweil EK, Mooseker MS. Myosin 1E interacts with synaptojanin-1 and dynamin via its SH3 domain. FEBS Lett. 2008;581:644–650. doi: 10.1016/j.febslet.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W, Schorb M, Kaksonen M, Briggs J. a G. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 2012;150:508–520. doi: 10.1016/j.cell.2012.05.046. [DOI] [PubMed] [Google Scholar]
- Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Shevchenko A, Shevchenko A, Li R. Direct Involvement of Yeast Type I Myosins in Cdc42-dependent Actin Polymerization. J. Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Iii AMK, Demarini DJ, Shah NG. Additional Modules for Versatile and Economical PCR-based Gene Deletion and Modification in Saccharomyces cerevisiae. 1998;961:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Drubin DG. Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J. Cell Biol. 2010;188:769–777. doi: 10.1083/jcb.200909176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco A, Ries J, Nedelec F, Kaksonen M. Visualizing the functional architecture of the endocytic machinery. Elife. 2015 doi: 10.7554/eLife.04535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skruzny M, Brach T, Ciuffa R, Rybina S, Wachsmuth M, Kaksonen M. Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2533–E2542. doi: 10.1073/pnas.1207011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A, Lechler T, Spiridonov V, Shevchenko A, Li R, Winsor B. Saccharomyces cerevisiae Bzz1p Is Implicated with Type I Myosins in Actin Patch Polarization and Is Able To Recruit Actin-Polymerizing Machinery In Vitro. Mol. Cell. Biol. 2002;22:7889–7906. doi: 10.1128/MCB.22.22.7889-7906.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez C, Carroll RT, Burke T. a, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell. 2015;32:43–53. doi: 10.1016/j.devcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Takenawa T. Dynamic Shaping of Cellular Membranes by Phospholipids and Membrane-Deforming Proteins. Physiol. Rev. 2014;94:1219–1248. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- Sun Y, Kaksonen M, Madden DT, Schekman R, Drubin DG. Interaction of Sla2p’s ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol. Biol. Cell. 2005;16:717–730. doi: 10.1091/mbc.E04-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG. Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev. Cell. 2006;11:33–46. doi: 10.1016/j.devcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Sun Y, Leong NT, Wong T, Drubin DG. A Pan1/End3/Sla1 complex links Arp2/3-mediated actin assembly to sites of clathrin-mediated endocytosis. Mol. Biol. Cell. 2015 doi: 10.1091/mbc.E15-04-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonikian R, Xin X, Toret CP, Gfeller D, Landgraf C, Panni S, Paoluzi S, Castagnoli L, Currell B, Seshagiri S, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MH, Meng L, Rajmohan R, Yu S, Thanabalu T. Vrp1p-Las17p interaction is critical for actin patch polarization but is not essential for growth or fluid phase endocytosis in S. cerevisiae. Biochim. Biophys. Acta. 2010;1803:1332–1346. doi: 10.1016/j.bbamcr.2010.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.