Abstract
Signal transduction pathways activated by receptor tyrosine kinases (RTK) play a critical role in many aspects of cell function. Adaptor proteins serve an important scaffolding function that facilitates key signaling transduction events downstream of RTKs. Recent work integrating both structural and functional genomic approaches has identified several adaptor proteins as new oncogenes. In this review, we focus on the discovery, structure and function, and therapeutic implication of three of these adaptor oncogenes, CRKL, GAB2, and FRS2. Each of the three genes is recurrently amplified in lung adenocarcinoma or ovarian cancer, and is essential to cancer cell lines that harbor such amplification. Overexpression of each gene is able to transform immortalized human cell lines in in vitro or in vivo models. These observations identify adaptor protein as a distinct class of oncogenes and potential therapeutic targets.
Introduction
Receptor tyrosine kinase (RTK) signaling plays key roles in development and cell physiology. Inappropriate activation of these signaling pathways contributes to the genesis and progression of many types of cancer. A prototypical RTK signaling pathway starts with activation by growth factor ligand binding which induces receptor dimerization (Lemmon and Schlessinger, 2010). This dimerization event facilitates the trans-phosphorylation of RTKs, which recruits and activates downstream signaling molecules. These molecules may be directly recruited by binding to RTKs, or indirectly recruited by adaptor proteins that form specific complexes with both the molecules and the associated RTKs. Adaptor proteins lack enzymatic activity but provide an important scaffolding function that facilitates key signaling transduction events and regulates signal specificity and amplification (Pawson and Scott, 1997). As such, adaptor proteins exert temporal control over activated signaling pathways by and varying expression level and phosphorylation status.
The adaptor proteins are classified into two groups based on the structure and function (Gotoh, 2008). First group is comprised of docking proteins that have multiple tyrosine phosphorylation sites to bind downstream signaling proteins. Examples of this group include GRB2-associated binding protein (GAB), fibroblast growth factor receptor substrate 2 (FRS2), insulin receptor substrate (IRS), Src homology 2-containing protein (SHC), and downstream of kinase (DOK)-family proteins (Fig. 1). This group of adaptor proteins often contains membrane localization domains and are also called membrane-linked docking proteins (MLDP). The second group is comprised of adaptor proteins with only Src homology 3 (SH3) and/or SH2 domains to bind signaling proteins, without a membrane localization structure or phosphorylation sites. Examples of the second group include GRB2, CRK, and NCK.
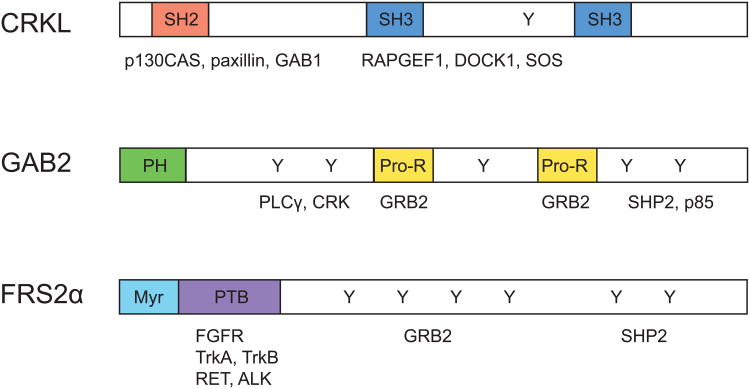
Fig. 1.
Schematic representation of CRKL, GAB2, and FRS2.
The CRKL protein structure contains one SH2 (Src homology 2) domain that binds to p130CAS (also known as BCAR1), paxillin, and GAB1, and two SH3 (Src homology 3) domains (SH3N and SH3C) that interact with RAPGEF1, DOCK1, and SOS. Structural domains of GAB2 include an N-terminal PH (Pleckstrin homology) domain critical for membrane localization, central proline-rich (Pro-H) domains that interact with SH2 and SH3 domain-containing proteins such as GRB2, and multiple phosphorylation sites are also present to bind to signaling partners such as PLC-γ, CRK, SHP2, and p85. FRS2α contains an N-terminal myristoylation site for membrane anchoring, a C-terminal phosphotyrosine binding (PTB) domain that interacts with limited species of receptors. It also includes multiple tyrosine phosphorylation sites that bind to SH2 domains of GRB2 and GAB1.
We and others have shown that several genes encoding adaptor proteins are recurrently amplified in lung adenocarcinomas and primary ovarian cancers and essential to proliferation and survival in cancer cells that harbor those amplifications (Brown et al., 2008; Luo et al., 2008, 2015; Kim et al., 2010; Cheung et al., 2011a, 2011b; Wang et al., 2012; Dunn et al., 2014). Overexpression of these genes in immortalized human cell lines promoted anchorage-independent growth and tumorigenesis. These studies suggest that adaptor protein-encoding genes are oncogenes in a subset of lung adenocarcinomas and ovarian cancers. In this review, we will focus on the discovery, structure and function, and therapeutic implications of CRKL, GAB2, and FRS2.
Structure and function of adaptor proteins
The CRK family consists of three members, CRKI, CRKII, and CRK-like protein (CRKL). CRKI and CRKII are alternative transcripts of CRK. The CRK family proteins integrate signals from multiple sources (Fig. 2) including growth factor receptors (Birge et al, 2009), integrin receptors (Cabodi et al, 2010), bacterial and viral pathogens (Weidow et al, 2000; Heikkinen et al., 2008), and apoptotic cells (Albert et al., 2000). Examples of the growth factor receptors that mediate signaling through CRK or CRKL include EGFRs, neurotrophin growth factor receptors (TrkA), IGF-R, PDGFRα, VEGFR, Met, and EphB2 receptor (Birge et al., 2009). CRK family also belongs to the category of intracellular integrin signaling adaptor proteins that include the Cas family, the IPP complex, and the Cap family (Cabodi et al, 2010). The integrin receptors, such as β1 integrin receptor, are enzymatically inactive receptors that upon binding the extracellular matrix (ECM), undergo conformational change and initiate signal transduction through intracellular adaptor proteins.
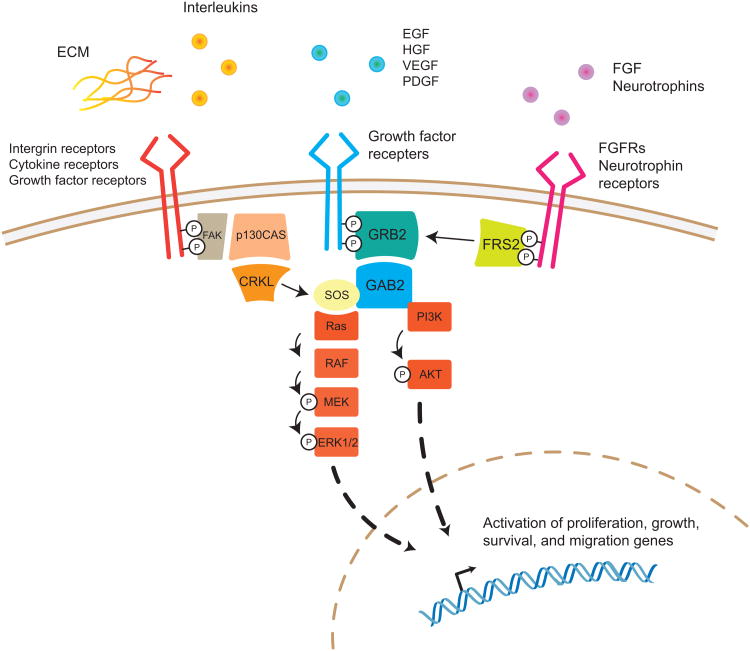
Fig. 2.
Overview of the signaling pathways associated with CRKL, GAB2, and FRS2.
CRKL is activated by multiple types of cell surface receptors including integrin receptors upon binding to extracellular matrix (ECM), cytokine receptors, and growth factor receptors. GAB2 and FRS2 are primarily activated by growth factor receptors. FRS2 exhibits more specificity towards fibroblast growth factor receptors and neutrophin receptors. All three adaptor proteins lead to Ras/MAPK and PI3K/AKT pathway activation that promote cancer initiation and progression.
CRKL consists of an N-terminal SH2 domain followed by two SH3 domains (SH2-SH3N-SH3C) (Fig. 1) (Feller, 2001). The SH2 domains of CRKL bind to a specific motif (Y-x-x-P) present within the docking proteins such as p130CAS (also known as BCAR1), paxillin, and GRB2-associated binding protein (GAB). The N-terminal SH3 domain (SH3N) binds to proteins that contain a proline-rich motif (P-x-x-P-x-K), such as Son of Sevenless (SOS), RAPGEF1 (also known as C3G), p85, and BCR-ABL1 (Nichols et al., 1994; Oda et al, 1994; Feller et al., 1995; Sattler et al., 1997). The protein complexes formed by CRKL and these binding partners are important for many biological processes such as cell proliferation, survival, adhesion and migration (Feller, 2001; Birge et al, 2009).
GAB2 belongs to an evolutionary conserved family of three proteins, GAB 1, GAB2, and GAB3. Three family members share 40%–50% sequence homology but each member also has unique structural motifs that allow specific signaling to downstream receptors (Gu and Neel, 2003). GAB2 protein contains highly conserved structural motifs that include an N-terminal Pleckstrin homology (PH) domain, central proline-rich domains, and multiple tyrosine residues (Fig. 1) (Gu et al, 1998). The PH domain plays a role in membrane localization of GAB2 through binding to cell membrane phospholipids. The central proline-rich domains serve as a docking site for SH3 domain-containing proteins such as GRB2. GRB2 is the primary upstream regulator of GAB2 and binds to GAB2 through its C-terminal “canonical” (P-x-x-P-x-R) or “atypical” (P-x-x-x-R-x-x-K-P) SH3-binding motifs (Lock et al, 2000).
GRB2 is the main upstream regulator of GAB2 and indirectly recruits GAB2 to a wide variety of activated membrane receptors. These receptors include receptor tyrosine kinases such as EGFR, KIT, cytokine receptors (interleukin receptors, erythropoietin, thrombopoietin), Fc receptors, T- and B-cell antigen receptors, and G-protein coupled receptors (Nishida and Hirano, 2003). Certain receptors, such as IL-2 and IL-3 receptors that do not have GRB2 binding sites, require an additional bridging protein such as SHC to activate the GRB2-GAB2 complex. Multiple tyrosine residues on GAB2 are phosphorylated by activated cell membrane receptors, then interact with SH2 domain-containing downstream proteins such as SHP2 and p85 that propagate downstream signaling pathways, such as the RAS-MAPK and PI3K-AKT signaling pathways. GAB2 also form complexes with other downstream effectors including phospholipase C-γ (PLC- γ), CRK, SHC, and SHIP (Gu et al, 1998). However, the functional significance of these complexes is not well characterized.
FRS2 (FGF receptor substrate 2) family of adaptor proteins has two members, FRS2α and FRS2β Two proteins are similar in structure, both contain an N-terminal myristoylation site, a C-terminal phosphotyrosine binding (PTB) domain, and tyrosine phosphorylation sites (Fig. 1) (Gotoh, 2008). The N-terminal myristoylation site involves a consensus sequence (M-G-x-x-x-S/T) for constitutive binding to the membrane lipids (Eswarakumar et al., 2005). The PTB domain is able to bind to phosphorylated tyrosine residues on cell surface receptors.
FRS2 serves as a docking protein for a limited species of receptor tyrosine kinases (Fig. 2), including FGF receptors, neurotrophin receptors (TrkA and TrkB), RET, and ALK receptors (Gotoh, 2008). The specificity of receptor binding relies on the PTB domain that binds to the juxtamembrane domains of different RTKs. In crystallographic studies, the highly conserved peptide sequence of the FRS2 PTB domain was shown to form two distinct conformations when binding to either FGFR receptor or TrkA receptor (Yan et al, 2002). FRS2a contains six tyrosine phosphorylation sites and FRS2β contains five tyrosine phosphorylation sites. Upon activation by the RTK, the phosphorylated tyrosine residues on FRS2 can bind to the SH2 domain of GRB2 (Gotoh et al., 2004; Kouhara et al, 1997). As described previously, GRB2 binds and recruit downstream effectors such as GAB1 to activate PI3K-AKT pathway, and SOS, a guanine nucleotide exchange factor, to activate RAS-MAPK pathway. These signaling pathways lead to expression of target genes that control cellular functions such as proliferation, survival, migration, and differentiation.
Identifying therapeutic targets through integrated genomics approach
The Cancer Genome Atlas (TCGA) project has provided an initial survey of genetic and epigenetic alterations that occur in various cancer types. However, only a subset of the large number of these alterations contributes to the cancer phenotype, mixed with other benign alterations as results of the genomic instability in cancer cells. To decipher which molecular events are important in driving cancer initiation and/or progression, understanding the function of these altered alleles in cancer is necessary.
In parallel to the genome characterization efforts, our laboratory initiated Project Achilles, a systematic effort to identify cancer dependencies (Luo et al., 2008; Cheung et al., 2011a). We have performed a genome-scale, pooled short hairpin RNA (shRNA) screen in over 200 cancer cell lines to look for genes that are essential to cancer cell survival. By integrating genetic and lineage background information in cell lines and primary tumors with functional data (Bell et al., 2011; Barretina et al., 2012), this complementary approach has identified previously unknown ovarian cancer lineage-specific dependencies such as PAX8 (Cheung et al., 2011a). PAX8 is focally amplified in 16% of HGSOCs and expressed at a higher level in ovarian cancer cell lines than non-ovarian cell lines. Through Project Achilles, PAX8 has found to be an essential gene for ovarian cancer cell survival. Suppression of PAX8 with individual shRNAs targeting PAX8 induced apoptosis selectively in ovarian cancer cells. The evidence strongly suggests PAX8 as a lineage-specific oncogene and a potential therapeutic target in HGSOC.
RNA interference represents one of the many approaches to functionally annotate the cancer genome (Boehm and Hahn, 2011). Gain-of-function approaches with overexpression systems can also be used to explore gene functions in cancer. An increasingly complete collection of human ORFs (open reading frames) has become available (Park et al., 2005; Lamesch et al., 2007; Yang et al., 2011) for genome-scale high-throughput screen in mammalian cells. The most recent collection includes 16,100 fully sequenced human ORFs, representing over 13,500 human genes (Yang et al., 2011). Gain of function screens using human ORF expression libraries has provided insights into oncogenic RAS signaling and resistance to MAPK pathway inhibition (Johannessen et al., 2013; Shao et al., 2014).
Recently discovered adaptor proteins as oncogenes
CRKL
Previous studies have identified a recurrent high-level and focal amplification peak on chromosome 22q11.21 in 3% of 371 primary lung adenocarcinomas (Weir et al., 2007; Kim et al., 2010). Broader copy number gain spanning that region was found in another 13% of tumors. The focal amplicon contains five genes including CRKL, PI4KA, ZNF74, THAP7, and LZTR1. Out of 84 non-small cell lung cancer (NSCLC) cell lines, three cell lines with CRKL amplification confirmed by fluorescence in situ hybridization (FISH) were selected to study the functional significance. Interestingly, a mutually exclusive relationship between amplifications of CRKL and EGFR was observed in both primary lung adenocarcinomas and the 84 NSCLC cell line collection at the Dana-Farber Cancer Institute (Cheung et al., 2011b).
We and others showed that NSCLC cells that harbor CRKL amplifications are dependent on CRKL expression for proliferation and survival and suppression of CRKL induces apoptosis (Cheung et al., 2011b; Kim et al., 2010). In addition, suppression of CRKL in tumors derived from CRKL-amplified NSCLC cell lines induced tumor regression in vivo. Overexpression of CRKL in immortalized but nontumorigenic human airway epithelial cells (AALE) induced anchorage-independent growth and cooperated with NF1 to induce tumor formation in vivo (Cheung et al., 2011b). The ability of CRKL overexpression to induce cell transformation in AALE cells is dependent on the integrity of the N-terminal SH3 domain (SH3N). These experimental observations demonstrate CRKL as a NSCLC oncogene and modulating CRKL activity by targeting the SH3N domain may carry therapeutic potential in a subset of NSCLC patients.
CRKL is known to regulate signaling through interactions of its SH3 domain with proline-rich motif containing proteins, such as SOS, C3G, and p85 (Tanaka et al., 1994; Gotoh et al., 1995; Sattler et al., 1997). In the case of CRKL-induced cell transformation, CRKL forms complexes with SOS1 and C3G and activates SOS1-RAS-RAF-ERK and SRC-C3G-RAP1 signaling pathways (Cheung et al., 2011b). The authors found that overexpression of CRKL in AALE cells consistently increased phosphor-T185/Y187 ERK1/2 levels and the expression of constitutively active RAP1 could partially rescue NSCLC cells from proliferation inhibition induced by CRKL suppression. These findings suggest RAS and RAP1 signaling plays an important role in proliferation of NSCLC cells with CRKL amplifications.
Besides its role in de novo oncogenesis, CRKL amplification may contribute to acquired resistance to targeted therapy in NSCLC. In a patient whose tumors exhibited initial response to EGFR inhibitor therapy but subsequently developed acquired resistance, CRKL amplification was found in the resistant tumor sample but not in the pretreatment sample (Cheung et al., 2011b). Further studies showed SOS1-dependent MAPK signaling contributes to CRKL-induced gefitinib resistance (Cheung et al., 2011b). These observations implicate CRKL copy number gain as a new genetic event after treatment and a mechanism of acquired resistance to EGFR inhibitors in NSCLCs.
CRK-family proteins have been studied using genetically engineered mouse models. Homozygous Crkl knockout in 129/Sv and C57BL/6 backgrounds results in embryonic lethality that resembles 22q11 deletion syndrome, also known as DiGeorge syndrome (Guris et al., 2001). In a transgenic mouse model for breast cancer, the mammary tumor virus (MMTV)-CRK transgene induced focal mammary tumor development with a 15-month latency. The finding suggests a role for CRK in breast cancer progression in vivo (Fathers et al., 2010). Others have shown that in mice harboring null mutation in the Crkl locus, CRKL is required for the transformation function of BCR-ABL fusion protein (p210) in chronic myeloid leukemia (Seo et al., 2010).
CRKL and several interacting proteins have been implicated in human cancer. When overexpressed in fibroblasts, CRKL is phosphorylated and transforms fibroblasts in a RAS-dependent fashion (Senechal et al., 1996). CRKL is also a well validated substrate of the BCR-ABL tyrosine kinase in patients with chronic myelogenous leukemia. Mutations in the CRKL-binding site in BCR-ABL suppresses transformation activity by 2–3 fold, while a double mutation in CRKL- and GRB2-binding sites reduced the transforming ability of BCR-ABL protein by 15-fold. In neuroblastomas, ALK activating mutations have shown to activate RAP1 through CRKL-C3G complexes (Schonherr et al., 2010).
Other members of the CRK family of adaptor proteins have been shown to be overexpressed in lung adenocarcinoma, human colon cancers, malignant glioblastoma (Nishihara et al., 2002; Miller et al., 2003; Takino et al., 2003). High level of CRK expression is associated with an aggressive phenotype, poor prognosis and shorter survival in lung adenocarcinoma patients (Miller et al., 2003). The CRK family proteins have been reported to induce transformation, migration and invasion (Cabodi et al., 2010). CRK mediates transformation through increasing p130CAS-associated activity of Src family kinases (Sakai et al., 1997). In glioblastoma cells, CRK overexpression increases cell migration and invasion through DOCK1-associated early attachment to laminin, cell motility, and growth (Takino et al., 2003). Together these studies suggest that CRKL contributes to the pathogenesis of several cancers, both as a direct oncogene and as a key effector of other oncogenic events.
GAB2
The oncogenic potential of GAB2 was found through an in vivo screen of recurrently amplified genes in ovarian cancer to identify genes that induce tumors (Dunn et al., 2014). Using an immortalized human embryonic kidney cell line, we screened a total of 587 open reading frames (ORFs) representing 455 genes amplified in ovarian cancers were introduced to nude mice in a pooled format. Out of the 455 genes screened, 25 unique ovarian cancer ORF sequences were recovered and GAB2 was the only ORF that was present in all three tumors formed from the starting pool. In addition, GAB2 was significantly enriched in the tumors compared to the starting pool, representing over 95% of ORF sequences in two out of the three tumors.
The 11q14 chromosomal region of focal copy number gain containing GAB2 was observed in 24.2% of 562 primary ovarian cancers characterized by TCGA project (Bell et al., 2011). The 11q14 amplicon is also the fourth most frequently amplified region in ovarian cancer. The peak of the amplified region contains GAB2, KCTD21 and USP35 and is telomeric to CCND1 and PAK1. GAB2 expression has been shown to be required for the survival of ovarian cancer cell lines that overexpress GAB2 (Dunn et al., 2014).
GAB2-associated transformation and dependency in ovarian cancer is mediated by the activation of the PI3K-AKT signaling pathway (Dunn et al., 2014). We observed that GAB2 overexpression in IOSE cells induced serine 473 phosphorylation of AKT1. In addition, ovarian cancer cell lines that have GAB2 overexpression or amplification are sensitive to PI3K pathway inhibition. This finding is consistent with the screen results using the HA1E-M model, which facilitates identification of oncogenes that activates the PI3K pathway. The mean IC50 of GDC-0941, a PI3K small molecular inhibitor, in cell lines that harbor GAB2 amplification/overexpression is comparable to cell lines with activating mutations of PIK3CA or loss of PTEN (Dunn et al., 2014). Although GAB2 also binds SHP2, which in turn leads to the activation of ERK signaling, GAB2-amplified or overexpressed ovarian cancer cell lines do not exhibit increased sensitivity to MEK inhibitors (Dunn et al., 2014). These observations implicate GAB2 as a potential target in the subset of ovarian cancers that harbor 11q14 amplifications.
Amplification of chromosomal region 11q13-14.1 which contains GAB2 has been observed in several human malignancies (Schwab, 1998). CCND1, located on 11q13.2, has long been considered as the driving oncogene on the amplicon. However, the large size of the amplified region and narrow peaks that are telomeric and distinct from CCND1 suggest that several genes may be selected in this amplicon (Adams et al., 2012). Thus amplifications of 11q13-14.1 may involve both CCND1 and GAB2.
Beyond these studies in high grade serous ovarian cancers, several studies suggest that GAB2 acts as a proto-oncogene in breast cancer, ovarian cancer, and melanoma (Bentires-Alj et al., 2006; Brown et al., 2008; Horst et al., 2009). GAB2 overexpression in an immortalized human mammary epithelial cell line (MCF10A) induced increased proliferation and altered dependency on EGF and other growth factors (Brummer et al., 2006). GAB2 cooperates with ERBB2 to transform primary mammary epithelial cells through activation of downstream GAB2-SHP2-ERK signaling (Bentires-Alj et al., 2006). In MMTV-ErbB2 transgenic model models, homozygous deletion of Gab2 had a modest effect on the initiation and progression of mammary tumors but significantly suppressed the development of lung metastases. The GAB2-deficient tumor cells showed decreased migration in vitro, which is mediated by the impaired activation of Mek/Erk signaling but not the PI3K-Akt pathway (Ke et al., 2007). The study suggests a possible role of GAB2 in breast cancer metastasis.
In ovarian cancer, GAB2 overexpression has also been observed in serous cystadenocarcinomas (Wang et al., 2012). In addition, silencing GAB2 in GAB2-overexpressed cell lines inhibits migration and invasion and causes upregulation of E-cadherin, and this effect is mediated by activating the GAB2-PI3K-ZEB1 signaling pathway and epithelial-to-mesenchymal (EMT) transition (Wang et al., 2012). These studies suggest GAB2 as a key regulator in the GAB2-PI3K-ZEB1 pathway in the initiation and progression of ovarian cancer and other cancer types, and serves as a potential therapeutic target with existing PI3K inhibitors.
FRS2
FRS2 was identified as one of the 50 genes that are recurrently amplified in high-grade serous ovarian cancer (HGSOC) and essential for ovarian cancer cell line survival (Luo et al., 2015). Analysis of 489 HGSOC primary tumors identified 31 focal amplifications that encode 1825 genes, including known ovarian oncogenes such as CCNE1 and MYC (Bell et al., 2011). In parallel, 582 genes were identified from a genome-scale, loss-of-function study (Project Achilles) as genes that are essential to ovarian cancer cell line proliferation and survival. Among 50 genes that are both amplified and essential in ovarian cancer, FRS2 exhibited unique features such as high frequency of amplification and scored highly among the 55,000 short hairpin RNAs tested in Project Achilles. The 12q15 chromosomal region containing FRS2 is focally amplified in 12.5% of 559 primary HGSOCs characterized by the Cancer Genome Atlas project. A structurally similar chromosomal region is amplified in other cancer types such as breast adenocarcinoma, lung adenocarcinoma, lung squamous carcinoma, and head and neck squamous cell carcinomas. Similar to CRKL and GAB2, we observed a mutually exclusive relationship between FRS2 amplification and FGFR1, FGFR2, FGFR3 amplifications in primary ovarian tumors.
We found that FRS2-amplified ovarian cancer cell lines are dependent on FRS2 expression and FRS2 suppression in 12q15 amplified cell lines induced apoptotic cell death. Furthermore, FRS2 overexpression in immortalized human embryonic kidney and ovarian epithelial cell lines conferred the ability to grow in an anchorage independent manner and as tumors in nude mice. FRS2, an adaptor protein predominantly in the FGFR pathway, facilitates downstream activation of RAS-MAPK pathway in immortalized ovarian epithelial cells. These observations identify FRS2 as an oncogene in a subset of HGSOC that harbor FRS2 amplifications.
Frs2 homozygous deletion results in early embryonic lethality in mice, likely due to the ubiquitous expression of Frs2 and the crucial role of FGF signaling in development (Gotoh et al., 2005). In FGF19-induced hepatocellular carcinoma transgenic model, an anti-FGF19 antibody (1A6) was shown to inhibit FGF19 binding to FGFR4 and FGF19-induced FRS2 and MAPK phosphorylation (Desnoyers et al., 2008).
Besides ovarian cancer, several recent studies have linked FRS2 and 12q15 amplification to high-grade liposarcomas. (Wang et al., 2011; Zhang et al., 2013). An amplified region containing FRS2 gene was found via high-resolution SNP/copy number variation microarray of 47 well- differentiated and dedifferentiated liposarcomas (Wang et al., 2011). Furthermore, a separate study demonstrated sensitivity of FRS2-amplified high-grade liposarcoma cell lines to FRS2 suppression through shRNAs (Zhang et al., 2013). In 40 glioma tumor samples, FRS2 is among 10 genes that are recurrently amplified and overexpressed (Fischer et al., 2008). These studies suggest that FRS2 has an emerging role in several cancer types through regulating key signaling pathways downstream of FGFRs.
Adaptor protein as a new class of oncogenes
Here we described the identification, structure and function, and therapeutic implication of three oncogenes that encode separate adaptor proteins in signal transduction pathways. Each gene was discovered through comprehensive approaches of cancer genome characterization, identification of cancer dependencies through loss-of-function genetics, and gain-of-function study of genes that induce transformation.
This new class of oncogenes represents a functionally distinct group from the well-known kinase oncogenes. Unlike kinases, the intrinsic activity of the adaptor protein comes from its ability to form complexes with upstream and downstream proteins and facilitate signal transduction. Therefore the oncogenic potential of the adaptor protein is limited to the stoichiometric ratio between its binding partners and itself. To date, these adaptor proteins are overexpressed, often by increased copy number, but not mutated. Other genetic alterations such as mutations or translocations involving CRKL, GAB2, and FRS2 exist based on the TCGA analysis of over 20 cancer types (unpublished data), but the functional significance of these sequence alterations has not been reported and remains obscure.
A common feature of three oncogenes described in this paper is the mutual exclusive relationship with their upstream or downstream signaling partners. A statistically significant mutually exclusive relationship has been observed between amplifications of CRKL and EGFR, amplification of GAB2 and either PIK3CA amplification or PTEN loss, and amplifications of FRS2 and FGFR1, FGFR2, or FGFR3 (Cheung et al., 2011a; Dunn et al., 2014; Luo et al., 2015). This pattern of mutual exclusivity is also observed among other oncogenes within the same signaling pathway, such as KRAS and EGFR mutations, or TP53 and MDM2 mutations. These observations implicate a level of functional redundancy between two genetic alterations and suggest that adaptor protein can independently drive oncogenesis without aberrant upstream or downstream kinase activity.
Therapeutic approaches to targeting adaptor proteins
Adaptor proteins lack tyrosine or serine/threonine kinase domains and present a unique challenge to rationalized drug design. Traditional approaches for targeting activated RTK pathway fall into two categories: small molecule inhibitors that target the ATP-binding site and monoclonal antibodies that bind to the extracellular domain of RTKs. However, neither of these approaches can be applied to adaptor proteins. A new approach to cancer therapy has been the development of small molecules targeting protein-protein interactions (Wells and McClendon, 2007). Targeting protein-protein interface has been difficult in the past due to several reasons. The interaction surface is often smooth and void of any clefts and pockets for drug binding. The surface often comprises non-contiguous amino acid residues in the polymer chain. Peptides derived from short contiguous chain are poor chemical starting sequences. Furthermore, protein-protein interaction surfaces are relatively large (1500-3000 square angstroms) compared to protein-small molecule interactions (300-1000 square angstroms) (Sato and Gotoh, 2009).
Despite the challenges, several studies have discovered small subsets of contact surface amino acid residues, named “hotspots”, are critical for the protein-protein interaction (Clackson and Wells, 1995; Muller et al., 1997; Thanos et al., 2006; Moreira et al., 2007). ABT-737 is an example of newly emerged protein-protein interaction inhibitors that targets members of the B-cell lymphoma 2 (BCL-2) family. ABT-737 and its derivative ABT-263 bind to the hydrophobic helical domains of BCL-XL, BCL-2 and BCL-W, which are important regulators of apoptotic cell death (Oltersdorf et al., 2005). ABT-263 (Navitoclax, Abbott Laboratories, USA) is currently undergoing Phase I/II trial for multiple lymphoid and solid malignancies.
Targeting adaptor proteins with protein-protein interaction inhibitor adds another layer of specificity toward targeted therapy. Conformations of the tyrosine kinase domain and serine/threonine kinase domain are highly conserved across species, especially in the substrate-binding region. Therefore kinase inhibitors are often non-specific and present with wide range of toxicities. Since certain adaptor proteins such as FRS2 demonstrate specificity toward tyrosine kinase receptors and adaptor proteins, inhibition of these adaptors may have limited toxicity and increased efficacy for patients compared to kinase inhibition.
Acknowledgments
This work was supported in part by grants from the U.S. NIH (U01 CA176058), the H.L. Snyder Medical Research Foundation and a HHMI Medical Student Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams SJ, Aydin IT, Celebi JT. GAB2--a scaffolding protein in cancer. Mol Cancer Res. 2012;10:1265–1270. doi: 10.1158/1541-7786.MCR-12-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert ML, Kim JI, Birge RB. alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol. 2000;2:899–905. doi: 10.1038/35046549. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, Godwin AK, Gross J, Hartmann L, Huang M, Huntsman DG, Iacocca M, Imielinski M, Kalloger S, Karlan BY, Levine DA, Mills GB, Morrison C, Mutch D, Olvera N, Orsulic S, Park K, Petrelli N, Rabeno B, Rader JS, Sikic BI, Smith-McCune K, Sood AK, Bowtell D, Penny R, Testa JR, Chang K, Dinh HH, Drummond JA, Fowler G, Gunaratne P, Hawes AC, Kovar CL, Lewis LR, Morgan MB, Newsham IF, Santibanez J, Reid JG, Trevino LR, Wu YQ, Wang M, Muzny DM, Wheeler DA, Gibbs RA, Getz G, Lawrence MS, Cibulskis K, Sivachenko AY, Sougnez C, Voet D, Wilkinson J, Bloom T, Ardlie K, Fennell T, Baldwin J, Gabriel S, Lander ES, Ding L, Fulton RS, Koboldt DC, McLellan MD, Wylie T, Walker J, O'Laughlin M, Dooling DJ, Fulton L, Abbott R, Dees ND, Zhang Q, Kandoth C, Wendl M, Schierding W, Shen D, Harris CC, Schmidt H, Kalicki J, Delehaunty KD, Fronick CC, Demeter R, Cook L, Wallis JW, Lin L, Magrini VJ, Hodges JS, Eldred JM, Smith SM, Pohl CS, Vandin F, Raphael BJ, Weinstock GM, Mardis R, Wilson RK, Meyerson M, Winckler W, Getz G, Verhaak RGW, Carter SL, Mermel CH, Saksena G, Nguyen H, Onofrio RC, Lawrence MS, Hubbard D, Gupta S, Crenshaw A, Ramos AH, Ardlie K, Chin L, Protopopov A, Zhang JH, Kim TM, Perna I, Xiao Y, Zhang H, Ren G, Sathiamoorthy N, Park RW, Lee E, Park PJ, Kucherlapati R, Absher DM, Waite L, Sherlock G, Brooks JD, Li JZ, Xu J, Myers RM, Laird PW, Cope L, Herman JG, Shen H, Weisenberger DJ, Noushmehr H, Pan F, Triche T, Berman BP, Van den Berg DJ, Buckley J, Baylin SB, Spellman PT, Purdom E, Neuvial P, Bengtsson H, Jakkula LR, Durinck S, Han J, Dorton S, Marr H, Choi YG, Wang V, Wang NJ, Ngai J, Conboy JG, Parvin B, Feiler HS, Speed TP, Gray JW, Levine DA, Socci ND, Liang Y, Taylor BS, Schultz N, Borsu L, Lash AE, Brennan C, Viale A, Sander C, Ladanyi M, Hoadley KA, Meng S, Du Y, Shi Y, Li L, Turman YJ, Zang D, Helms EB, Balu S, Zhou X, Wu J, Topal MD, Hayes DN, Perou CM, Getz G, Voet D, Saksena G, Zhang JNH, Zhang H, Wu CJ, Shukla S, Cibulskis K, Lawrence MS, Sivachenko A, Jing R, Park RW, Liu Y, Park PJ, Noble M, Chin L, Carter H, Kim D, Karchin R, Spellman PT, Purdom E, Neuvial P, Bengtsson H, Durinck S, Han J, Korkola JE, Heiser LM, Cho RJ, Hu Z, Parvin B, Speed TP, Gray JW, Schultz N, Cerami E, Taylor BS, Olshen A, Reva B, Antipin Y, Shen R, Mankoo P, Sheridan R, Ciriello G, Chang WK, Bernanke JA, Borsu L, Levine DA, Ladanyi M, Sander C, Haussler D, Benz CC, Stuart JM, Benz SC, Sanborn JZ, Vaske CJ, Zhu J, Szeto C, Scott GK, Yau C, Hoadley KA, Du Y, Balu S, Hayes DN, Perou CM, Wilkerson MD, Zhang N, Akbani R, Baggerly KA, Yung WK, Mills GB, Weinstein JN, Penny R, Shelton T, Grimm D, Hatfield M, Morris S, Yena P, Rhodes P, Sherman M, Paulauskis J, Millis S, Kahn A, Greene JM, Sfeir R, Jensen MA, Chen J, Whitmore J, Alonso S, Jordan J, Chu A, Zhang JH, Barker A, Compton C, Eley G, Ferguson M, Fielding P, Gerhard DS, Myles R, Schaefer C, Shaw KRM, Vaught J, Vockley JB, Good PJ, Guyer MS, Ozenberger B, Peterson J, Thomson E, Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentires-Alj M, Gil SG, Chan R, Wang ZC, Wang Y, Imanaka N, Harris LN, Richardson A, Neel BG, Gu H. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006;12:114–121. doi: 10.1038/nm1341. [DOI] [PubMed] [Google Scholar]
- Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13. doi: 10.1186/1478-811X-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Hahn WC. Towards systematic functional characterization of cancer genomes. Nat Rev Genet. 2011;12:487–498. doi: 10.1038/nrg3013. [DOI] [PubMed] [Google Scholar]
- Brown LA, Kalloger SE, Miller MA, Shih Ie M, McKinney SE, Santos JL, Swenerton K, Spellman PT, Gray J, Gilks CB, Huntsman DG. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47:481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- Brummer T, Schramek D, Hayes VM, Bennett HL, Caldon CE, Musgrove EA, Daly RJ. Increased proliferation and altered growth factor dependence of human mammary epithelial cells overexpressing the Gab2 docking protein. J Biol Chem. 2006;281:626–637. doi: 10.1074/jbc.M509567200. [DOI] [PubMed] [Google Scholar]
- Cabodi S, del Pilar Camacho-Leal M, Di Stefano P, Defilippi P. Integrin signalling adaptors: not only figurants in the cancer story. Nat Rev Cancer. 2010;10:858–870. doi: 10.1038/nrc2967. [DOI] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, Jiang G, Hsiao J, Mermel CH, Getz G, Barretina J, Gopal S, Tamayo P, Gould J, Tsherniak A, Stransky N, Luo B, Ren Y, Drapkin R, Bhatia SN, Mesirov JP, Garraway LA, Meyerson M, Lander ES, Root DE, Hahn WC. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011a;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HW, Du JY, Boehm JS, He F, Weir BA, Wang XX, Butaney M, Sequist LV, Luo BA, Engelman JA, Root DE, Meyerson M, Golub TR, Janne PA, Hahn WC. Amplification of CRKL Induces Transformation and Epidermal Growth Factor Receptor Inhibitor Resistance in Human Non-Small Cell Lung Cancers. Cancer Discov. 2011b;1:608–625. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, Carano R, D'Souza A, Qing J, Mohtashemi I, Ashkenazi A, French DM. Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene. 2008;27:85–97. doi: 10.1038/sj.onc.1210623. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Cheung HW, Agarwalla PK, Thomas S, Zektser Y, Karst AM, Boehm JS, Weir BA, Berlin AM, Zou L, Getz G, Liu JF, Hirsch M, Vazquez F, Root DE, Beroukhim R, Drapkin R, Hahn WC. In vivo multiplexed interrogation of amplified genes identifies GAB2 as an ovarian cancer oncogene. Proc Natl Acad Sci USA. 2014;111:1102–1107. doi: 10.1073/pnas.1311909111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Fathers KE, Rodrigues S, Zuo D, Murthy IV, Hallett M, Cardiff R, Park M. CrkII transgene induces atypical mammary gland development and tumorigenesis. Amer J Pathol. 2010;176:446–460. doi: 10.2353/ajpath.2010.090383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- Feller SM, Knudsen B, Hanafusa H. Cellular proteins binding to the first Src homology 3 (SH3) domain of the proto-oncogene product c-Crk indicate Crk-specific signaling pathways. Oncogene. 1995;10:1465–1473. [PubMed] [Google Scholar]
- Fischer U, Keller A, Leidinger P, Deutscher S, Heisel S, Urbschat S, Lenhof HP, Meese E. A different view on DNA amplifications indicates frequent, highly complex, and stable amplicons on 12q13-21 in glioma. Mol Cancer Res. 2008;6:576–584. doi: 10.1158/1541-7786.MCR-07-0283. [DOI] [PubMed] [Google Scholar]
- Gotoh N. Regulation of growth factor signaling by FRS2 family docking/scaffold adaptor proteins. Cancer Sci. 2008;99:1319–1325. doi: 10.1111/j.1349-7006.2008.00840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh N, Laks S, Nakashima M, Lax I, Schlessinger J. FRS2 family docking proteins with overlapping roles in activation of MAP kinase have distinct spatial-temporal patterns of expression of their transcripts. FEBS Lett. 2004;564:14–18. doi: 10.1016/S0014-5793(04)00287-X. [DOI] [PubMed] [Google Scholar]
- Gotoh N, Manova K, Tanaka S, Murohashi M, Hadari Y, Lee A, Hamada Y, Hiroe T, Ito M, Kurihara T, Nakazato H, Shibuya M, Lax I, Lacy E, Schlessinger J. The docking protein FRS2alpha is an essential component of multiple fibroblast growth factor responses during early mouse development. Mol Cell Biol. 2005;25:4105–4116. doi: 10.1128/MCB.25.10.4105-4116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T, Hattori S, Nakamura S, Kitayama H, Noda M, Takai Y, Kaibuchi K, Matsui H, Hatase O, Takahashi H, et al. Identification of Rap1 as a target for the Crk SH3 domain-binding guanine nucleotide-releasing factor C3G. Mol Cell Biol. 1995;15:6746–6753. doi: 10.1128/mcb.15.12.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Neel BG. The “Gab” in signal transduction. Trends in cell biology. 2003;13:122–130. doi: 10.1016/s0962-8924(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Gu H, Pratt JC, Burakoff SJ, Neel BG. Cloning of p97/Gab2, the major SHP2-binding protein in hematopoietic cells, reveals a novel pathway for cytokine-induced gene activation. Mol Cell. 1998;2:729–740. doi: 10.1016/s1097-2765(00)80288-9. [DOI] [PubMed] [Google Scholar]
- Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27:293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- Heikkinen LS, Kazlauskas A, Melen K, Wagner R, Ziegler T, Julkunen I, Saksela K. Avian and 1918 Spanish influenza a virus NS1 proteins bind to Crk/CrkL Src homology 3 domains to activate host cell signaling. The Journal of biological chemistry. 2008;283:5719–5727. doi: 10.1074/jbc.M707195200. [DOI] [PubMed] [Google Scholar]
- Horst B, Gruvberger-Saal SK, Hopkins BD, Bordone L, Yang Y, Chernoff KA, Uzoma I, Schwipper V, Liebau J, Nowak NJ, Brunner G, Owens D, Rimm DL, Parsons R, Celebi JT. Gab2-mediated signaling promotes melanoma metastasis. Amer J Pathol. 2009;174:1524–1533. doi: 10.2353/ajpath.2009.080543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE, Garraway LA. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Wu D, Princen F, Nguyen T, Pang Y, Lesperance J, Muller WJ, Oshima RG, Feng GS. Role of Gab2 in mammary tumorigenesis and metastasis. Oncogene. 2007;26:4951–4960. doi: 10.1038/sj.onc.1210315. [DOI] [PubMed] [Google Scholar]
- Kim YH, Kwei KA, Girard L, Salari K, Kao J, Pacyna-Gengelbach M, Wang P, Hernandez-Boussard T, Gazdar AF, Petersen I, Minna JD, Pollack JR. Genomic and functional analysis identifies CRKL as an oncogene amplified in lung cancer. Oncogene. 2010;29:1421–1430. doi: 10.1038/onc.2009.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, Rogers J, Lawlor S, McLaren S, Dricot A, Borick H, Cusick ME, Vandenhaute J, Dunham I, Hill DE, Vidal M. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock LS, Royal I, Naujokas MA, Park M. Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J Biol Chem. 2000;275:31536–31545. doi: 10.1074/jbc.M003597200. [DOI] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang XP, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou XC, Nguyen TY, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LY, Kim E, Cheung HW, Weir BA, Dunn GP, Shen RR, Hahn WC. The tyrosine kinase adaptor protein FRS2 is oncogenic and amplified in high-grade serous ovarian cancer. Mol Cancer Res. 2015;13:502–509. doi: 10.1158/1541-7786.MCR-14-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Chen G, Gharib TG, Wang H, Thomas DG, Misek DE, Giordano TJ, Yee J, Orringer MB, Hanash SM, Beer DG. Increased C-CRK proto-oncogene expression is associated with an aggressive phenotype in lung adenocarcinomas. Oncogene. 2003;22:7950–7957. doi: 10.1038/sj.onc.1206529. [DOI] [PubMed] [Google Scholar]
- Moreira IS, Fernandes PA, Ramos MJ. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- Muller YA, Li B, Christinger HW, Wells JA, Cunningham BC, de Vos AM. Vascular endothelial growth factor: crystal structure and functional mapping of the kinase domain receptor binding site. Proc Natl Acad Sci USA. 1997;94:7192–7197. doi: 10.1073/pnas.94.14.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols GL, Raines MA, Vera JC, Lacomis L, Tempst P, Golde DW. Identification of CRKL as the constitutively phosphorylated 39-kD tyrosine phosphoprotein in chronic myelogenous leukemia cells. Blood. 1994;84:2912–2918. [PubMed] [Google Scholar]
- Nishida K, Hirano T. The role of Gab family scaffolding adapter proteins in the signal transduction of cytokine and growth factor receptors. Cancer Sci. 2003;94:1029–1033. doi: 10.1111/j.1349-7006.2003.tb01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Tanaka S, Tsuda M, Oikawa S, Maeda M, Shimizu M, Shinomiya H, Tanigami A, Sawa H, Nagashima K. Molecular and immunohistochemical analysis of signaling adaptor protein Crk in human cancers. Cancer Lett. 2002;180:55–61. doi: 10.1016/s0304-3835(01)00763-7. [DOI] [PubMed] [Google Scholar]
- Oda T, Heaney C, Hagopian JR, Okuda K, Griffin JD, Druker BJ. Crkl is the major tyrosine-phosphorylated protein in neutrophils from patients with chronic myelogenous leukemia. J Biol Chem. 1994;269:22925–22928. [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O'Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- Park J, Hu Y, Murthy TV, Vannberg F, Shen B, Rolfs A, Hutti JE, Cantley LC, Labaer J, Harlow E, Brizuela L. Building a human kinase gene repository: bioinformatics, molecular cloning, and functional validation. Proc Natl Acad Sci USA. 2005;102:8114–8119. doi: 10.1073/pnas.0503141102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Sakai R, Nakamoto T, Ozawa K, Aizawa S, Hirai H. Characterization of the kinase activity essential for tyrosine phosphorylation of p130Cas in fibroblasts. Oncogene. 1997;14:1419–1426. doi: 10.1038/sj.onc.1200954. [DOI] [PubMed] [Google Scholar]
- Sato T, Gotoh N. The FRS2 family of docking/scaffolding adaptor proteins as therapeutic targets of cancer treatment. Expert Opin Ther Targets. 2009;13:689–700. doi: 10.1517/14728220902942330. [DOI] [PubMed] [Google Scholar]
- Sattler M, Salgia R, Shrikhande G, Verma S, Pisick E, Prasad KV, Griffin JD. Steel factor induces tyrosine phosphorylation of CRKL and binding of CRKL to a complex containing c-kit, phosphatidylinositol 3-kinase, and p120(CBL) J Biol Chem. 1997;272:10248–10253. doi: 10.1074/jbc.272.15.10248. [DOI] [PubMed] [Google Scholar]
- Schonherr C, Yang HL, Vigny M, Palmer RH, Hallberg B. Anaplastic lymphoma kinase activates the small GTPase Rap1 via the Rap1-specific GEF C3G in both neuroblastoma and PC12 cells. Oncogene. 2010;29:2817–2830. doi: 10.1038/onc.2010.27. [DOI] [PubMed] [Google Scholar]
- Schwab M. Amplification of oncogenes in human cancer cells. BioEssays. 1998;20:473–479. doi: 10.1002/(SICI)1521-1878(199806)20:6<473::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Senechal K, Halpern J, Sawyers CL. The CRKL adaptor protein transforms fibroblasts and functions in transformation by the BCR-ABL oncogene. J Biol Chem. 1996;271:23255–23261. doi: 10.1074/jbc.271.38.23255. [DOI] [PubMed] [Google Scholar]
- Seo JH, Wood LJ, Agarwal A, O'Hare T, Elsea CR, Griswold IJ, Deininger MW, Imamoto A, Druker BJ. A specific need for CRKL in p210BCR-ABL-induced transformation of mouse hematopoietic progenitors. Cancer Res. 2010;70:7325–7335. doi: 10.1158/0008-5472.CAN-10-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, Zwang Y, Roberts TM, Root DE, Jacks T, Hahn WC. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takino T, Nakada M, Miyamori H, Yamashita J, Yamada KM, Sato H. CrkI adapter protein modulates cell migration and invasion in glioblastoma. Cancer Res. 2003;63:2335–2337. [PubMed] [Google Scholar]
- Tanaka S, Morishita T, Hashimoto Y, Hattori S, Nakamura S, Shibuya M, Matuoka K, Takenawa T, Kurata T, Nagashima K, et al. C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc Natl Acad Sci USA. 1994;91:3443–3447. doi: 10.1073/pnas.91.8.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos CD, DeLano WL, Wells JA. Hot-spot mimicry of a cytokine receptor by a small molecule. Proc Natl Acad Sci USA. 2006;103:15422–15427. doi: 10.1073/pnas.0607058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XK, Asmann YW, Erickson-Johnson MR, Oliveira JL, Zhang HY, Moura RD, Lazar AJ, Lev D, Bill K, Lloyd RV, Yaszemski MJ, Maran A, Oliveira AM. High-resolution genomic mapping reveals consistent amplification of the fibroblast growth factor receptor substrate 2 gene in well-differentiated and dedifferentiated liposarcoma. Gene Chromosome Cancer. 2011;50:849–858. doi: 10.1002/gcc.20906. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sheng Q, Spillman MA, Behbakht K, Gu H. Gab2 regulates the migratory behaviors and E-cadherin expression via activation of the PI3K pathway in ovarian cancer cells. Oncogene. 2012;31:2512–2520. doi: 10.1038/onc.2011.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidow CL, Black DS, Bliska JB, Bouton AH. CAS/Crk signalling mediates uptake of Yersinia into human epithelial cells. Cell Microbiol. 2000;2:549–560. doi: 10.1046/j.1462-5822.2000.00079.x. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba II, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Yan KS, Kuti M, Yan S, Mujtaba S, Farooq A, Goldfarb MP, Zhou MM. FRS2 PTB domain conformation regulates interactions with divergent neurotrophic receptors. J Biol Chem. 2002;277:17088–17094. doi: 10.1074/jbc.M107963200. [DOI] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, Johannessen CM, Silver SJ, Nguyen C, Murray RR, Hieronymus H, Balcha D, Fan C, Lin C, Ghamsari L, Vidal M, Hahn WC, Hill DE, Root DE. A public genome-scale lentiviral expression library of human ORFs. Nat Meth. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chu K, Wu X, Gao H, Wang J, Yuan YC, Loera S, Ho K, Wang Y, Chow W, Un F, Chu P, Yen Y. Amplification of FRS2 and activation of FGFR/FRS2 signaling pathway in high-grade liposarcoma. Cancer Res. 2013;73:1298–1307. doi: 10.1158/0008-5472.CAN-12-2086. [DOI] [PubMed] [Google Scholar]