Understanding the genetic architecture of gene expression is an intermediate step in understanding the genetic architecture of complex diseases. RNA-seq technologies have improved the quantification of gene expression and allow measurement of allelic specific expression (ASE). ASE is hypothesized to result from the direct effect of cis regulatory variants, but a proper estimation of the causes of ASE has not been performed to date. In this study we take advantage of a sample of twins to measure the relative contribution of genetic and environmental effects on ASE and we found substantial effects of gene × gene (G×G) and gene × environment (G×E) interactions. We propose a model where ASE requires genetic variability in cis, a difference in the sequence of both alleles, but where the magnitude of the ASE effect depends on trans genetic and environmental factors that interact with the cis genetic variants.
Gene expression is a cellular phenotype that informs about the functional state of the cell. It is used as an intermediate phenotype between genetic variants and complex traits to help in the identification of causal genes affecting the variation of complex traits. Gene expression by itself is a complex trait that depends on genetic and environmental causes. Many researchers have studied the genetics of gene expression and thousands of expression quantitative loci (eQTLs) have been identified within different populations and tissues1-3. Recently, epistatic interactions affecting gene expression have been described4, adding more complexity to the genetic architecture of gene expression. The use of RNAseq technologies to measure gene expression allows the estimation of allelic specific expression (ASE). ASE measures the difference in expression of two haplotypes of an individual at a specific genetic locus5-7 (Supplementary Fig. 1-A). While eQTLs are population based measures of the effect of genetics on gene expression, ASE is a more direct measure of how gene expression changes at the individual level. In addition, ASE is much less sensitive to technical parameters since such effects would affect both alleles equally. While ASE may occur in a stochastic way within each single cell, measurements from a population of cells for each individual represent the average behavior of the two alleles and, theoretically, are expected to result from the direct effect of genetic regulatory variants in cis. ASE is therefore expected to be much less influenced by environmental and experimental variability, accounting for approximately 70% of the variance1, which allows us to dissect in more detail the remaining 30% of genetic variability. In this study we dissect the underlying causes of ASE, by measuring the relative contribution of genetic and environmental factors and propose biological models of ASE action. To achieve these goals we sequenced the mRNA fraction of the transcriptome of ~400 female twin pairs (~800 individuals) from the TwinUK cohort in four tissues: fat, skin, blood and lymphoblastoid cell lines (LCL) using 49bp paired-end sequencing in an Illumina HiSeq2000. We sequenced 766 fat samples, 814 LCL samples, 716 skin samples and 384 blood samples and obtained 28M exonic reads per sample on average. Genotype information was imputed into the 1000 Genomes Phase 1 reference panel. By constructing a quantitative measure of ASE and exploiting the twin structure, we can dissect the proportions of its variation which are due to distinct genetic and non-genetic causes.
Since our expectations are that cis eQTLs play an important role in ASE, we looked for cis eQTLs in the four tissues. We used a linear regression approach with SNPs in a 1Mb window each side of the TSS for each gene (see Methods). We identified 9166 significant ciseQTLs in fat, 9551 in LCLs, 8731 in skin and 5313 in blood (1% FDR).
We used the RNAseq data to estimate ASE for every individual at every transcribed heterozygous SNP in the four tissues separately. First we ran a test to localize statistically significant ASE sites; then we defined a quantitative phenotype that measures the amount of ASE at a site and looked for the variance components of that phenotype.
To assess if a heterozygous site shows statistically significant ASE we used a binomial test on the proportion of reference alleles versus total counts (see methods). Since ASE estimates are sensitive to read coverage and mapping bias, we restricted our analysis to sites with at least 30 reads that passed a rigorous filtering process to control for mapping bias and other confounders7 (see methods). We tested an average of 1582 sites per individual, 8% of which were statistically significant at FDR 10% (Supplementary Table 1). We identified 8013 ASE sites in fat, 10751 in LCL, 9538 in skin and 6827 in blood. About 80% of the ASE sites are in genes for which we also identified a cis eQTL (Supplementary Table 1). We assume that the genes with ASE without observed cis eQTL also have genetic variants in cis causing the allelic imbalance, but we did not have the power to find them due to small effect sizes or the variants having low frequency in the population or being involved in epistatic or gene-environment interactions. We cannot exclude the possibility that, in some cases, homeostatic / feedback mechanisms act to constrain total expression so that an imbalance in allelic expression does not change the total output.
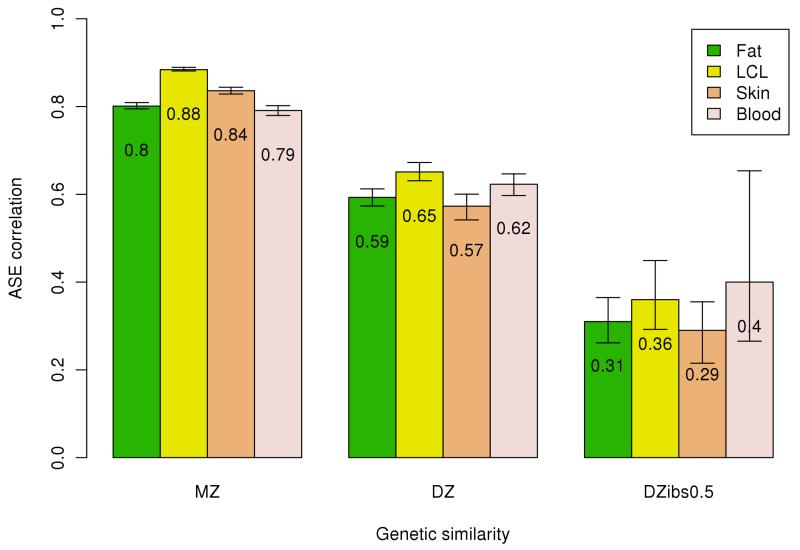
To quantify genetic and environmental sources of variation in ASE we developed an extension of the classical variance components approach based on the correlations within MZ and DZ twin pairs. We defined a quantitative phenotype of ASE as the logit of the proportion of reference alleles. This measure is not dependent on the overall gene expression level and is not susceptible to give false interactions due to trans or environmental effects that increase the overall level of expression. We jointly analyzed all the sites in genes with at least one eQTL, where both siblings have at least 30 reads overlapping the site and ASE is statistically significant for at least one of the siblings. We estimated the correlation of the ASE phenotype within MZ and DZ twins and observed that the correlation among DZs was higher than half of the correlation among MZs (Figure 1). That could indicate a potential shared environment component, but in our case, it is more likely to be due to the fact that the cis eQTL has a large effect on ASE and our DZ twins are genetically more similar than random mating predicts at the ASE locus (mean Identical By State (IBS) coefficient at the eQTL for DZ twins is 0.9). Indeed, when we looked at correlation between DZ twins that are IBS=0.5 (and hence share half of the contribution of the additive eQTL of MZ twins) we observed that this correlation is less than half of the correlation between MZ twins (Figure 1), indicating the potential presence of non additive genetic effects.
Figure 1.
ASE correlation among twin pairs for different categories of genetic similarity. MZ: monozygotic twins; DZ: dizygotic twins; DZibs05: DZ twins with identical by state (IBS) equal to 0.5 at the eQTL locus. The 95% confidence intervals were calculated by using 1000 bootstrap permutations.
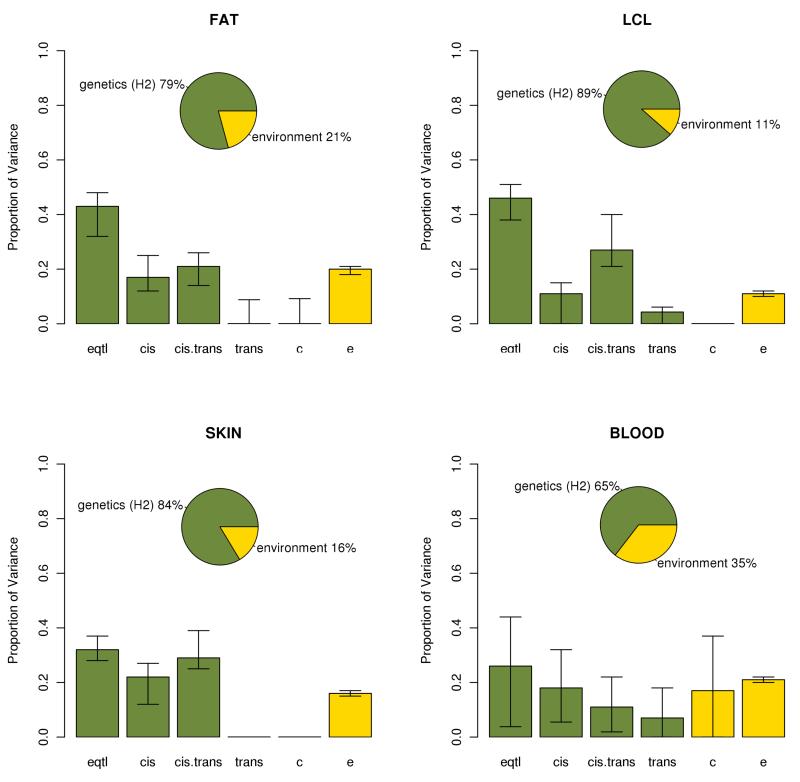
To incorporate these complexities in the model, we separated the twin pairs depending on the average genetic similarity genome wide (1 for MZ, 0.5 for DZ), genetic similarity at the locus based on the Identity By Descent (IBD) status in the cis region surrounding the gene, and the genetic similarity of the eQTL based on the IBS status at the eQTL locus. We estimated the correlations of the ASE phenotype for each category of twins and modeled these correlations as a function of six variance components (see methods).These components represent the proportions of variance in ASE that could be explained by environmental variation, by the top eQTL, by other variants in cis, by variants in trans, and by genetic interactions. As recombination is unlikely to have occurred within the cis window, cis epistatic interactions are generally not broken up within twin pairs and thus their contribution to variance is effectively additive. Instead we looked at calculating the proportion of variance explained by cis-trans interactions. We found that the heritability (the sum of all the genetic components, eQTL + cis + trans + interactions) of ASE ranges from 62% to 88% (Figure 2). The effect of the best cis eQTL per gene accounts for 26% to 46% of the variance on ASE. That means that nearly half of the heritability of ASE is due to a common ciseQTL. The remaining is due to other genetic effects in cis (11% to 22% of the ASE variance) and genetic interaction effects (11% to 29% of the ASE variance). As expected by the biological assumptions, we did not observe significant additive trans effects. We found a significant effect of the shared environment only in blood (11% of the ASE variance). That could be due to the fact that blood is more heterogeneous than the other tissues, with variable proportions of different cell types in individuals and shared environment affecting the counts of different cell types. In the shared environment component we are likely picking up cell-type specific effects. We used 1000 bootstrap permutations to calculate confidence intervals of our variance components estimates (Figure 2). This approach is robust to different coverage thresholds and the presence of several ASE sites in the same gene (Supplementary Figs. 2 and 3). In summary, the main causes of ASE are genetic variants in cis, as expected, but between 38% and 49% of the variance in the ASE ratio is due to genetic interactions and environmental factors.
Figure 2.
Variance components for ASE for the model with cis × trans interaction: ‘H2’ (equal to eqtl + cis + cis.trans + trans) is the heritability of ASE, ‘eqtl’ is the proportion of variance explained by a common eQTL, ‘cis’ is the proportion of variance explained by other variants in cis, ‘cis.trans’ is the proportion of variance explained by interactions between cis and trans genetic variants, ‘trans’ in the proportion of variance explained by genetic variants in trans, ‘c’ is the proportion of variance explained by the shared environment and ‘e’ is the proportion of variance explained by the individual environment. The 95% confidence intervals where calculated using 1000 bootstrap permutations.
Our variance components model shows that genetic effects do not explain all the observed variance in ASE and that environmental factors can have an effect on ASE. Given the nature of the ASE which, contrary to total gene expression, is internally controlled, these environmental effects should be mainly mediated by true biological mechanisms mediated by epigenetic mechanisms and much less likely by technical and experimental effects. However, environmental/epigenetic effects alone cannot create allelic imbalance as ASE is averaged over a large population of cells so stochastic effect are equally distributed between the two alleles; to observe ASE a cisDNA sequence effect is required. We therefore postulated the existence of gene × environment (G×E) interactions affecting ASE.
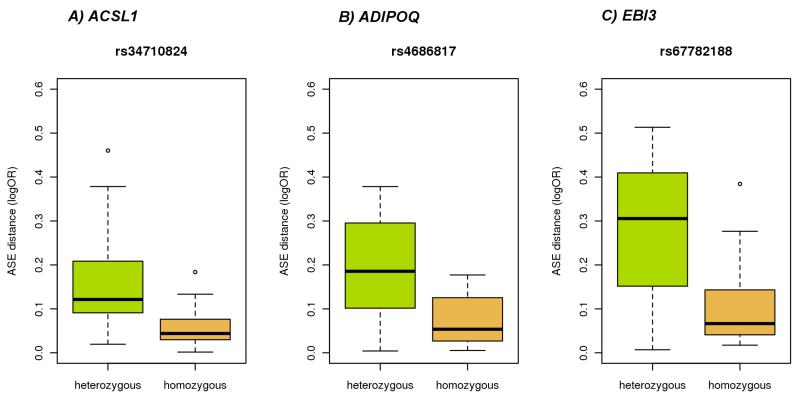
To identify cases of G×E we used an analysis inspired by the classical discordant MZ twins analysis8-10. We defined the phenotype as the absolute difference in measured ASE between MZ twins, and looked for SNPs around the ASE site that were associated with this phenotype. Significant associations suggest the influence of environmental factors affecting ASE in a genotype dependent manner. After multiple testing correction, we found evidence of G×E in fat and LCLs but not conclusive results in skin and blood (Supplementary Table 2, Supplementary Table 3, Supplementary Table 4 and Supplementary Table 5). One of the top hits in LCLs is the Epstein-Barr virus induced 3 gene (EBI3) (Figure 3 and Supplementary Fig. 4). That means that ASE at the EBI3 gene depends on the interaction of cis genetic variants and an environmental factor likely, in this case, to be related to the transformation process of the B cells with EBV. The two top hits in fat are ADIPOQ and ACSL1, two genes that code for Adiponectin and Long-chain-fatty-acid—CoA ligase 1 proteins respectively (Figure 3 and Supplementary Fig. 4). These two proteins are functionally related: both participate in the gene ontology biological processes ‘response to fatty acid’ and ‘response to nutrient’, and both are known to be regulated by environmental factors such as diet and exercise in a genotype dependent manner11-14. Attempts to link it to environmentally affected phenotypes (BMI, glucose levels, insulin levels) did not show any significant associations, which is not surprising since these are phenotypes affected by the environment and not direct environmental measures. The analysis above suggests that environment can modulate the effect of SNPs on gene expression.
Figure 3.
G×E examples discovered using discordant MZ twins analysis. MZ twin pairs show a different ASE effect on some genes depending on the genotype of specific SNPs. The Y axis shows the ASE difference between MZ twins. Since MZ twins are genetically identical, this association reflects the interaction of the SNP with an unknown environment. Boxplots represent the difference in ASE in MZ twin pairs that are heterozygous (in green) and homozygous (in orange) at the SNP of interest. A) ASE in gene ACSL1 shows G×E interaction with SNP r334710824 in Fat; B) ASE in gene ADIPOQ shows G×E interaction with SNP rs4686817 in Fat; C) ASE in gene EBI3 shows G×E interaction with SNP rs67782188 in LCLs.
In conclusion, these results show a complex genetic architecture for cis-regulation of gene expression measured through ASE. We propose a model where the allelic imbalance in expression (ASE) requires genetic variability in cis, however, the magnitude of the ASE effect depends on trans genetic and environmental factors that interact with the cis genetic variants (Supplementary Fig. 1-B and Supplementary Fig. 1-C). Examples of interactions between cis and trans genetic variants affecting gene expression have been described recently4. Here we provide a global quantification of the magnitude of these effects. About 38% to 49% of the variance of the observed ASE is not explained by additive genetic effects. This means that a substantial amount of the variance observed in ASE, and therefore in genetic regulation of gene expression, is due to G×G and G×E interactions. It is worth noting that our results show no additive trans effects on ASE. This does not mean that there are no additive trans effects affecting gene expression; it means that the trans effects affecting ASE are not additive. We found an example of G×E interaction on gene expression that has been widely described in the literature (Adiponectin), supporting the validity of our approach. However, the limitation on power due to the sample size prevented us discovering specific associations in most other cases. Allelic gene expression is the molecular phenotype closest to the action of genetic variation. The presence of widespread G×G and G×E interactions affecting this phenotype implies that G×G and G×E can be important in other complex phenotypes, including diseases. The proposed model has implications for the interpretation of the effect of GWAS genetic variants on complex diseases. About 80% of the GWAS signals are estimated to be regulatory variants. The search for G×G and G×E interactions conditioning on relevant biological models rather than whole genome agnostic searches is likely to recover a substantial fraction of genetic and non-genetic variance associated with disease risk.
Online Methods
Sample collection
The study included 856 Caucasian female individuals recruited from the TwinsUK Adult twin registry. Punch biopsies (8mm) were taken from a photo-protected area adjacent and inferior to the umbilicus. Subcutaneous adipose tissue was dissected from each biopsy, weighed and immediately stored in liquid nitrogen. Similarly, the remaining skin tissue was weighed and stored in liquid nitrogen. Peripheral blood samples were collected and lymphoblastoid cell lines (LCLs) were generated by Epstein Barr Virus transformation of the B-lymphocyte component by the European Collection of Cell Cultures agency.
The St. Thomas' Research Ethics Committee (REC) approved on 20th September 2007 the protocol for dissemination of data, including DNA, with the REC reference number RE04/015. On 12th of March of 2008, the St Thomas' REC confirmed this approval extends to expression data. Volunteers gave informed consent and signed an approved consent form prior to the biopsy procedure. Volunteers were supplied with an appropriate detailed information sheet regarding the research project and biopsy procedure by post prior to attending for the biopsy.
Genotying and imputation
Samples were genotyped on a combination of the HumanHap300, HumanHap610Q, 1M-Duo and 1.2MDuo 1M Illumnia arrays. Samples were imputed into the 1000 Genomes Phase 1 reference panel (data freeze, 10/11/2010)15 using IMPUTE216 and filtered (MAF<0.01, IMPUTE info value < 0.8).
RNA processing
Samples were prepared for sequencing with the Illumina TruSeq sample preparation kit (Illumina, San Diego, CA) according to manufacturer’s instructions and were sequenced on a HiSeq2000 machine. Afterwards, the 49-bp sequenced paired-end reads were mapped to the GRCh37 reference genome17 with BWA v0.5.918. We use genes defined as protein coding in the GENCODE 10 annotation19. We excluded samples that failed in the library prep or sequence process. We also excluded samples with less than 10 million reads sequenced and mapped to the exons. Finally we excluded samples in which the sequence data did not correspond with the actual genotype data. We ended with 766 samples for fat, 814 for LCL, 716 for skin and 384 for blood (we had blood samples for only half of the individuals).
eQTL discovery
Exon quantifications
All overlapping exons of a gene were merged into meta-exons with identifier of the form “geneID_start.pos_end.pos”. We counted a read in a meta-exon if either its start or end coordinate overlapped a meta-exon.
Normalization
All read count quantifications were corrected for variation in sequencing depth between samples by normalizing the reads to the median number of well-mapped reads. We used only exons quantified in more than 90% of the individuals. We removed the effects of technical covariates regressing out the first 50 factors from PEER 20, including BMI and age in the model to preserve important biological sources of variation.
eQTL association
Since our data samples are twins, they are not independent observations and we needed to take that into account in our models. We used the two-steps strategy described in Aulchenko et al.21 First we kept the residuals of a mixed model that removed the effects of the family structure using the implementation in GenAbel R package. We then transformed those residuals using a rank normal transformation. Finally, we performed a linear regression of the transformed residuals on the SNPs in a 1Mb window around the transcription start site for each gene, using MatrixeQTL R package 22. We did the association at the exon level and we kept the best association per gene.
Permutations
We permuted the quantifications of each exon 2000 times, keeping the best p-value per exon from each round. From these data, we adjusted the empirical FDR to 1% according to the most stringent exon of each gene, stratifying the analysis on the number of exons for a given gene.
Sites Filtering in ASE
In all the ASE analyses we excluded sites that are susceptible to allelic mapping bias: 1) sites with 50bp mapability < 1 based on the UCSC mapability track, implying that the 50bp flanking region of the site is non-unique in the genome, and 2) simulated RNA-seq reads overlapping the site that show >5% difference in the mapping of reads that carry the reference or non-reference allele. To verify that the genotype is a true heterozygous, we used only sites with >=30 reads, and sites where both alleles are observed in RNAseq data7.
Binomial test for ASE
We assessed statistically significant ASE sites using a binomial test. We did a test for each heterozygous SNP in every individual to detect the presence of statistically significant allelic imbalance. For each site-individual we counted the number of reads covering each allele and calculated a binomial test comparing the observed proportion of reference allele counts with the expected proportion. In theory, this expected proportion should be 0.5 but mapping bias can change it a little bit. To correct for systematic bias in allelic ratios we calculated the overall reference to total allele ratio for each individual for each SNP base combination. These ratios were then used as the expected ratios in the binomial test. We called significant ASE sites using a 10% FDR threshold. We assess the robustness of our significant ASE calls in four ways. First, we evaluated the concordance of ASE among tissues by measuring ASE of ASE significant sites from one tissue in another tissue in the same individual and observed a replication rate about 70% in the three tissues with complete sample size (Supplementary Fig. 5). We then analyzed 5 samples of the GEUVADIS project that where sequenced between 2 and 7 times in different laboratories7,23. We observed that the ASE ratio is quite stable for coverage of 30 reads or more (Supplementary Fig. 6). We also observed that the agreement in ASE significant calls is stable for different coverages (Supplementary Fig. 7). Finally, we analyzed two LCL samples (from the Geuvadis Project7) following the same protocol and analysis pipeline as the one described in the present paper and compared the results to the ASE ratios obtained from a new technique that uses microfluidic multiplex PCR and sequencing (mmPCR)24. The experimental and statistical analysis of the two samples was independently performed in the two different laboratories. We found a very good agreement between the results we obtained using RNAseq and the new mmPCR technique. The replication rate is about 80-82% and the correlation among the ASE ratios for sites that are significant using RNAseq is 0.86 (Supplementary Fig. 8). These observations show a high degree of replicability of ASE measures.
Quantification of ASE
The measure we used for the variance components analysis of ASE is the logit of the percentage of reference alleles. Being p = REF _COUNT /TOTAL_COUNT the percentage of reference allele at a site for an individual, the measure of ASE is:
This measure is not dependent on the overall gene expression level and thus is not susceptible to give false interactions due to trans effects or environmental effects that increase the overall level of expression (Supplementary Fig. 9 and Supplementary Fig. 10).
IBD and IBS calculations
IBD
We calculated the haplotypes in a 1Mb window around the TSS of each gene and counted the number of haplotype alleles that are shared between the twin pairs at each locus.
IBS
We estimated IBS for each twin pair at each locus based on the eQTL-ASE site haplotype. For each site, we counted the number of alleles in the eQTL-ASE site haplotype that are equal between the pair.
The difference between the IBS an the IBD estimates is that for the IBD we take into account the information of a 1Mb haplotype and for the IBS estimates we use only the haplotype with two SNPs: the eQTL and the ASE site.
Variance Components Models
Classical variance components models in twins model the phenotypic correlation between MZ twins and DZ twins as a function of the additive genetic variance and the shared environment variance25:
and
where A represents the additive genetic effects and C the effects due to the common environment between the twin pair (events that affect each member of a twin pair in the same way). The individual environmental effect (events that occur to one twin but not the other) would be E = 1 –cormz. From the two equations above, we get that heritability can be estimated as:
Here, we extend this model to incorporate new sources of variation:
-
-
Aqtl: additive effect due to the best eQTL
-
-
Acis: other genetic additive effects in cis
-
-
Atrans: additive genetic effects in trans
-
-
I: epistatic interaction between trans and cisgenetic effects
Where A = Aqtl + Acis + Atrans + I
Then, our model has six variance component: 1) variance due to the effect of the major cis eQTL (the IBS status at this locus), 2) variance due to the rest of the genetic variants in cis (including the effect of rare variants, captured by the IBD status), 3) variance due to genetic variants in trans (the genome-wide IBD), 4) variance due to non-additive genetic effects (genetic interactions), 5) variance due to the shared environmental effect and 6) variance due to the individual environmental effect.
The equations of the extended model are:
Where:
| cormz | is the correlation within MZ twins |
| cordz_ibd1 | is the correlation within DZ twins that are IBD=1 at the gene |
| cordz_ibd05_ibs1 | is the correlation within DZ twins that are IBD=0.5 at the gene and IBS=1 at the eQTL |
| cordz_ibd05_ibs05 | is the correlation within DZ twins that are IBD=0.5 at the gene and IBS=0.5 at the eQTL |
| cordz_ibd0_ibs1 | is the correlation within DZ twins that are IBD=0 at the gene and IBS=1 at the eQTL |
| cordz_ibd0_ibs05 | is the correlation within DZ twins that are IBD=0 at the gene and IBS=0.5 at the eQTL |
| cordz_ibd0_ibs0 | is the correlation within DZ twins that are IBD=0 at the gene and IBS=0 at the eQTL |
To calculate these correlations we used sites covered by at least 30 reads, showing significant ASE in genes with at least one cis eQTL. Since the number of individuals that have ASE at a given site is small, we analyzed all the sites together to get a global estimate of the variance components. This strategy has been used previously with gene expression data1,26.
To solve the system of equations we used the non-linear optimization package Rsolnp from the R statistical environment 27. We estimated the solution that minimizes the quadratic errors, forcing the variances components to be positive.
Genotype by Environment Interaction
For every ASE site with data for at least 50 MZ twin pairs we calculated the Mann-Whitney test:
for all the SNPs in a 1Mb window around the transcription start site of the gene holding the ASE site. ASE_distance is the absolute value of the difference in the ASE phenotype between the two siblings of the MZ pair and snpi represents the genotype of one SNP. Since we are looking for an effect on ASE we expect a similar behavior for the two homozygous genotypes. Therefore, for the association analysis we coded the genotypes in two categories: homozygous and heterozygous. To correct for multiple testing we calculated the number of effective tests and applied the Bonferroni correction based on these number of tests. Since MZ twins are genetically identical, a difference in ASE between two MZ siblings has to be caused by environmental/epigenetic causes. A significant association in our tests suggests the existence a G×E interaction affecting ASE. It is worth noting that the associated SNP genotype is not equivalent to the existence of ASE as other variants may be contributing to ASE as well. There are cases of homozygous pairs with ASE and heterozygous pairs without ASE and that, in all cases, the difference in ASE is larger for the heterozygous (Supplementary Fig. 11). Finally, the existence of ASE does not imply a significant G×E interaction as shown in Supplementary Fig. 12.
Supplementary Material
Acknowledgements
This work has been funded by the EU FP7 grant EuroBATS (No. 259749) which also supports AAB, AB, MND, DG, AV, TDS. AAB is also supported by a grant from the South-Eastern Norway Health Authority, (No. 2011060). RD is supported by the Wellcome Trust (No. 098051). The Louis-Jeantet Foundation, Swiss National Science Foundation, European Research Council and the NIH-NIMH GTEx grant supports ETD. TS is an NIHR senior Investigator and holder of an ERC Advanced Principal Investigator award. JBR and HZ are supported by the Canadian Institutes of Health Research, Fonds de Recherche Sante du Quebec, and the Quebec Consortium for Drug Discovery. Most computations were performed at the Vital-IT(http://www.vital-it.ch) Center for high-performance computing of the SIB Swiss Institute of Bioinformatics. The TwinsUK study was funded by the Wellcome Trust; EC FP7(2007-2013) and the National Institute for Health Research (NIHR) Clinical Research Facility at Guy’s & St Thomas’ NHS Foundation Trust and NIHR Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. SNP Genotyping was performed by The Wellcome Trust Sanger Institute and National Eye Institute via NIH/CIDR. We thank the twins for their voluntary contribution to this project.
Footnotes
Data accession numbers
RNAseq data are available under EGA accession number EGAS00001000805.
The authors declare no competing financial interest.
References
- 1.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–9. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stranger BE, et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemani G, et al. Detection and replication of epistasis influencing transcription in humans. Nature. 2014 doi: 10.1038/nature13005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–7. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013 doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essaoui M, et al. Monozygotic twins discordant for 18q21.2qter deletion detected by array CGH in amniotic fluid. Eur J Med Genet. 2013 doi: 10.1016/j.ejmg.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Souren NY, et al. Adult monozygotic twins discordant for intra-uterine growth have indistinguishable genome-wide DNA methylation profiles. Genome Biol. 2013;14:R44. doi: 10.1186/gb-2013-14-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surakka I, et al. A genome-wide association study of monozygotic twin-pairs suggests a locus related to variability of serum high-density lipoprotein cholesterol. Twin Res Hum Genet. 2012;15:691–9. doi: 10.1017/thg.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson JF, et al. Gene-nutrient interactions in the metabolic syndrome: single nucleotide polymorphisms in ADIPOQ and ADIPOR1 interact with plasma saturated fatty acids to modulate insulin resistance. Am J Clin Nutr. 2010;91:794–801. doi: 10.3945/ajcn.2009.28255. [DOI] [PubMed] [Google Scholar]
- 12.Joseph PG, Pare G, Anand SS. Exploring gene-environment relationships in cardiovascular disease. Can J Cardiol. 2013;29:37–45. doi: 10.1016/j.cjca.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Martinez P, et al. Adiponectin gene variants are associated with insulin sensitivity in response to dietary fat consumption in Caucasian men. J Nutr. 2008;138:1609–14. doi: 10.1093/jn/138.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warodomwichit D, et al. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring) 2009;17:510–7. doi: 10.1038/oby.2008.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parts L, Stegle O, Winn J, Durbin R. Joint genetic analysis of gene expression data with inferred cellular phenotypes. PLoS Genet. 2011;7:e1001276. doi: 10.1371/journal.pgen.1001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 22.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–8. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.t Hoen PA, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31:1015–22. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 24.Zhang R, et al. Quantifying RNA allelic ratios by microfluidic multiplex PCR and sequencing. Nat Methods. 2014;11:51–4. doi: 10.1038/nmeth.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falconer DS, MacKay TFC. Introduction to Quantitative Genetics. Longmans Green; Harlow, Essex, UK: 1996. [Google Scholar]
- 26.Price AL, et al. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet. 2011;7:e1001317. doi: 10.1371/journal.pgen.1001317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Team RDC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.