Abstract
The Parkinson’s Disease Biomarkers Program (PDBP) is a multi-site study designed to identify Parkinson’s Disease (PD) biomarkers that can be used to improve the understanding of PD pathophysiology and to develop tools that provide novel measures to evaluate PD clinical trials. The PDBP consortium comprises numerous individual projects of which two are specifically geared to the development of brain imaging markers for diagnosis, progression, and prognosis of PD or related disorders. All study data from PD patients, atypical parkinsonian patients, patients with essential tremor, and healthy controls collected from the sites are integrated in the PDBP database and will be publically available. All subjects are asked to submit blood samples, and undergo a battery of clinical evaluations that cover motor, cognitive, and other background information. In addition, a subset of subjects contributed cerebrospinal fluid samples. A restricted access, web-based Data Management Resource facilitates rapid sharing of data and biosamples across the entire PD research community. The PDBP consortium is a useful resource for research and collaboration aimed at the discovery of biomarkers and their use in understanding the pathophysiology of PD.
1. Introduction
The Parkinson’s Disease Biomarkers Program (PDBP) is an ongoing effort funded by the National Institute of Neurological Diseases and Stroke (NINDS) of the National Institutes of Health (NIH) that provides an infrastructure for research aimed at discovering diagnostic and progression biomarkers of PD. Such biomarkers are critical in the assessment and design of more effective interventional phase II and III trials, and would facilitate application of existing treatment options. For example, current disease-modifying drug trials typically focus on early PD, yet enrollment of patients with atypical forms of parkinsonism or essential tremor (ET) can confound the interpretation of trial results. Developing quantitative brain-based markers that differentiate PD from other forms of atypical parkinsonism and PD from ET would be a major advance in the field of movement disorders.
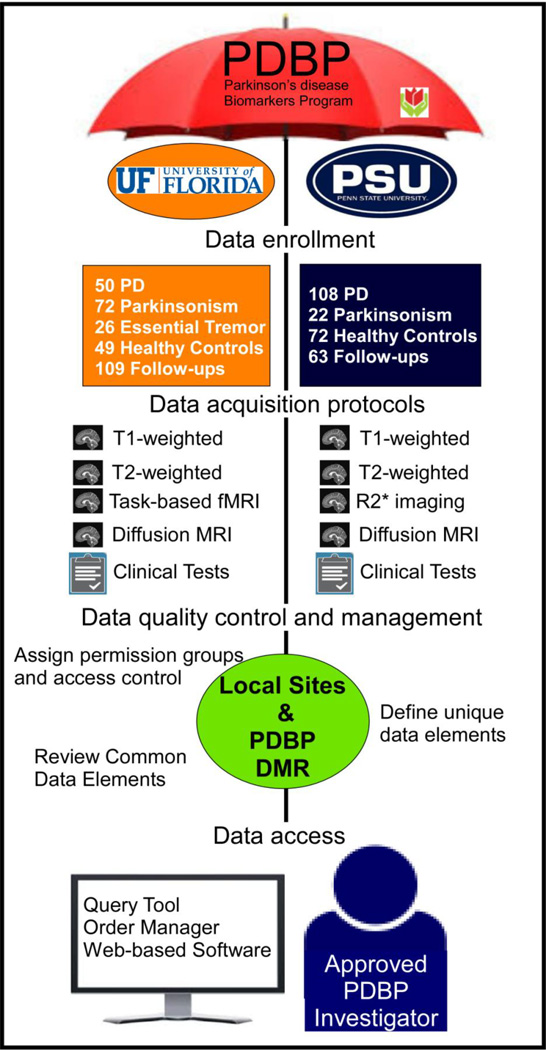
The PDBP is a new consortium model in which the NINDS works with numerous investigators to accelerate PD biomarker discovery and details of the goals and projects are summarized at the PDBP website (https://pdbp.ninds.nih.gov/index.jsp). In brief, the ten projects in the PDBP consortium contribute data and biospecimens every six months, based on a protocol established by the consortium members. Data and biospecimens then may be requested (via the Data Management Resource) by any interested researcher in academia or industry. All requests are evaluated by data access and/or biospecimen access committees that meet on a regular basis. PDBP projects are supported through a centralized PDBP Data Management Resource (DMR) and biosample repository. The DMR was developed, and is maintained, by the NIH Center for Information Technology, and serves stakeholders by providing electronic data capture, data quality assessment, and data access. Thus, the PDBP is building a resource of longitudinally-collected data and biosamples from well-characterized PD and control participants that, at its completion, will include more than 1,500 individuals.
The PDBP also receives, processes, stores, and distributes structural and functional magnetic resonance data that can be shared by the neuroscience research community. In addition to established researchers, the goal is to draw into this arena junior investigators and scientists with novel and innovative approaches. As such, the imaging data are unique to each site with different protocols, and allow for different modalities to be compared within a site, and then validated at the other site or in future studies. These PDBP data also will permit comparative and meta-analyses with other multi-site efforts such as the Parkinson’s Progression Markers Initiative of the Michael J. Fox Foundation. The PDBP neuroimaging repository is an essential part of the infrastructure for the PDBP. The PDBP includes single site cohorts, and scientists who will work closely with the PDBP DMR to assure standardization of brain data with biological sample collection, protection of subject privacy and rights, and transparency regarding availability of samples and brain imaging data for PD biomarker discovery research.
1.1. Data Contribution Requirements
Contributing de-identified clinical data to the PDBP DMR occurs through a web-based data entry system, using electronic clinical research forms for general and PD-specific NINDS common data elements (CDEs). All subjects sign consent for broad-sharing of their de-identified data. A Global Unique Identifier (GUID) is used as the subject ID permitting researchers to share data specific to a study participant without exposing personally identifiable information (PII). The GUIDs are random numbers that have no relationship to PII and PII is never shared with the DMR. Table 1 indicates the common behavioral and biospecimen data available from the PDBP DMR. Clinical and cognitive assessment forms include: Demographics, Behavioral History; Family History, Prior and Concomitant Medications; Vital Signs, Movement Disorders Society-Unified Parkinson’s Disease Rating Scale (MDS-UPDRS); Montreal Cognitive Assessment (MoCA); Rapid Eye Movement Behavior Disorder Questionnaire; Modified Schwab and England Scale, Parkinson’s Disease Quality of Life Questionnaire-39; Epworth Sleepiness Scale; University of Pittsburg Smell Identification Test (UPSIT); and Hamilton Depression and Anxiety Scales (HAM-D & HAM-A). Following quality assessment, all clinical data are deposited in a secure password-protected database.
Table 1.
PDBP DMR Elements
| Behavioral PDBP Data Elements | Biospecimen PDBP Data Available |
|---|---|
| Behavioral History | Blood (plasma, serum, whole blood, |
| Demographic Information | DNA, RNA) |
| MDS-UPDRS (Section III) | NeuroX genotyping |
| Neurological Exam (partial) | Structural Imaging |
| Prior and Concomitant Medications | Function Imaging |
| Vital Signs | CSF |
At each visit, subjects are asked to contribute whole blood (e.g., DNA, RNA for genomic), plasma, serum and CSF for biochemical and metabolomics studies. Sample collection is performed according to a strict protocol (https://pdbp.ninds.nih.gov/jsp/biorepository.jsp) and samples are shipped to the NINDS Human Genetics Repository for storage. All samples are available for analysis, and can be ordered through the PDBP DMR. Genomic sequencing involves NeuroX, a full exome array along with neurologic and neurodegenerative disease focused content1.
Imaging data are entered using the Medical Image Processing, Analysis, and Visualization (MIPAV) application that runs on all Java-enabled platforms (http://mipav.cit.nih.gov/). MIPAV can read and write numerous formats (e.g., DICOM, NIfTI-, etc), and is freely available. MIPAV is used for image entry and analysis by other data repositories (e.g., NDAR, FITBIR) as well as individual researchers. The MIPAV tool supports the receipt of unprocessed brain images in DICOM, MINC 1.0 and 2.0, Analyze, NIfTI-1, AFNI, and SPM (See Figure 1). Uploaded data are provided in either unprocessed DICOM or NifTI-1 formatted data.
Figure 1.
1.2. Data Access
PDBP was designed to provide open access to imaging, biospecimen, and clinical data. Investigators accessing PDBP data must complete the online registration process and Program Data Use Certification Agreement. This material is then reviewed first by the DAC and/or BRAC, and then by the PDBP DMR which in turn will provide material transfer agreement information after which biomaterials will be sent back to the researcher upon approval. There are policy documents for publications that use these clinical and imaging data including: a user code of conduct, and guidance for informed consent that enable de-identified data sharing and biospecimen sharing. In addition, a researcher may request to access modules such as the Query, Order Manager, or NINDS Bio Repository Group.
1.3. Query Tool
It is anticipated that most of the request for access to data will be approved expeditiously. Once approved, the website has a “query” tool (i.e., a data informatics program) within the PDBP DMR that allows researchers to search datasets collected across PDBP projects and other NINDS-funded PD clinical studies. The Query tool is based on NINDS PD common data elements, as well as unique elements created within the PDBP DMR. The Query tool allows searches to be done across studies, clinical forms and/or data elements. Detailed study synopses and the recently developed Data Dictionary provide information on the collection parameters and variables associated with PDBP data, so that researchers can better define data searches and corresponding results. For example, research using the query tool would be able to see the title of the study, status, date modified and created, and a variable name that corresponds to a data element. Use of the Query tool requires PDBP data access permission.
1.4. Order Manager
The order manager is used to create and submit orders for biosamples. Researchers will use the query tool in order to add items to a data basket. The data basket will have a table that contains all the data that was requested. Next, the researcher will find the sample information for those samples they would like to order, and then finalize their order and upload scanned copies of the NINDS Human Genetics Repository Material Transfer Agreement. Once approved, biosamples will be sent to the researcher.
1.5. Available Data
Currently, the PDBP supports ten projects for PD Biomarker Discovery. All projects are collecting clinical data and biosamples. Of these ten projects, two project sites are collecting brain magnetic resonance imaging (MRI) data (The Pennsylvania State University and the University of Florida). As will be described later, the imaging acquired at each site is not standardized and is not setup to be combined across sites. The PDBP goal is to develop novel and innovative markers of PD pathophysiology that can be evaluated and validated in currently available or in future multi-site studies. An example of this approach was in a recent paper that found that the posterior substantia nigra has elevated free-water in the University of Florida PDBP cohort, and this finding was then validated on the multi-site cohort from the Michael J. Fox Foundation Parkinson Progressive Marker Initiative2.
Both commercial and academic researchers can request access to data by completing a Data Use Request form and agreeing to comply with the PDBP Data Use Certification Agreement. The PDBP Data Access Committee (DAC), composed of NINDS staff and PDBP operation staff, reviews all requests for clinical data access. Requests for biosamples require submission of: 1) a lay abstract describing the research for which the biosamples will be used; 2) a detailed experimental design and power analysis; and 3) NINDS Human Genetics Repository Material Transfer Agreement for Biospecimens signed (by the investigator and the institutional official). Requests for biospecimens are reviewed by the PDBP Biospecimen Resource Committee (BRAC) that is composed of NINDS staff and subject matter experts. All data generated with PDBP biospecimens must be submitted to the PDBP DMR for broad data sharing. Details on data access policies, and forms for requesting data, can be found at: https://pdbp.ninds.nih.gov/jsp/researchers.jsp. The PDBP DMR provides a query tool that enables qualified researchers to conveniently search, merge, and analyze clinical, imaging, genetic, and biologic data. These data can also be downloaded to common separated values (CSV) files for further analysis. PDBP operations staff are available to assist users with questions, and respond to all queries within 24 hours.
2. Materials and Methods
2.1. Subject Populations
Imaging data are being acquired from healthy controls and patients. Healthy controls are individuals who did not have prior history of major psychiatric or neurological disease. Patients were diagnosed by movement disorders specialists at either the Milton S. Hershey Medical Center or the UF Health Center for Movement Disorders and Neurorestoration. The UK PD Society Brain Bank criteria3 were used to diagnosis PD, American Academy of Neurology and American Autonomic Society criteria4 were used for probable multiple systems atrophy (MSA), NINDS-PSP criteria5 were used for probable progressive supranuclear palsy (PSP), and the Statement of the Movement Disorders Society criteria6 were used for essential (ET). All subtypes within each disease diagnosis are being collected. Each patient’s diagnosis is updated within the DMR during follow up visits.
2.2. Penn State College of Medicine and Milton S. Hershey Medical Center (PSHMC)
The PSHMC site is implementing multimodal MRI techniques in combination with fluid-based iron protein profiles to evaluate in vivo markers for diagnosing PD and predicting progression. The research from this team is focused on using diffusion imaging, susceptible-weighted imaging (R2*) and other MRI measures as biomarker(s) for PD-related pathology in nigrostriatal pathways7–9. The imaging modalities from this project will determine whether these specific images are potential in vivo markers.
The cohort from PSHMC includes healthy controls, Parkinson’s disease, and atypical parkinsonian populations. Imaging data available from this site include T1- and T2-weighted data, R2* data, and diffusion imaging data that are collected at three time points (i.e., baseline, 18, and 36 months). Clinical evaluations, lumbar puncture, and blood samples are collected every 18 months, which includes the neuroimaging time points to 36 months.
2.2.1. PSHMC Enrollment Data
As of December 1, 2014, the PSHMC site has baseline data on 108 individuals with PD, 22 with PSP or MSA, and 72 healthy controls. The site has longitudinal data on 36 individuals with PD, two with parkinsonism, and 25 healthy controls. All imaging is performed while patients take their normally prescribed medications.
2.2.2. PSHMC Imaging Protocols
For PSHMC individuals, all imaging data are obtained from a 3.0-Tesla MRI system (Trio; Siemens Magnetom; Erlangen, Germany) with an 8-channel phased-array head coil. The diffusion MRI acquisition echo planar imaging (EPI) sequence consists of the following parameters: diffusion gradient directions = 42 and 7, repetition time = 8,300 ms, echo time = 82 ms, b-values: 0, 1000 s/mm2, field of view = 256 × 256 mm, in-plane resolution = 2 mm isotropic, flip angle = 90°, number of contiguous slices = 65, slice thickness = 2 mm, and acceleration factor p = 2 (see Table 1).
Susceptibility weighted images are acquired using a multi-gradient echo (1.0 mm isotropic voxels) with 6 echoes with an interval of 8 ms ending at 47 ms, repetition time = 54 ms, 80 × 80 matrix, 20 degree flip angle, 64 contiguous slices with a 2 mm slice thickness. The structural data are acquired with magnetization prepared-rapid acquisition gradient echo (MP-RAGE sequence (1.0 mm isotropic voxels) with 176 contiguous slices, field of view = 256 × 256, slice thickness = 1; repetition time = 1,540 ms; echo time = 2.34 ms; inversion time = 807 ms; flip angle = 9° for T1-weighted images; and repetition time = 2,500; echo time = 315; flip angle = 120° fast spin echo sequence for T2-weighted images.
2.3. University of Florida
The University of Florida (UF) site is focused on developing non-invasive techniques to understand differences in the pathophysiology and structural degeneration of the brain at baseline and following one year of progression across different movement disorders. The focus is on motor task-based functional magnetic resonance imaging using an established, and reliable motor task10–12, and diffusion magnetic resonance imaging using bi-tensor analytic methods2, 13. The overall goal is to develop candidate markers of progression in PD that are unique to those in other movement disorders14, 15.
The UF research team is enrolling individuals with PD, MSA, PSP, ET, and healthy controls, and is acquiring a complete clinical assessment of participants combined with the PDBP blood draw specifications. Imaging data available from this site include T1- and T2-weighted data, diffusion MRI, and motor task-based functional MRI. Imaging and clinical data are collected at baseline and one year.
2.3.1. UF Enrollment Data
As of December 1, 2014, the UF site had baseline data on 50 individuals with PD, 72 with atypical parkinsonism (i.e., MSA and PSP), 26 with ET and 49 healthy controls. The UF site has longitudinal data on 36 individuals with PD, 45 with parkinsonism, and 28 healthy controls. The clinical and behavioral data are collected at enrollment and follow-up. All imaging is performed following overnight withdrawal of anti-Parkinsonian medications.
2.3.2. UF Imaging Protocol
For the UF individuals, diffusion-weighted images are acquired on a 3T Philips Medical Systems MRI scanner (Achieva, Best, The Netherlands) using a 32-channel head coil. The whole-brain diffusion MRI acquisition sequence consists of the following parameters: diffusion gradient directions = 64; repetition time = 7,748 ms; echo time = 86 ms; b-values: 0; 1000 s/mm2; field of view = 224 × 224 mm; in-plane resolution = 2 mm isotropic; flip angle = 90°; number of contiguous slices = 60; slice thickness = 2 mm; and acceleration factor p = 2 (see Table 2).
Table 2.
Image Scanning Parameters for PSHMC
| T1-weighted | T2-weighted | Susceptibility weighted (R2*) |
Diffusion weighted | |
|---|---|---|---|---|
| Sequence | MP-RAGE | FSE | Multi-gradient-echo | EPI |
| TR (ms) | 1540 | 2500 | 54 | 8300 |
| TE (ms) | 2.34 | 316 | 6 echoes:7–47ms Interval: 8 | 82 |
| Flip Angle | 9 | 120 | 20 | 90 |
| Inversion time (ms) | 807 | |||
| b value (s/mm2) | - | - | - | 1000 |
| Diffusion directions | - | - | - | 42 & 7 |
| Resolution (mm3) | 1×1×1 | 1×1×1 | 1×1×1 | 2×2×2 |
| Thickness (mm) | 1 | 1 | 2 | 2 |
| Field of View | 256 × 256 | 256 × 256 | 256 × 256 | 256 × 256 |
| Number of Slices | 176 | 176 | 64 | 65 |
| Imaging direction | Sagittal | Sagittal | Axial | Axial |
MP-RAGE = magnetization prepared-rapid acquisition gradient echo; TE = echo time; TR = repetition time; EPI = echo planar image
Functional data are acquired using a single shot gradient echo EPI (3.0 mm isotropic voxels, TE = 30ms, TR = 2500ms, field of view = 240 × 240 matrix, 80 degree flip angle, 46 contiguous axial slices, zero gap. The functional MRI task is consistent with prior published work10, 16.
The structural data are acquired with a 3D Turbo field echo (TFE) sequence: repetition time = 8.2 ms; echo time = 3.7 ms; inversion time = 1012 ms; flip angle = 8°; field of view = 240 mm2; acquisition matrix = 240 × 240; voxel size = 1 mm isotropic with no gap between slices (n = 170) for T1-weighted images. A single TFE sequence was implemented for T2-weighted images with 6 echoes with an interval of 8 ms from 7 to 47 ms and the following scan parameters: flip angle = 22, 1 mm isotropic resolution, 224 × 224 acquisition matrix with 170 between slices.
2.4. Imaging Quality Control and Management
The data undergo quality control inspection at the local site and also by NIH quality control. The imaging quality control procedure first involves a check of parameters by a staff member of the MR center, and then the use of a custom program to monitor head motion and rotations in all six directions. Data are also inspected for any abnormalities in the brain (e.g., calcifications, stroke, and vascular malformation) by MR technicians and doctoral-level investigators. Any abnormalities that are detected are consulted and confirmed by a neuroradiologist and conveyed to the participants. The data are then backed-up locally to a secure server, and uploaded to the DMR.
3. Future directions for the PDBP Resource
The NINDS Parkinson Disease Biomarker Program is a novel model that combines hypothesis-driven research, creation of a research resource, a unique consortium model for collaboration, and a flexible data management system. With regard to brain imaging, it has had a successful beginning with data from both imaging sites now available for download. Published reports using PDBP data have just been disseminated to the scientific community. The PDBP, along with other multi-site efforts, will be well-positioned to test viable, non-invasive markers for PD progression using functional, structural, and diffusion MRI data.
Future directions for the PDBP resource will include continued enhancements to the DMR, Query tool, and Order manager to improve their capabilities and user-friendliness. Continued data collection by the ten consortium projects, and increased data analysis efforts, are underway. These include integrating genetic sequence data with brain imaging and clinical classification. The PDBP also plans to expand its available data and biosamples by incorporating data from additional NINDS-supported PD research projects including NINDS Udall Centers and the National Brain and Tissue Resource for PD and related disorders (NBTR-PD). As these examples demonstrate, the PDBP consortium has become an important resource for research and collaboration efforts for the discovery of biomarkers and obtaining further knowledge of the pathophysiology of PD.
Table 3.
Image Scanning Parameters for UF
| T1-weighted | T2-weighted | Functional | Diffusion weighted | |
|---|---|---|---|---|
| Sequence | Multi-shot TFE | Single-shot TFE | Single-echo EPI | EPI |
| TR (ms) | 8.2 | 54 | 2500 | 7748 |
| TE (ms) | 3.7 | 6 echoes: 7–47ms Interval: 8 | 30 | 86 |
| Flip Angle | 8 | 22 | 80 | 90 |
| Inversion time | 1017 | |||
| b value (s/mm2) | - | - | - | 1000 |
| Diffusion directions | - | - | - | 64 |
| Resolution (mm3) | 1×1×1 | 1×1×1 | 3 × 3 × 3 | 2×2×2 |
| Thickness (mm) | 1 | 1 | 3 | 2 |
| Field of View | 240 × 240 | 224 × 224 | 240 × 240 | 224 × 224 |
| Number of Slices | 170 | 170 | 46 | 60 |
| Imaging direction | Axial | Axial | Axial | Axial |
TE = echo time; TR = repetition time; EPI = echo planar image; TFE = turbo field echo
Highlights.
The PDBP supports research and resource building for PD biomarker discovery
The PDBP contains biospecimen and clinical data on all patients enrolled at multiple sites
The PDBP has designated sites responsible for the collection of structural and functional imaging data
The PDBP consortium is a useful resource for research and collaboration in understanding movement disorders
Acknowledgements
This work was supported by the National Institutes of Health (U01 NS082151, R01 NS075012) and the Parkinson’s Disease Biomarker Program and the National Institute of Neurological Disorders and Stroke.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nalls MA, Bras J, Hernandez DG, et al. NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiology of Aging. 2015;36:1605.e1607–1605.e1612. doi: 10.1016/j.neurobiolaging.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ofori E, Pasternak O, Planetta PJ, et al. Increased free water in the substantia nigra of Parkinson's disease: a single-site and multi-site study. Neurobiology of Aging. 2015;36:1097–1104. doi: 10.1016/j.neurobiolaging.2014.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes AJ, Ben-Shlomo Y, Daniel SE, Lees AJ. What features improve the accuracy of clinical diagnosis in Parkinson's disease: a clinicopathologic study. 1992. Neurology. 2001;57:S34–S38. [PubMed] [Google Scholar]
- 4.Gilman S, Low PA, Quinn N, et al. Consensus statement on the diagnosis of multiple system atrophy. J Neurol Sci. 1999;163:94–98. doi: 10.1016/s0022-510x(98)00304-9. [DOI] [PubMed] [Google Scholar]
- 5.Litvan I, Agid Y, Calne D, et al. Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome): report of the NINDS-SPSP international workshop. Neurology. 1996;47:1–9. doi: 10.1212/wnl.47.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Deuschl G, Bain P, Brin M. Consensus statement of the Movement Disorder Society on Tremor. Ad Hoc Scientific Committee. Mov Disord. 1998;13(Suppl 3):2–23. doi: 10.1002/mds.870131303. [DOI] [PubMed] [Google Scholar]
- 7.Du G, Lewis MM, Styner M, et al. Combined R2* and Diffusion Tensor Imaging Changes in the Substantia Nigra in Parkinson's Disease. Movement Disorders. 2011;26:1627–1632. doi: 10.1002/mds.23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du G, Lewis MM, Sen S, et al. Imaging nigral pathology and clinical progression in Parkinson's disease. Movement Disorders. 2012;27:1636–1643. doi: 10.1002/mds.25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Péran P, Cherubini A, Assogna F, et al. Magnetic resonance imaging markers of Parkinson’s disease nigrostriatal signature. Brain. 2010;133:3423–3433. doi: 10.1093/brain/awq212. [DOI] [PubMed] [Google Scholar]
- 10.Spraker MB, Prodoehl J, Corcos DM, Comella CL, Vaillancourt DE. Basal ganglia hypoactivity during grip force in drug naive Parkinson's disease. Hum Brain Mapp. 2010;31:1928–1941. doi: 10.1002/hbm.20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neely KA, Kurani AS, Shukla P, et al. Functional Brain Activity Relates to 0–3 and 3–8 Hz Force Oscillations in Essential Tremor. Cereb Cortex. 2014:24. doi: 10.1093/cercor/bhu142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Planetta PJ, Kurani AS, Shukla P, et al. Distinct functional and macrostructural brain changes in Parkinson's disease and multiple system atrophy. Hum Brain Mapp. 2014 doi: 10.1002/hbm.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasternak O, Sochen N, Gur Y, Intrator N, Assaf Y. Free water elimination and mapping from diffusion MRI. Magnetic Resonance in Medicine. 2009;62:717–730. doi: 10.1002/mrm.22055. [DOI] [PubMed] [Google Scholar]
- 14.Prodoehl J, Li H, Planetta PJ, et al. Diffusion tensor imaging of Parkinson's disease, atypical parkinsonism, and essential tremor. Mov Disord. 2013;28:1816–1822. doi: 10.1002/mds.25491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaillancourt DE, Spraker MB, Prodoehl J, et al. High-resolution diffusion tensor imaging in the substantia nigra of de novo Parkinson disease. Neurology. 2009;72:1378–1384. doi: 10.1212/01.wnl.0000340982.01727.6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prodoehl J, Spraker M, Corcos D, Comella C, Vaillancourt D. Blood oxygenation level-dependent activation in basal ganglia nuclei relates to specific symptoms in de novo Parkinson's disease. Mov Disord. 2010;25:2035–2043. doi: 10.1002/mds.23360. [DOI] [PMC free article] [PubMed] [Google Scholar]