Abstract
A rapid, sensitive and quantitative immunoassay for the targeted detection and decontamination of E. coli based on Fe3O4 magnetic nanoparticles (MNPs) and plasmonic popcorn-shaped gold nanostructures attached single-walled carbon nanotubes (AuNP@SWCNT) is presented. The MNPs were synthesized as the support for the monoclonal antibody (mAb@MNP). E. coli (49979) was captured and rapidly preconcentrated from sample with the mAb@MNP, followed by binding with Raman-tagged concanavalin A-AuNP@SWCNT (Con A-AuNP@SWCNT) as detector nanoprobes. A Raman tag 5,5'-dithiobis-(2-nitrobenzoic acid) (DTNB) generated Raman signal upon 670 nm Laser excitation enabling detection and quantification of E. coli concentration with a limit of detection of 102 CFU/mL and a linear logarithmic response range of 1.0 × 102 to 1.0 × 107 CFU/mL. The mAb@MNP could remove more than 98% of E. coli (initial concentration of 1.3 × 104 CFU/mL) from water. The potential of the immunoassay to detect E. coli bacteria in real water samples was investigated and the results were compared with the experimental results from classical count method. There was no statistically significant difference between the two methods (p > 0.05). Furthermore, the MNP/AuNP@SWCNT hybrid system exhibits enhanced photothermal killing effect. The sandwich-like immunoassay possesses the potential for rapid bioanalysis and simultaneous biosensing of multiple pathogenic agents.
1 Introduction
Escherichia coli (E. coli) is a bacterium commonly causing infections in hospitals and communities, and displays resistance to 3rd generation cephalosporin and fluoroquinolones.1 The rapid detection, quantification and eradication of pathogens is considered a powerful tool for disease diagnosis, drug discovery research, food safety, environmental monitoring, and biodefense. Traditionally, culture is the gold standard for detection of many microorganisms.2, 3 However, this method is protracted, and faces the possibility of reduced viability due to extraneous environmental factors. Various types of molecular immunoassay tests have been developed to detect E.coli, especially methods involving enzyme-linked immunosorbent assay (ELISA) and Polymerase chain reaction-PCR techniques.4–6 ELISA and PCR are very sensitive and yield qualitative information of the tested microorganisms. Nonetheless, these conventional techniques are expensive and require skilled staff and complex sample pretreatment.4–9 Other studies have used microfluidic devices to combine the separation-concentration capabilities for bacteria detection,10 and SERS-based optical analysis.11
Surface enhanced Raman scattering (SERS)-based detection has recently attracted significant interest because of its potential as a highly sensitive immunoassay, unique photostability and potential for multiplex detection.12–16 Multi-branched sharp tip and edge metal nanostructures have been shown to cause increased electric field enhancement. However, the non-uniform attachment of these plasmonic nanostructures on the target analyte makes it difficult to conduct quantitative analysis. New approaches have been developed for solving this problem such as SERS-based ‘convective assembly’ for bacteria identification17 and electron-beam lithography (EBL) assemblage of nanoparticles in prefabricated nanohole arrays.18 A notable drawback with these methods is the high concentration of bacteria solution required to obtain detectable SERS signals. Practically, the number of pathogens in contaminated samples is always in low concentration. Therefore, methods that can concentrate bacteria samples and significantly increase the intensity of the SERS signal to rapidly detect and quantify pathogens at low concentrations are highly desired.
Heterogeneous sandwich-like SERS-based immunoassay assemblages for sensitive detection of pathogens have been reported.19–21 In these systems the primary antibodies are usually immobilized on a solid-state support and the sandwich immunocomplex is formed between the immobilized primary antibodies and the secondary signal antibody.19–21 However, immobilization of antibody on solid surfaces in air compromises their orientation that affects their binding abilities and consequently compromise quantitative bioanalysis. Homogeneous immunoassays have been reported to overcome diffusion-limited kinetics associated with heterogeneous immunoassays, and are characterized by shorter incubation times that make such immunoassays rapid.22,23 Recently noble metal nanoparticle attached-SWCNTs were shown to have enhanced NIR absorption, improved biocompatibility, and surface enhanced Raman scattering imaging.14,24,25 Compared to the frequently-used gold nanoshells and nanorods, these gold-SWCNTs hybrids may act as a novel platform for multimodality analyte diagnosis and therapy. In this work, we report a sandwich-like multifunctional magnetic–plasmonic immunoassay using mAb@MNP as capture substrates and DTNB-tagged Con A-AuNP@SWCNTs as SERS probes. We hypothesized that the as-assembled Con A-AuNP@SWCNT immunoassay can potentially serve as multifunctional immunoassay for rapid separation, detection and quantification of pathogens, and a better photothermal agent when combined with magnetic performance.
We demonstrate that they can be used for fast concentration, selective separation, detection, and quantification of bacteria. To facilitate rapid concentration and selective separation, mAb conjugated magnetic APTES@MNP core-shell was used. The SWCNTs are selected as useful templates to generate uniform attachment of the multi-branched popcorn-shaped gold nanostructures thus creating numerous so-called “hot spots” with greater electric field enhancement. The mannose-modified AuNP@SWCNTs is used for loading reporter probes DTNB-tagged Con A. Given the significant binding affinity of α-D-mannose to the lectin Con A,26 but its limited binding affinity for E. coli,27 its biological association with the pathogens can be enhanced by using the multivalent Con A which will increase the binding sites of E. coli on the nanoprobes surface.28–29 At pH≥7, Con A exists as a tetramer with four binding sites possessing high specificity for their cognate sugar moieties D-mannosyl and D-glucosyl.30–32 The analytical performance of the sandwich-like SERS-based assay was evaluated according to limit of detection, the linear range of detection and response time. Further, we demonstrate the comparison of the photothermal killing effect of the different nanostructure systems.
2 Experimental sections
2.1 Materials and reagents
Hexadecyltrimethylammonium bromide (CTAB, ≥96%) and hydrogen tetrachloroaurate (III) trihydrate (HAuCl4·3H2O, 99.99%), iron (III) chloride hexahydrate (FeCl3∙6H2O), iron (II) chloride tetrahydrate (FeCl2∙4H2O), aqueous ammonium hydroxide (28.0%), L-ascorbic acid (AA, ≥ 99.7%), ethanol (≥99.7%), tetraethyl orthosilicate (TEOS), (3-amino)propyltriethoxysilane (APTES), 5,5-dithiobis-(2-nitrobenzoic acid) (DTNB), SWCNTs (diameter 0.7–1.1 nm and length 300–2300 nm), 70% HNO3, silver nitrate (99.9%), sodium borohydride, trisodium citrate dehydrate, glutaraldehyde (GA), Concanavalin A (Con A), PEG-SH (MW 1k) and Bovine serum albumin (BSA) were purchased from Sigma-Aldrich. The E. coli antibody mAb13622 were purchased from the American Type Culture Collection (ATCC, Rockville, MD). E. coli 49979 and Salmonella DT104 bacteria were kindly donated by Dr. Huey-Min Hwang, Environmental Science Department- Jackson State University. All reagents were used as received. All solutions were prepared by ultrapure Millipore water (18.2 MΩ cm−1) from a Millipore system. PBS buffer solution (PBS 2 mM, pH 7.4) served as the buffer solution and PBS with 0.05% (v/v) Tween 20 (PBST) was used as the washing buffer.
2.2 Bacterial cultures
Pure colony cultures of E. coli 49979 and Salmonella DT104 were grown for 12 h at 37 °C in Luria broth and Tryptic soy broth respectively as instructed by ATCC. Calculation of CFU/mL was done according to published protocols.33 Cells were resuspended in PBS to the required concentration. For safety considerations, all of the bacterial samples were placed in an autoclave at 121 °C for 20 min to kill the bacteria.
2.3 Preparation of monoclonal antibody conjugated magnetic nanoparticles (mAb-MNPs)
(a) Magnetic nanoparticles (MNPs)
First, magnetic nanoparticles (MNPs) were precipitated in alkali solution of Fe2+ and Fe3+ (molar ratio 1:2) at 85°C via the co-precipitation method.34 Typically, FeCl3∙6H2O (2.92 g, 0.0108 mol) and FeCl2∙4H2O (1.074 g, 0.0054 mol) were dissolved in 50 mL deoxygenated water at 85°C under N2 protection and vigorous mechanical stirring. Next, 4.5 mL of ammonium hydroxide (28%) was quickly injected into the reaction mixture in one portion. The addition of the base to the Fe2+/Fe3+ salt solution resulted in the formation of the black precipitate of MNPs immediately by the reaction Fe2+ + 2Fe3+ + 8OH− → Fe3O4 + 4H2O. The reaction continued for another 25 min and the mixture was cooled to room temperature. The black precipitate was washed 3 times with distilled water, and 2 times with ethanol through magnetic decantation. This was dried in a vacuum oven for 24 hrs.
(b) Monoclonal antibody conjugated magnetic nanoparticles (mAb-MNPs)
The resulting MNPs (~0.8 g) were dispersed in a mixture of 4 mL of DI water and 20 mL of absolute ethanol by sonication for 10 min. Next, 0.05 g of TEOS, and 0.8 mL of ammonia (28%–30%) solution was added and the reaction was performed at room temperature for 1 h under mechanical agitation. After that, 30 µL of amino propyl triethoxysilane (APTES) was added and the mixed solution was agitated for an additional 1 ½ h to obtain the APTES@MNP core-shell NPs functionalized with amino groups.35 The resultant products were washed three times with ethanol and water to eliminate excess reagent. The product was dried under vacuum at room temperature. Monoclonal capture antibodies against the target pathogen were immobilized onto the amine-terminated APTES@MNPs via the well-established glutaraldehyde (GA) linker method. Briefly, 0.02g of APTES@MNPs was dispersed into 5.0 mL of PBS solution containing 150µL of 2.5% GA for about 2 h with gentle shaking at room temperature. The GA-modified magnetic nanoparticles were washed with PBS, isolated by an external permanent magnet and re-dispersed in 10 mL of PBS. Next, 2.0 mL of diluted GA-modified MNPs was incubated with pathogen-specific antibodies (50 μL 0.1 mg/mL anti-E. coli antibody) in PBS buffer, for 12 h at 4°C. The antibody-modified nanoparticles were next blocked with BSA and washed with PBS to remove excess unbound antibodies. This was kept at 4°C in PBS for future use.
2.4 Thiol-modified f-SWCNT
To attach the AuNPs onto the surface of the f-SWCNTs, the pristine SWCNTs were first oxidized by using nitric acid to produce carboxyl groups as has been reported.36 Briefly, pristine SWCNTs (30 mg) were dispersed in 30 mL of 3 M HNO3 solution with the aid of sonication for 1 h, followed by refluxing at 105°C for 2 ½ h. The oxidized SWCNTs were obtained by vacuum filtration, re-dispersed in 12 mL 1 M HCl solution, washed with copious amounts of deionized water (DI), and dried in a vacuum oven at 60°C overnight. Next, the oxidized SWCNTs were dispersed in 10 mL SOCl2 / 1 mL DMF under sonication for 3 min in a seal tube. The reaction mixture was refluxed at 70 °C for 3 h. The acyl chloride activated SWCNTs (SWCNTs-COCl) were obtained by rotation vaporization under vacuum and immediately reacted with 30 mM cysteamine in DMF at 110°C for 2 ½ h. The thiol terminated SWCNT (f-SWCNTs) were obtained by vacuum filtration, washed with ethanol and copious amounts of DI water.
2.5 Attachment of gold nanopopcorns
The popcorn-shaped gold nanostructures were prepared as previously described.14 Attachment of popcorn-shaped AuNPs onto the f-SWCNTs was carried out by adding an excess of AuNP colloids in order to achieve the highest possible surface attachment onto the f-SWCNTs. Typically, 10 mg of f-SWCNTs were dispersed in 10 mL of nanopure water via sonication for 5 min. AuNP colloids were added to the f-SWCNTs dispersion dropwise, and the mixture was agitated until the mixture retained the bluish color, characteristic of the popcorn shaped AuNPs. The mixture was left at rest for 12 h. The mixture was centrifuged at 2200 × g force for 45 min to remove the excess unbound AuNPs from the mixture and finally re-dispersed in 10 mL of water.
2.6 Synthesis of (3-mercaptopropyl)-D-mannopyranoside
The synthesis of (3-mercaptopropyl)-D-mannopyranoside was achieved according to modified literature procedures.37–39 Briefly, the thiolated mannose was easily prepared in two steps from 1,2,3,4,6-penta-O-acetate mannopyranoside by initial condensation with 3-bromopropan-1-ol, under Lewis acid mediated conditions. Isolation of the resultant glycoside as its tetraacetate, and displacement of the bromide ion in this compound by treatment with potassium thioacetate gave the thiopropyl mannoside acetate, which on de-esterification by treatment with sodium methoxide and methanol afforded the thiolated mannose. The thiolated mannose was characterized using NMR and MS. Spectral data were in agreement with literature values.53 (See SI-1 and Figures S1a–c for experimental details and characterization).
2.7 Preparation of SERS Con A-AuNP@SWCNTs nanoprobes
Initially, 1.5 mL of AuNP@SWCNTs was incubated with 100 µL of 5% of thiolated-D-mannose for 12 h at room temperature and centrifuged twice at 1740 × g force for 30 min. The precipitate was re-dispersed in PBS. The thiol-terminated D-mannose bond on the AuNPs surfaces via well-known Au-S chemistry.40 Next, 150 µL of 0.04 mM DTNB labeled Con A (52.0 µg/mL) in PBS supplemented with Ca2+ and Mn2+, (which are both required as co-factors for binding), was added to 1.5 mL of mannose functionalized AuNP@SWCNTs and the resulting mixture was allowed to conjugate at 4°C for 12 h (S2).41 PEG-SH was added to a final concentration of 0.5% (w/v) and incubated for 20 min at 25 °C. The mixture was centrifuged (1740 × g force, 15 min, 4 °C) and supernatant decanted. The DTNB-tagged Con A-AuNP@SWCNTs was obtained by re-dispersing the precipitate in 2 mL PBS.
2.8 Capture of E. coli using mAb-MNP
30 µL of diluted antibody-conjugated MNP was added to 0.5 mL of bacterial samples containing different serial dilutions of E. coli and slowly agitated at room temperature for 20 min to achieve binding equilibrium. Next, the MNP-bacteria immunocomplex were magnetically collected and rinsed in washing buffer two times. This immunocomplex was subsequently used in the next step of SERS-based detection and quantification.
2.9 Sandwich-like immunoassay and SERS measurements
120 µL Con A-loaded AuNP@SWCNT SERS probes were added into 0.5 mL of the MNP-bacteria and kept at room temperature for 30 min with gentle shaking to form the sandwich-like immunocomplex. After incubation for 30 min under shaking, the mixture was separated by a magnet and washed two times with PBST. The purified composites were dispersed in 200 μL of PBS for SERS detection. SERS spectroscopy was used to detect and quantify the immobilized pathogens. Laser excitation of the samples was performed using 670 nm Laser (power 20 mW, acquisition time 20 s). For Photothermal studies, the cells were exposed to a 670 nm (2.5 W/cm2) laser source for varied lengths of time. The cell viability was assessed by plate-count technique (SI-3).
2.10. Characterization techniques and Raman measurements
UV-Vis absorption spectra were acquired using Shimadzu UV-2600/2700 spectrophotometer, which is run by Varian’s Cary Win UV software version 2.0. The sample solutions (1 mL) were placed in a cell, and spectral analysis was performed in the 300 to 800 nm range at room temperature. The morphology and distribution of samples were examined using the Transmission Electron Microscope (TEM) JEM-2100F (JEOL, Tokyo Japan) at 200 kV. FTIR spectra were acquired using a Nexus 670 FTIR (Thermo Nicolet, Madison, WI) equipped with a universal attenuated total reflection (UATR) accessory, detector, and a KBr beam splitter. All spectra were averaged over 128 scans at a resolution of 4 cm−1. Peak information was obtained using OMNIC software provided by Thermo Nicolet. The sizes and zeta potentials of the MNP and modified MNPs were determined using Zetasizer ZEN3600 instrument from Malvern Instruments Ltd, (Worcestershire, UK). The mean hydrodynamic diameters of the nanoparticles were determined by DLS. Dual angle detection mode based on 173° backscattered and 13° forward scattered light was applied and the average value of three scans with an interval of 1 min was calculated and evaluated. For the SERS experiment, we used a continuous wavelength diode-pumped solid-state (DPSS) laser operating at 670 nm as an excitation light source with a power output of 20 mW on the sample. For excitation and data collection, we used InPhotonics 670 nm Raman fiber optic probe, which is a combination of two single fiber-optic cables, (90 micron excitation, 200 micron collection fiber) with filtering and steering micro-optics; N.A. 0.22. For Raman signal collection, we used a miniaturized QE65000 Scientific-grade Spectrometer from Ocean Optics, with a response range of 220–3600 cm−1. The Hamamatsu FFT-CCD detector used in the QE65000 provides 90% quantum efficiency, with high signal-to-noise and rapid signal processing speed as well as remarkable sensitivity for low-light level applications. The Raman spectrum was analyzed with Ocean Optics data acquisition Spectra Suite spectroscopy software. We have used 20 s acquisition time and 5 scan averaging. SERS data were collected from three different areas on each sample at room temperature.
3 Results and discussion
3.1. Characterization of mAb-MNP
In the present study, MNPs were chosen as the capture and decontamination tools based on their well-documented biological applications. MNPs were synthesized by following an established protocol described earlier.34 Representative transmission electron microscopy (TEM) image of MNPs is shown in Figure 1A. The particle size of MNPs was in the range of 10 – 20 nm. The MNPs were initially reacted with TEOS to form a thin coat of silica shell for further amination with APTES. The TEM image of APTES@MNPs reveal small clusters of MNPs within the silica shell (Figure 1B). The average particle size of APTES@MNPs was in the range of 20 – 50 nm as revealed by the Hydrodynamic size histogram of APTES@MNPs obtained by DLS measurements (Figure 1C). The FTIR measurements were carried out to investigate the interaction between MNPs and silica. Figure 1D shows the FTIR spectra of MNPs and APTES@MNPs core-shell nanostructures.
Figure 1.
TEM images of (A) MNP, (B) APTES@MNP core−shell structures. (C) Histogram of hydrodynamic diameter of MNP (D) FTIR spectra of MNPs before (black) and after (red) APTES modification
The FTIR band at 592 cm−1 is characteristic of the Fe-O stretching mode due to Fe3O4, and the band at 1030 cm−1 correspond to Si-O-Si or Si-O-Fe stretching vibrations of the silica shell (in red). The IR spectrum of the APTES@MNP shows an increase in the band at 1131 cm−1, which is associated with C-N stretching modes. The increase in the band at 3450 cm−1 confirms the existence of N-H stretching modes of the APTES molecule. The amine-terminated silica shell provide the MNP core several advantages, including biocompatibility, aqueous stability, and offer adaptable linkage points for antibody conjugation.
3.2 Characterization of the binding affinity and selectivity of mAb-MNP
For selective binding of E. coli, non-specific binding and selectivity was investigated using binding efficiency (BE %) and surface charge measurement studies. The surface charge characteristics of the MNPs were verified by the zeta potentials measured by Malvern nano Zetasizer. The surface charge characteristic accompanying the stepwise addition of additional layer components on the MNP is shown in Figure 2A. When the MNP was modified with silica shell, an obvious decrease in the surface charge value was observed because the TEOS undergoes hydrolysis reaction to form the silica shell and introduced the -OH groups on the surface of the MNPs. When modified with APTES, the amine groups introduced causes the peak zeta potential to become more positive relative to the silica shell-modified MNPs which indicate the successful modification with the amine group. Zeta potential studies further revealed that there was reversal in charge after GA and BSA layering. BSA layering introduced a zeta potential of −30 mV which suggests that the core-shell structures have a low probability of aggregation, and helps prevent non-specific binding. The binding affinity and selectivity of mAb-MNP was further evaluated by binding efficiency (BE %) determination using E. coli and Salmonella DT104 (Figure 2B).
Figure 2.
(A) Surface charge characteristics of additional layer components on the MNP (B) Effect of number of washing cycles on binding efficiency. (Error bars indicate the standard deviation obtained from three measurements).
Initially, to optimize the magnetic separation, the amount of mAb-MNP and washing cycles were varied and their effects on the binding efficiency (BE %) for E. coli evaluated. To determine the binding efficiency (BE %), 104 CFU/mL of pathogens were incubated with 50µL of mAb-MNP at room temperature for 20 min and separated using an external magnet. Three consecutive wash cycles (with PBS, PBST, and PBS) were performed, and the unbound cells in the supernatant was plated onto the designated agar for each bacterium and incubated at 37 °C for 18 h. Then, the binding efficiency (BE %) was calculated by dividing the number of the bacterial cells immobilized by the magnetic nanoparticles, which is the difference between the total number of the bacterial cells (Pc) and supernatant (Ps), over the total number of bacterial cells in the control.
The effect of the washing cycles on binding affinity and selectivity of mAb-MNP are shown in Figure 2B. After three washing cycles, the binding efficiency was reduced for all bacteria. However, after the three washing cycles, the percentage of bound E. coli was still over 62 %, but only 5% for salmonella DT 104. Non-specific binding was decreased with washing cycles as shown in Figure 2B. These results show that mAb-MNP has a high affinity and selectivity for E. coli.
3.3. Characterization of reporter nanoprobes (DTNB-tagged Con A-AuNP@SWCNTs)
AuNP@SWCNTs possess a high surface-to-volume ratio that allow the loading of multiple kinds of molecules and the design of stable multifunctional nanoprobes.43,44 DTNB, a SERS active molecule was used as Raman tag to increase the sensitivity and reproducibility of the detection method.45,46 Hence 150 µL DTNB-labeled Con A mixture in the ratio 1:2 (v/v) was observed to be the optimal loading ratio, allowing mannose-modified AuNP@SWCNTs nanoprobes to be stable in aqueous solution with good SERS activity and NIR absorption (SI-2 and Figure S2A, C). TEM, UV-Vis and Raman spectroscopy were used to characterize the AuNPs, f-SWCNTs, and mannose-modified AuNP@SWCNT before and after loading with DTNB-labeled con A. Figure 3A is the TEM image of the popcorn-shaped gold nanostructures with the average diameter in the range of 28 – 38 nm, showing rough surfaces consisting of many sharp tips and edges.
Figure 3.
TEM images of (A) AuNPs, (B) f-SWCNTs, (C) mannose-AuNP@SWCNTs
Such rough surface gold nanostructures in high yield have been shown to increase electric-field enhancement which is important for applications involving metal nanoparticles as SERS-based sensors.47 Figure 3B is the TEM image showing the well-dispersed f-SWCNTs after thiol-modification. Figure 3C displays the morphology of mannose modified-AuNP@SWCNT hybrid nanostructure. The popcorn-shaped gold nanostructures decorated the f-SWCNTs, an evidence for abundant presence of SH-terminated functional groups on the nanotube walls. AuNPs attached along SWCNTs with very minimal level of aggregation and at a minimum inter-particle distance. The uniform attachment of the AuNPs on the SWCNTs result in effective plasmon coupling that has been reported to cause signal enhancement via the “hot-spot phenomenon.”48 Figure S2A is the TEM image of DTNB-tagged Con A-AuNP@SWCNT. UV-vis spectroscopy was used to monitor the attachment of AuNP on AuNP@SWCNT nanohybrid and the binding of E. coli (Figures 4 and S2B). As shown in the UV-vis spectra of AuNP@SWCNT nanohybrid (Figure 4A-blue), the attachment of AuNP was indicated by the surface plasmon resonance peak of AuNP at around 570 nm in the AuNP@SWCNT nanohybrid spectrum, which is similar to previous studies.14 Moreover, it is reported that the gold nanostructures appear popcorn-shaped when its sharp peak is in the range of 550 – 600 nm.14 The UV-Vis spectrum of f-SWCNTs (Figure 4A-black) was largely featureless and free of Van Hove singularities, a characteristic associated with successful modification of SWCNTs. The mannose-presenting AuNP@SWCNT did not show a significant shift from AuNP@SWCNT hybrid. However, on loading DTNB-labeled con A, the appearance of a shoulder around 340 nm is observed (Figure 4A-red) which is ascribed to the π-π* transition of the C=O bonds of DTNB-labelled con A. When DTNB-tagged Con A-AuNP@SWCNT was added to E. coli samples, significant binding of E. coli occurred (Figure 4 B black and S2D). The surface plasmon absorption band shifts to longer wavelengths with maximum absorbance at ~640 nm and shows a broadening which is consistent with binding of E. coli (Figure 4B black).
Figure 4.
Absorption spectra of (A) f-SWCNT (black), mannose-AuNP@SWCNTs (blue), Nanoprobes (red), and (B) Nanoprobes (red), Nanoprobes in the presence of E. coli (black).
This presented a good condition for excitation with a 670 nm Laser source and therefore could efficiently transform the absorbed NIR light-energy into heat.24 At pH≥7, the multivalent binding of Con A to the E. coli surface O-antigen supports the strong adhesion of E. coli to the mannose-modified AuNP@SWCNT surface by forming bridges between mannose and the E. coli surface via the lectin-carbohydrate interaction.27,28 The specific binding property of Con A to O-antigen and mannose promotes the specificity of the capture nanoprobes in response to Gram-negative bacteria.31,32 To establish that the binding was induced by con A recognition of the bacteria, rather than non-specific interactions, control experiments were carried out without the con A and UV-Vis investigated. No significant shift in wavelength or broadening in plasmon absorption band in the UV-visible absorption spectrum was observed, indicating that no significant interaction between the hybrid nanostructure and the E. coli occurred. The stability of the DTNB-tagged con A-AuNP@SWCNT was characterized by absorption spectra as shown in the Supporting Information Figure S2C. The blue curve shows the absorption spectrum of the DTNB-tagged Con A-AuNP@SWCNT stored at 4 °C for 1 week. Comparing with the absorption spectrum of the freshly prepared nanoprobes (red curve), only a very slight peak shift appears, which indicate that the hybrid nanoprobes have good stability.
3.4 SERS-based sandwich-like immunocomplex formation
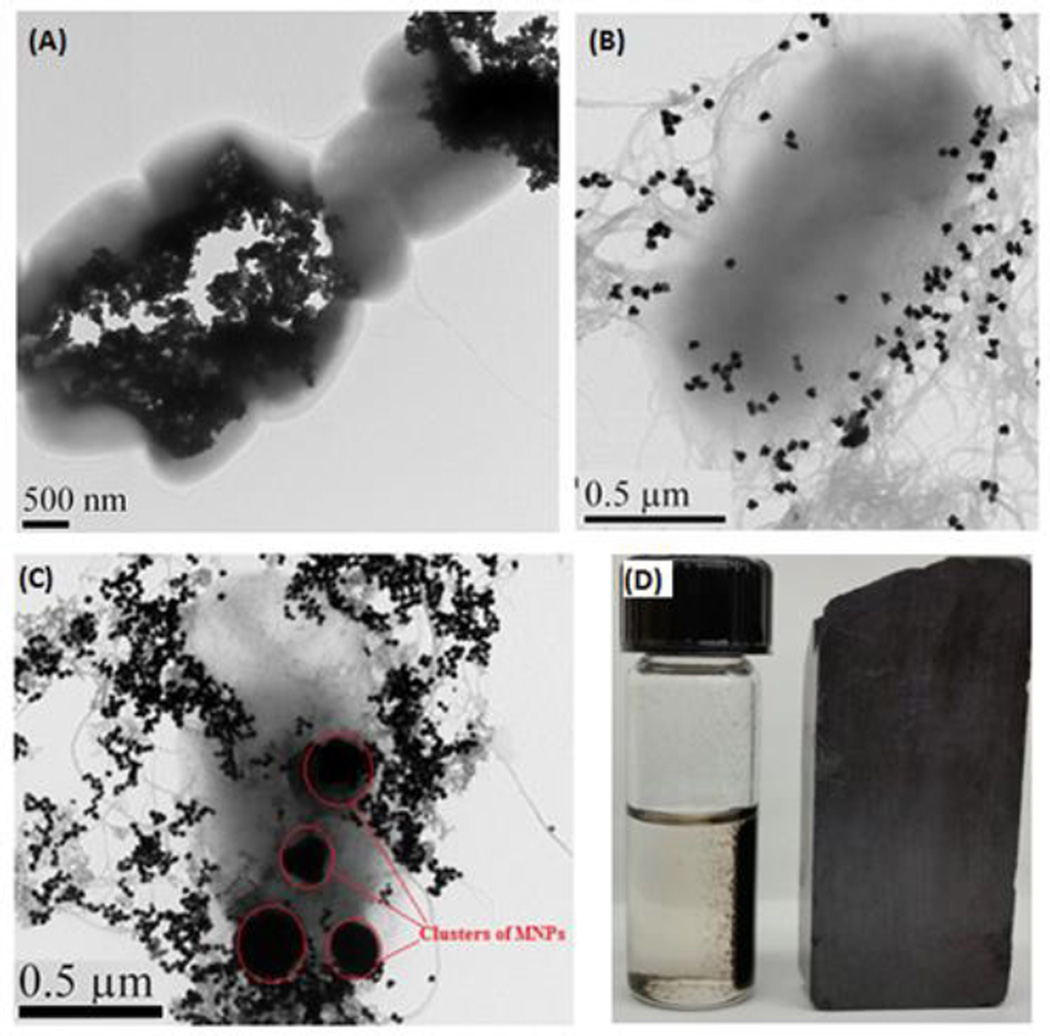
Using the mAb-MNP and the SERS-based nanoprobes, we performed a sandwich-like immunoassay to rapidly detect and quantify E. coli. The TEM images show the E. coli bacteria to be closely bound to the mAb-MNP (Figure 5A) forming clusters of E. coli. The close binding of E. coli is mediated by the E. coli monoclonal antibody conjugated on the MNP.
Figure 5.
TEM image of (a) E. coli captured by mAb-MNPs (b) E. coli captured by nanoprobes (c) immunocomplex (d) Digital photograph showing magnetic separation of E.coli bound MNP/Nanoprobe immunocomplex.
Figure 5B and Figure S2D are the TEMs showing the interactions of E. coli to con A-AuNP@SWCNT. Con A are multivalent, which results in higher binding avidity. Figure 5C is the sandwich-like immunocomplex of MNP/E. coli/AuNP@SWCNT. The homogenous phase of mAb-MNP and AuNP@SWCNT facilitates the solution-phase detection and overcomes the diffusion-limited kinetics associated with solid-phase immunoassays. An external magnetic field was used to concentrate and separate target bacteria from water sample. Figure 5D is the digital photograph showing the rapid separation of the bound E. coli by an external magnet. The results were also characterized by SERS measurements. Fig. 6(A) illustrates the average of three SERS readouts taken from different positions of E. coli immobilized on the nanoprobes at different concentrations of E. coli (107–102 CFU/mL). The E. coli immobilized onto the immuno-nanomaterials formed microbial clusters that resulted in significant SERS enhancement. Under magnetic pull-down, greater signal enhancement is obtained. The spectra are consistent in shape with significant variations in the intensity of the key peaks at 1348 cm−1 (DTNB nitro symmetric stretch),49 and ~1600 cm−1 (G-band of SWCNT) that was proportional to the concentration of the E. coli (Figure 6B). This implied good spectral reproducibility from our SERS-active hybrid probe which was important for biological sample measurements. Control experiments with mAb@MNP and AuNP@SWCNT probe in the absence of E. coli showed no enhancement (Figure 6B).
Figure 6.
SERS spectra of different concentrations of E. coli in (A) DTNB-tagged con A-AuNP@SWCNT (B) the sandwich immunoassay (20 mW excitation power, 30 s acquisition time), (C) Linear fitting of the peak intensities at 1348 cm−1 as a function of the logarithm of E. coli concentration (High resolution SERS spectral region between 1300–1420 cm−1). Error bars indicate the standard deviation obtained from three measurements.
The calibration curve was plotted with the changes of the DTNB peak at 1348 cm−1 vs. the different log concentrations of E. coli (Figure 6C). The calibration curve showed no plateau from 102–107 CFU/mL of E. coli, indicating that the sandwich-like immunoassay had a wider limit of quantitation (LOQ) than the one previously reported.14 This may be due to higher loading of the detection biomolecules on the nanostructures possessing large surface area-to-volume ratio. The impressive linear range can be used to quantify the number of microorganisms in per unit volume of a sample. The limit of detection (LOD) for E. coli is estimated to be 102 CFU/mL, as defined by the lowest concentration that produces a signal three times stronger than the standard deviation of the control at 1348 cm−1 DTNB peak.
To verify that the SERS signal change of the nitro stretch band was specific to recognition of E. coli, we tested our sandwich immunoassay with Salmonella DT104. The SERS response of the immunoassay at 105 CFU/mL of Salmonella DT104 showed a much lower intensity change in response to Salmonella DT104 than to E. coli at the same CFU/mL. This further confirmed that the immunoassay possessed low cross-reactivity with other bacteria and high selectivity towards E. coli.
The potential application of the developed SERS method was investigated with real water samples such as tap water, puddle and stream water. The results obtained were compared with the classical cultured-based plate count technique. The analysis results are presented in table 1.
Table 1.
Comparison of the results obtained for the analysis of real water samples by the proposed SERS method and classical culture-based method
| water sample | Classical count methoda |
Proposed method (SERS)a |
|---|---|---|
| Tap Water | 177 ± 8.22 | 178 ± 4.79 |
| puddle water | 238 ± 7.70 | 240 ± 4.35 |
| stream water | 270 ± 13.17 | 266 ± 9.00 |
CFU/mL unless otherwise stated
E. coli concentration was determined for tap, puddle and stream water as 178, 240, and 266 CFU/mL respectively. The results obtained by the proposed SERS method were close to those obtained by the classical culture-based method, indicating that the mean of the proposed SERS method is not statistically significantly different from the mean of classical count method (p > 0.05).
The analytical performance comparison of the proposed method to the other methods approved by the US Environmental Protection Agency (EPA) is summarized in table 2 below. The proposed method presents a rapid, sensitive and quantitative immunoassay for the targeted detection and decontamination of pathogens.
Table 2.
Figures of merit of the nanoparticle-based method for determination of E. coli
| Methods/Materials used | Analytical ranges/ LODsa |
Comments | Refs |
|---|---|---|---|
| Classical culture-based methods multiple-tube culture and membrane filter technique |
Prolonged incubation time (2 – 7 days) and Labor-intensive. Antagonistic organisms interference, Lack specificity, and a weak level of detection of slow-growing or stressed pathogens |
2 3 |
|
| Molecular methods (a) Immunological methods |
103–104 | Are expensive in terms of consumables Problem of cross-reactivity with non-target cells. Require skilled staff. |
4 5 |
| (b) PCR methods | 1–106 | Inhibitor interference and not quantitative. High risk of false negative/false positive results. Require highly skilled staff. Expensive laboratory instruments and reagents. |
6 8 9 |
| MNP/AuNP SWCNTb |
102—107 | Specific, sensitive, rapid, and cheap. No preprocessing steps. Quantitative. Decontaminating and Bactericidal. |
This work |
CFU/mL or g, unless otherwise stated.
Analytical range and LOD is improved by concentration of target organism
3.5 Cytotoxicity and antibacterial photothermal studies
In order to determine the cytotoxicity of the mAb-MNP and the nanoprobes toward E. coli, 1.3 × 104 CFU/mL of E. coli were immobilized on different amounts of mAb-MNP and nanoprobes for 3 h and incubated on agar plates at 37 °C for 18 h (SI-3). E. coli incubated in the absence of the nanomaterials was used as the control. Our results show over 97% bacterial viability in a 150 µL of the original nanoprobes and 50 µL of diluted mAb-MNP. Bacterial viability in a sandwich assay of 1:4 (v/v) mAb-MNP: nanoprobes were above 96% (Figure S3A). These results confirmed the non-toxicity of the nanomaterials and the sandwich-like assay. Preliminary results indicate that at higher concentrations of mAb-MNP in the sandwich-like immunoassay, both the bacterial viability and SERS signal intensity decreased while at a lower concentration, bacterial binding efficiency was decreased. If more mAb-MNPs are used in the immunoassay, after magnetic separation there are too many MNPs in the immunocomplex product which can obstruct laser irradiation onto the SERS nanoprobes. This condition can reduce the detection sensitivity. The hybrid of AuNP@SWCNT was investigated for photothermal properties (SI-4). Upon NIR laser excitation (670 nm OEM laser at 2.5 Wcm−2), the cell viability rate studies demonstrate that the magnetically pulled-down sandwich-like immunoassay provided rapid and effective killing of up to 90% of E. coli in 9 min but took more than 12 min when AuNP@SWCNT in dispersed state was used (Figure 7A and B, and Figure S3B). When only popcorn-shaped gold nanostructures (AuNPs) were used, viability rate was still above 60% after 12 min of exposure. Gold nanoparticles and SWCNTs have recently been reported to show strong light-induced heating properties.50–52 The synergetic effects enhance the light-to-heat conversion extent by AuNP@SWCNT upon NIR laser irradiation which results into improved killing of the captured bacteria. The magnetic pull-down further enhanced the killing efficiency of the concentrated bacteria more than in the dispersed state.32 Untreated E. coli were used as the control. When exposed to the red light for 15 min, the bacteria viability rate in the absence of nanomaterial was above 98%. This shows that NIR laser exposure alone was harmless to the bacteria (Figure S3B). The local temperature increases due to light-induced heating by the hybrid nanostructures produced sufficient heat for the killing of bacteria, which could therefore be the next avenue for exploration to target and destroy drug-resistant pathogens.
Figure 7.
(A) Digital photographs of colonies of sandwiched E. coli when exposed to NIR light for varied length of times (B) Scatter plots showing viability (%) of untreated and differently treated E. coli when exposed to NIR. (Error bars indicate the standard deviation obtained from three measurements).
4 Conclusions
In this study, we have developed rapid, selective and sensitive homogenous sandwich-like SERS-based immunoassay for concentration, detection, quantification and photothermal killing of E. coli 49979 using plasmonic DTNB-tagged con A-AuNP@SWCNT and target-specific mAb-MNP. The assembled DTNB–tagged con A-AuNP@SWCNT possesses unique optical properties of high SERS activity, and tunable NIR absorption that effectively transformed NIR laser light into heat, as well as low cytotoxicity and good biocompatibility. The effectiveness of these immuno-nanoprobes for pathogen detection was demonstrated by a sandwich-type immunoassay using target-specific mAb-MNP as capture substrates. A good linear relationship is found between the peak intensity at 1348 cm−1 and the logarithm of E. coli concentration in the range between 102 – 107 CFU/mL. The limit of detection (LOD) is 102 CFU/mL. In addition the immunoassay possesses great potential to detect E. coli bacteria in real water samples and the results obtained are in good agreement with the experimental results from classical the count method. The analysis time of the developed assay system took less than 70 min (20 min for magnetic capture, 30 min for sandwich formation, less than 5 min for SERS measurement and, 12 min for photothermal exposure). Such sandwich-like immunoassay possesses potential applications in the design of efficient multifunctional SERS probes for multiplex detection and photothermal killing of multi-drug resistant food and water-borne pathogens.
Supplementary Material
Acknowledgements
The work described was supported by the National Science Foundation (HBCU-RISE: HRD-1137763, PREM: DMR-1205194), the National Institutes of Health through the NCRR-RCMI (Award Number: G12RR013459), and NIH/NIMHD (Award Number: G12MD007581) for the use of the Analytical CORE Facilities.
Footnotes
Electronic Supplementary Information (ESI) available: See DOI: 10.1039/b000000x/
References
- 1. http/www.who.int/drugresistance/in/Antimicrobial Resistance (AMR) 2014
- 2.Yeni F, Acar S, Polat OG, Soyer Y, Alpas H. Food Control. 2014;40:359–367. [Google Scholar]
- 3.D’Aoust JY, Sewell AM, Warburton DW. Int. J. Food Microbiol. 1992;16:41–50. doi: 10.1016/0168-1605(92)90124-l. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Yan B, Chen L. Chem. Rev. 2013;113:1391–1428. doi: 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- 5.Wang N, He M, Shi HC. Anal. Chim. Acta. 2007;590:224–231. doi: 10.1016/j.aca.2007.03.041. [DOI] [PubMed] [Google Scholar]
- 6.Croci L, Delibato E, Volpe G, De Medici D, Palleschi G. Appl. Environ. Microbiol. 2004;70:1393–1396. doi: 10.1128/AEM.70.3.1393-1396.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Opet NJ, Levin RE. J Microbiol., Methods. 2013;94:69–72. doi: 10.1016/j.mimet.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Jain S, Chattopadhyay S, Jackeray R, Abid CKVZ, Kohli GS, Singh H. Biosens. Bioelectron. 2012;31:37–43. doi: 10.1016/j.bios.2011.09.031. [DOI] [PubMed] [Google Scholar]
- 9.Cho IH, Irudayaraj J. Int. J. Food. Microbiol. 2013;164:70–75. doi: 10.1016/j.ijfoodmicro.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 10.Diao J, Young L, Kim S, Fogarty EA, Heilman SM, Zhou P, Shuler ML, Wu M, DeLisa MP. Lab Chip. 2006;6:381–388. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- 11.Floriano PN, Christodoulides N, Romanovicz D, Bernard B, Simmons GW, Cavell M, McDevitt JT. Biosens. Bioelectron. 2005;20:2079–2088. doi: 10.1016/j.bios.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H, Yang D, Ivleva NP, Mircescu NE, Niessner R, Haisch C. Anal. Chem. 2014;86:1525–1533. doi: 10.1021/ac402935p. [DOI] [PubMed] [Google Scholar]
- 13.Gracie K, Correa E, Mabbott S, Dougan JA, Graham D, Goodacre R, Faulds K. Chem. Sci. 2014;5:1030–1040. [Google Scholar]
- 14.Ondera TJ, Hamme AT., II J Mater Chem. B. 2014;2:7534–7543. doi: 10.1039/C4TB01195C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Z, Senapati D, Khan SA, Singh AK, Hamme A, Yust B, Sardar D, Ray PC. Chem. Eur. J. 2013;19:2839–2847. doi: 10.1002/chem.201202948. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Cushing SK, Wu N. Analyst. 2015;140:386–406. doi: 10.1039/c4an01079e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahraman M, Yazici M, Sahin F, Çulha M. Langmuir. 2008;20:894–901. doi: 10.1021/la702240q. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Yan B, Premasiri WR, Ziegler LD, Negro LD, Reinhard BM. Adv. Funct. Mater. 2010;20:2619–2628. [Google Scholar]
- 19.Abell JL, Driskell JD, Dluhy RA, Tripp RA, Zhao YP. Biosens. Bioelectron. 2009;24:3663–3670. doi: 10.1016/j.bios.2009.05.039. [DOI] [PubMed] [Google Scholar]
- 20.Gaster RS, Hall DA, Wang SX. Nano Lett. 2011;11:2579–2583. doi: 10.1021/nl1026056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chon H, Lee S, Son SW, Oh CH, Choo J. Anal. Chem. 2009;81:3029–3034. doi: 10.1021/ac802722c. [DOI] [PubMed] [Google Scholar]
- 22.Pei X, Zhang B, Tang J, Liu B, Lai W, Tang D. Anal. Chim. Acta. 2013;758:1–18. doi: 10.1016/j.aca.2012.10.060. [DOI] [PubMed] [Google Scholar]
- 23.Pekdemir ME, Erturkan D, Kulah H, Boyaci IH, Ozgen C, Tamer U. Analyst. 2012;137:4834–4840. doi: 10.1039/c2an35471c. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Wang C, Cheng L, Lee ST, Liu Z. J. Am. Chem. Soc. 2012;134:7414–7422. doi: 10.1021/ja300140c. [DOI] [PubMed] [Google Scholar]
- 25.Meng L, Niu L, Li L, Lu Q, Fei Z, Dyson P. Chem. Eur. J. 2012;18:13314–13319. doi: 10.1002/chem.201201811. [DOI] [PubMed] [Google Scholar]
- 26.Smith E, Thomas W. J. Am. Chem. Soc. 2003;125:6140–6148. doi: 10.1021/ja034165u. [DOI] [PubMed] [Google Scholar]
- 27.Lu Q, Lin H, Ge S, Luo S, Cai Q, Grimes C. Anal. Chem. 2009;81:5846–5850. doi: 10.1021/ac9008572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Z, Huang M, Xiao C, Zhang Y, Zeng X, Wang PG. Anal. Chem. 2007;79:2312–2319. doi: 10.1021/ac061986j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mann DA, Kanai M, Maly DJ, Kiessling LL. J. Am. Chem. Soc. 1998;120:10575–10582. [Google Scholar]
- 30.Zhang H, Shi Y, Lan F, Pan Y, Lin Y, Lv J, Zhu Z, Jiang Q, Yi C. Chem. Commun. 2014;50:1848–1850. doi: 10.1039/c3cc48417c. [DOI] [PubMed] [Google Scholar]
- 31.Pieters RJ. Org. Biomol. Chem. 2009;7:2013–2025. doi: 10.1039/b901828j. [DOI] [PubMed] [Google Scholar]
- 32.Vedala H, Chen Y, Cecioni S, Imberty A, Vidal S, Star A. Nano Lett. 2011;11:170–175. doi: 10.1021/nl103286k. [DOI] [PubMed] [Google Scholar]
- 33.Salam F, Tothill IE. Biosens. Bioelectron. 2009;24:2630–2636. doi: 10.1016/j.bios.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 34.Massart R. IEEE Trans. Magn. 1981;17:1247–1248. [Google Scholar]
- 35.Stöber W, Fink A, Bohn E. J. Colloid Interface Sci. 1968;69:62–69. [Google Scholar]
- 36.Tchoul M, Ford W, Lolli G. Chem. Mater. 2007;19:5765–5772. [Google Scholar]
- 37.Kartha KPR, Field RA. Tetrahedron. 1997;53:11753–11766. [Google Scholar]
- 38.Revell DJ, Knight JR, Blyth DJ, Haines AH, Russell DA. Langmuir. 1998;14:4517–4524. [Google Scholar]
- 39.Skirtenko N, Richman M, Nitzan Y, Gedanken A, Rahimipour S. Chem. Commun. 2011;47:12277–12279. doi: 10.1039/c1cc13518j. [DOI] [PubMed] [Google Scholar]
- 40.Zeng S, Yong KT, Roy I, Dinh XQ, Yu X, Luan F. Plasmonics. 2011;6:491–506. [Google Scholar]
- 41.Hone DC, Haines AH, Russell DA. Langmuir. 2003;19:7141–7144. [Google Scholar]
- 42.Chen L, Hong W, Guo Z, Sa Y, Wang X, Jung YM, Zhao B. J. Colloid Interface Sci. 2012;368:282–286. doi: 10.1016/j.jcis.2011.10.069. [DOI] [PubMed] [Google Scholar]
- 43.Zheng M, Huang X. J. Am. Chem. Soc. 2004;126:12047–12054. doi: 10.1021/ja047029d. [DOI] [PubMed] [Google Scholar]
- 44.Yeh YC, Creran B, Rotello VM. Nanoscale. 2012;4:1871–1880. doi: 10.1039/c1nr11188d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Güçlü K, Ozyürek M, Güngör N, Baki S, Apak R. Anal. Chim. Acta. 2013;794:90–98. doi: 10.1016/j.aca.2013.07.041. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Zong S, Li W, Wang C, Xu S, Chen H, Cui Y. J. Am. Chem. Soc. 2012;134:2993–3000. doi: 10.1021/ja208154m. [DOI] [PubMed] [Google Scholar]
- 47.Sánchez-Iglesias A, Pastoriza-Santos I, Pérez-Juste J, Rodríguez-González B, García de Abajo FJ, Liz-Marzán LM. Adv. Mater. 2006;18:2529–2534. [Google Scholar]
- 48.Wei H, Xu H. Nanoscale. 2013;5:10794–10805. doi: 10.1039/c3nr02924g. [DOI] [PubMed] [Google Scholar]
- 49.Guven B, Dudak FC, Boyaci IH, Tamer U, Ozsoz M. Analyst. 2014;139:1141–1147. doi: 10.1039/c3an01600e. [DOI] [PubMed] [Google Scholar]
- 50.Ray PC, Khan SA, Singh AK, Senapati D, Fan Z. Chem. Soc. Rev. 2012;41:3193–3209. doi: 10.1039/c2cs15340h. [DOI] [PubMed] [Google Scholar]
- 51.Kam NWS, O’Connell M, Wisdom JA, Da H. Proc. Natl. Acad. Sci. U S A. 2005;102:11600–11605. doi: 10.1073/pnas.0502680102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain PK, Huang X, El-Sayed IH, El-Sayed MA. Acc. Chem. Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 53.Altamore TM, Fernandez-Garcia C, Gordon AH, Hubscher T, Promsawan N, Ryadnov MG, Doig AJ, Woolfson DN, Gallagher T. Angew. Chem. Int. Ed. 2011;50:11167–11171. doi: 10.1002/anie.201101055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.