Abstract
STUDY QUESTION
Do the luminal fluids of the epididymis and the vas deferens contribute to sperm chromatin fragmentation (SCF) in mice?
SUMMARY ANSWER
The luminal fluids of both organs are required for activating SCF in mice, but the vas deferens luminal fluid does this more efficiently than that of the epididymis.
WHAT IS KNOWN ALREADY
Mice sperm have the ability to degrade their DNA in an apoptotic-like fashion when treated with divalent cations in a process termed SCF. SCF has two steps: the induction of reversible double-strand DNA breaks at the nuclear matrix attachment sites, followed by the irreversible degradation of DNA by nuclease. Single stranded DNA breaks accompany SCF.
STUDY DESIGN, SIZE, DURATION
Luminal fluids from two reproductive organs of the mouse (B6D2F1 strain), the epididymis and vas deferens, were extracted and tested for SCF activation with divalent cations using four different combinations of the sperm and the surrounding luminal fluids: (i) in situ—sperm were kept in their luminal fluid and activated directly; (ii) reconstituted—sperm were centrifuged and resuspended in their luminal fluid before SCF activation; (iii) mixed—sperm were centrifuged and resuspended in the luminal fluid of the other organ; (iv) no luminal fluid—sperm were centrifuged and reconstituted in buffer. All four experiments were performed without (controls) and with divalent cations (resulting in SCF). For each experimental condition, two different mice were used and the analyses averaged.
PARTICIPANTS/MATERIALS, SETTING, METHODS
DNA damage by SCF was analyzed by three different methods, the sperm chromatin structure assay (SCSA), terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) analysis and field inversion gel electrophoresis.
MAIN RESULTS AND THE ROLE OF CHANCE
In all three assays that we used, the vas deferens luminal fluid was much more efficient in stimulating SCF in the sperm from either source than that of the epididymis (P < 0.0001). Vas deferens sperm were capable of initiating lower levels of SCF in the absence of luminal fluid (P < 0.0001).
LIMITATIONS, REASONS FOR CAUTION
Analyses were performed in only one species, the mouse, but we used three separate assays in our analysis.
WIDER IMPLICATIONS OF THE FINDINGS
The data suggest that the luminal fluid of the male reproductive tract interacts with sperm during their transit providing a mechanism to degrade the DNA. We hypothesize that this is part of an apoptotic-like mechanism that allows the reproductive tract to eliminate defective sperm. The SCF model also allowed us to identify differences in the types of DNA lesions that the three tests can identify, providing important background information for the use of these tests clinically.
STUDY FUNDING/COMPETING INTEREST(S)
Funding was obtained from the National Institutes of Health, USA Grant HD060722 to W.S.W. and SCSA Diagnostics, Brookings, SD, USA.
Two of the authors work for SCSA Diagnostics, and one owns the company and the patents.
TRIAL REGISTRATION NUMBER
Trial registration number is only required for clinical trials.
Keywords: sperm DNA damage, sperm chromatin structure assay, TUNEL assay, sperm chromatin
Introduction
Mammalian sperm DNA is the most compact eukaryotic chromatin known (Balhorn, 1982; Ward and Coffey, 1991), yet it is susceptible to damage during its formation in spermiogenesis (Evenson et al., 1980; Said et al., 2004; Perez-Crespo et al., 2006; Sakkas and Alvarez, 2010; Grenier et al., 2010) and by other agents, such as reactive oxygen species (ROS), in fully mature spermatozoa (Aitken et al., 2013). Some reports have suggested that mature sperm contain apoptotic-like mechanisms that respond to environmental stimuli (Taylor et al., 2004; Grunewald et al., 2005). Aitken et al. have proposed that sperm are in a perpetual state of near-apoptotic induced death that needs to be held at bay (Pujianto et al., 2010). They have also demonstrated that mammalian sperm have the first part of a DNA repair mechanism that could be completed after fertilization in the oocyte (Smith et al., 2013). When taken together, these data suggest that rather than being inert, the mature sperm cell retains some control over the integrity of its DNA.
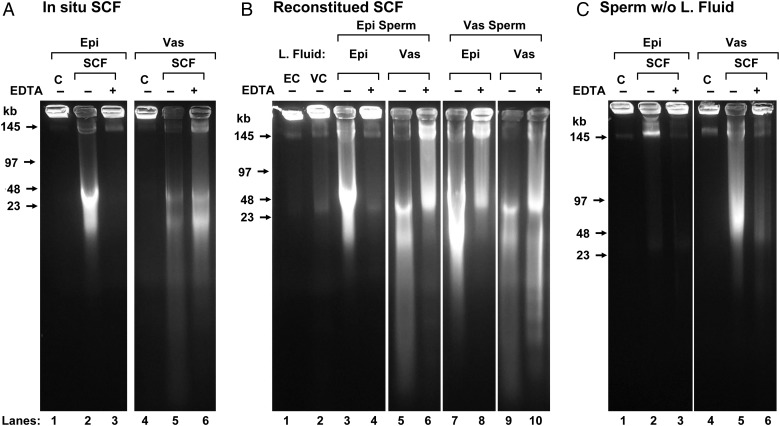
When mouse epididymal or vas deferens sperm are incubated with divalent cations in the presence of the luminal fluid in which they normally exist, they are induced to degrade their DNA into loop-sized fragments in a two-step process that we have termed sperm chromatin fragmentation (SCF) (Yamauchi et al., 2007a,b). The DNA is first degraded to 25–50 kb fragments, a process that can be reversed with EDTA, suggesting cleavage by topoisomerase II or a similar enzyme. Under certain conditions, this degradation proceeds to an irreversible digestion by another nuclease (Boaz et al., 2008). This is similar to apoptotic mechanisms that degrade eukaryotic DNA (Li et al., 1999; Widlak et al., 2000; Solovyan et al., 2002). The DNA degradation by SCF in sperm isolated from the vas deferens is greater than that of epididymal sperm, and is not fully reversible (Fig. 1A). We initially followed SCF by gel electrophoresis that can only detect double-stranded DNA breaks (dsDSBs), but recently we found that single-stranded DNA breaks (ssDSBs) are also induced by SCF, and for ssDSBs there was little difference between epididymal and vas deferens sperm (Ribas-Maynou et al., 2014).
Figure 1.
FIGE analysis of SCF. (A) In situ sperm chromatin fragmentation (SCF): epididymal (lanes 1–3) and vas deferens (lanes 4–6 sperm were induced to undergo SCF by incubation with Mn2+ and Ca2+ in the presence of their luminal fluid, without (lanes 2 and 5) or with subsequent EDTA treatment to reverse the double-stranded DNA breaks (dsDSBs) (lanes 3 and 6). Lanes 1–6 are reproduced from Yamauchi et al. (2007a). Control, untreated samples are shown in lanes 1 and 4 (B) reconstituted SCF: epididymal sperm were washed and resuspended in luminal fluid from the epididymis (lanes 3 and 4) or vas deferens (lanes 5 and 6) then induced to undergo SCF, without (lanes 3 and 5) or with subsequent EDTA (lanes 4 and 6). Vas deferens sperm were resuspended in luminal fluid from the epididymis (lanes 7 and 8) or vas deferens (lanes 9 and 10) then induced to undergo SCF, without (lanes 7 and 9) or with subsequent EDTA (lanes 8 and 10). Control, washed sperm from the epididymis (lane 1) and vas deferens (lane 2). (C) Sperm-induced to undergo SCF without luminal fluid: epididymal (lanes 1–3) and vas deferens (lanes 4–6) sperm were induced to undergo SCF without (lanes 2 and 5) or with (lanes 3 and 6) subsequent EDTA treatment. For each lane in the gel, two experiments were performed using one mouse each. Only one experiment is shown.
Here, we examined the role of the luminal fluids in the induction of SCF using three different analyses. We used field inversion gel electrophoresis (FIGE) to follow dsDSBs. We quantified the differences in DNA damage by SCF in mouse epididymal and vas deferens sperm using the sperm chromatin structure assay (SCSA®) (Evenson et al., 1999; Evenson, 2013) and the terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) assay (Smith et al., 2013). The data suggest that the luminal fluid is important for inducing SCF, and that sperm acquire the ability to initiate SCF as they mature in the male reproductive tract.
Methods
Animals
B6D2F1 (C57BL/6J X DBA/2) mice were obtained from the National Cancer Institute (Raleigh, NC, USA). Mice were kept in accordance with the guidelines of the Laboratory Animal Services at the University of Hawaii and those prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Resources National Research Council (DHEF publication no. [NIH] 80-23, revised 1985). The protocol for animal handling and the treatment procedures were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Hawaii. Mice were housed in standard cages with five or less mice per cage prior to being sacrificed for the experiments. In all experiments, male B6D2F1 mice that were 6 weeks to 2 months old were used. Mice were purchased from Charles River (Hollister, CA, USA).
Preparation of sperm
Mice were sacrificed by cervical dislocation, and epididymal and vas deferens sperm were collected separately in tris potassium cholride buffer (TKB) (100 mM KCl, 25 mM Tris, pH 7.5), or mHCZB (Yamauchi et al., 2007a,b) buffer (81.62 mM NaCl, 4.83 mM KCl, 1.18 mM KH2PO4, 5 mM CaCl2, 20 mM HEPES, 1 mM l-glutamine, 28 mM sodium lactate, 0.29 mM sodium pyruvate, 5.55 mM glucose), resuspended by gentle pipetting and incubated for 1 h at 37°C under four different conditions: controls were incubated without any additional chemicals; for SCF, 10 mM MnCl2 and 10 mM CaCl2 were added from stock solutions (1 M each); for SCF reversal (termed SCF-EDTA) 10 mM MnCl2 and 10 mM CaCl2 were added from stocks (as for SCF) and after the 1 h incubation EDTA was added from a 500 mM stock to100 mM final concentration and the sample was incubated for another 30 min 37°C. After treatment of sperm with divalent cations, all sperm were immotile. A separate series of sperm samples diluted in TKB were set aside for washed sperm samples. These samples were centrifuged briefly at 700 × g and their supernatants were collected into separate tubes. Then the pelleted sperm samples were washed two times with TKB, and resuspended in either TKB + 0.25% Triton X-100 (TX) (sperm, alone), or supernatant from the same (reconstituted) or other (mixed) preparations +0.25% TX and incubated for 1 h at 37°C.
Live/dead assay for SCF
Sperm from the epididymis or vas deferens were resuspended in mHCZB as described above, then treated without (control) or with 10 mM MgCl2 and 10 mM CaCl2 for 30 min at 37°C, then assayed using the Live/Dead Sperm Viability Kit (L-7011) from Invitrogen (Grand Island, NY, USA) according to manufacturer's specifications.
Analysis of sperm DNA degradation by FIGE
Plasma from the caudal epididymides and vas deferens of ∼8-week-old mice was extracted separately and suspended in mHCZB (Yamauchi et al., 2007a,b) to a final concentration of ∼108 sperm/ml. The suspension was mixed with agarose to a final concentration of 1% agarose and poured into molds making ∼5 mm thick plugs. The plugs were incubated at 37°C in TKB supplemented with 10 mM MnCl2 and 10 mM CaCl2 for 0, 15 min, 1 h or 4 h to initiate SCF or DNA degradation. For each time point, one plug was incubated in digestion buffer at 55°C to stop the reaction and one plug was incubated for 30 min with 30 mM EDTA to religate topoisomerase-induced strand breaks before stopping the reaction (Shaman et al., 2006). After the reaction, the plugs were incubated in digestion buffer (10 mM Tris, 5 mM EDTA, pH 7.8, 100 mM NaCl, 0.5% SDS and 20 mM dithiothreitol (DTT)) at 53°C for at least 30 min before placement in a 1% agarose gel for FIGE. For each experiment, sperm from one mouse was used. To reduce the number of mice used, sperm from the vas deferens and epididymis were taken from the same mouse. For this experiment, 22 mice were used because the experiment was repeated two times.
SCSA® protocol
After the different treatments described above, a small aliquot of each sample was taken for the analysis by gel electrophoresis and the rest of the sample was diluted to 500 μl TKB, flash frozen in an EtOH/dry ice bath and shipped to SCSA Diagnostics on dry ice overnight for analysis. After the arrival of the mouse sperm samples, they were transferred to a liquid nitrogen tank until time of measurements. Each individual sample was removed, thawed in a 37.0°C water bath for 30 s, and then an aliquot was transferred to TNE buffer (0.01 M Tris:0.15 M NaCl:0.001 M EDTA, pH 7.5) at 4°C to a final concentration of ∼1–2 × 106 sperm/ml. A total of 200 µl of this sperm suspension was admixed with 400 µl of a solution containing 0.08 N HCl:0.15 M NaCl:0.1% TX at 4°C. The sperm were stained 30 s later by adding 1.20 ml of staining solution containing 6 µg/ml AO (acridine orange, chromatographically purified, Polysciences, Inc., Warrington, PA, USA):0.2 M Na2HPO4 :0.1 M citric acid (pH 6.0):1 mM NaEDTA:0.15 M NaCl; so that the AO/DNA-P molar ratio was ≥2 (Darzynkiewicz et al., 1975). The acid/AO stained sample was placed in an Ortho Diagnostics, 2140 flow cytometer (FCM) sample chamber and sample flow was initiated to bring the sheath flow and sample flow to equilibrium by 2 min. Then 5000 sperm were analyzed at an event rate of 100–200 events/s. If the event rate was above 250 events/s, a new sample was prepared to ensure full equilibrium between the AO dye and sperm. The data were analyzed for the percentage of sperm with measurably increased red fluorescence (sperm with fragmented DNA) and those with high DNA stainability (HDS). For this experiment, one mouse was used for each of nine groups, and the experiment was repeated one time. Each preparation was analyzed two times.
SCSA® raw and SCSAsoft® computer re-oriented data
Sperm DNA data are seen as dot plots on the FCM oscilloscope screen. Each of the 5000 dots represents a single sperm that is characterized by the amount of green fluorescence in increments of 1/1024 (Y axis on a scale of 0–1024 units) and the amount of red fluorescence (X axis on a scale of 1–1024 units). The total fluorescent population on the X/Y axes is seen as cigar shaped due to the asymmetric shape and high density of the sperm head and measurements in a flow cytometer with orthogonal axes of laser beam and collecting lenses. This presents a potential problem in determining the exact amount of green and red fluorescence per sperm. Therefore, we process the sample file through our SCSAsoft® software (SCSA Diagnostics, Brookings, SD, USA) to re-orientate the data as total DNA stainability versus DNA fragmentation index (DFI: red/red + green fluorescence) that produces the total sperm signals as a vertical population from which a frequency histogram is accurately derived. Importantly, a ‘reference sperm sample constant’ is first established. For this sample and its replicates, the mean red fluorescence values are set at ∼125/1024 channels and green fluorescence values at ∼425/1024 channels. Then all experimental samples are measured at those same photomultiplier tube values (+5 channels). All experimental data are relative to this constant. The histogram signal is then divided up to represent sperm with non-denatured DNA (Box 1), moderate level of DNA denaturation (Box 2), high level of DNA denaturation (Box 3) and total %DFI (Box 2 + Box 3). The horizontal line at the top of the cigar shaped sperm signal is the threshold for the HDS population that represents sperm with an increase of dsDNA staining due to abnormally altered chromatin structure (see (Evenson et al., 2000) for example). All samples were measured four times and the mean and SD of those two values are calculated.
TUNEL assay
The protocol was based loosely on that by Smith et al. (2013). Epididymal and vas deferens spermatozoa were collected separately in TKB with 0.25% TX, resuspended by gentle pipetting and treated for 1 h at 37°C, as described above for SCF. After the treatments, sperm were centrifuged briefly, the supernatants were removed and the sperm were washed one time with TKB. Because we modified previous methods for TUNEL analysis, we treated mouse sperm with DNAse I to induce DNA breaks as a positive control. Sperm were suspended in TKB or 0.25% TX. The solution was brought to 20 mM MgSO4, 0.5 mM CaCl2 and 40 U/ml of DNAseI (New England Biolabs, Ipswich, MA, USA) was added. The suspension was incubated for 1 h at 37°C. All control and experimental samples were then resuspended in TKB with 2 mM DTT and incubated for 45 min at room temperature. After washing one time with phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, 1.8 mM KH2PO4: PBS), the sperm were fixed with 2% paraformaldehyde for 15 min on ice, washed three times with PBS and either stored in PBS with 0.1 M glycine at 4°C or processed immediately. The sperm were centrifuged briefly (700 g) and resuspended in fresh permeabilization buffer (0.1% TX in 0.1% sodium citrate) for 2 min on ice. The TUNEL staining was then performed using the In Situ Cell Death Detection Kit (Roche, Indianapolis, IN, USA) following manufacturer's instructions. Briefly, the cells were split into three separate tubes and treated as follows: no stain controls were incubated in PBS; label-only controls were incubated in label solution and the experimental samples were incubated in enzyme with label solution for 1 h at 37°C in a humidified chamber. The sperm were then washed three times for 5 min in PBS and analyzed by a fluorescence-activated cell sorter (FACS). At least 100 000 cells were assessed for TUNEL-positive (fluorescein isothiocyanate (FITC)) signal on BD FACSAria and analyzed using FACSDiva software (BD Biosciences, San Jose, CA, USA). Each experiment was performed at least three times. For this experiment, one mouse was used for each of nine groups, and the experiment was repeated two times. Results are shown as a TUNEL:label ratio of percentage FITC-positive cells.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. Each group in the SCSA and TUNEL assays was compared with every other group using Student's t-test. (GraphPad is a web-based resource, available at GraphPad.com). A value of P < 0.05 was considered statistically significant.
Results
Sperm viability when SCF is induced
The SCF that we induce in mouse sperm with divalent cations most likely activates a pathway in the majority of healthy sperm that is normally reserved for the degradation of damaged cells in the male reproductive tract. We tested whether the induction of SCF results in sperm cell death using the live/dead assay. We found that both epididymal (18.0 ± 8.4%, mean ± SD) and vas deferens sperm (16.7 ± 6.7%) had statistically significantly higher percentages of dead sperm (P < 0.0001) than the controls (78.0 ± 7.4% and 64.6± 6.5%, respectively). There was no difference between the two controls, or between the two SCF treated samples (Supplementary data, Table SI).
SCF in vas deferens sperm is more severe than that of epididymal sperm
Our previous analysis of SCF by FIGE suggested that there were many more DNA breaks in SCF-induced vas deferens sperm than in SCF-induced epididymal sperm (Fig. 1A, Lanes 1–6 are reproduced from Yamauchi et al. (2007a,b). However, because FIGE only identifies dsDSBs and SCF also induces ssDSBs (Ribas-Maynou et al., 2014), we used SCSA and the TUNEL assay to quantify the extent of DNA damage in SCF. Both techniques identify ssDSB and dsDSB. For each assay, treated sperm for controls and SCF-induction were prepared two times from different mice, and each sample was analyzed two times.
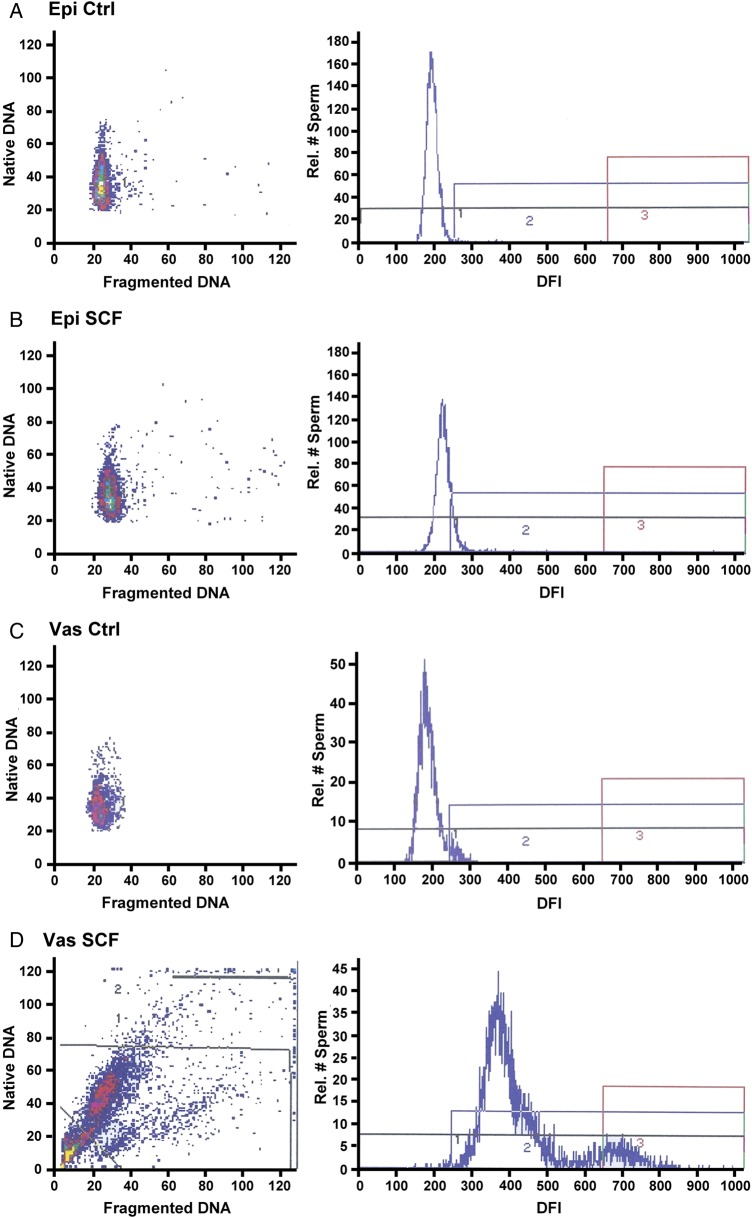
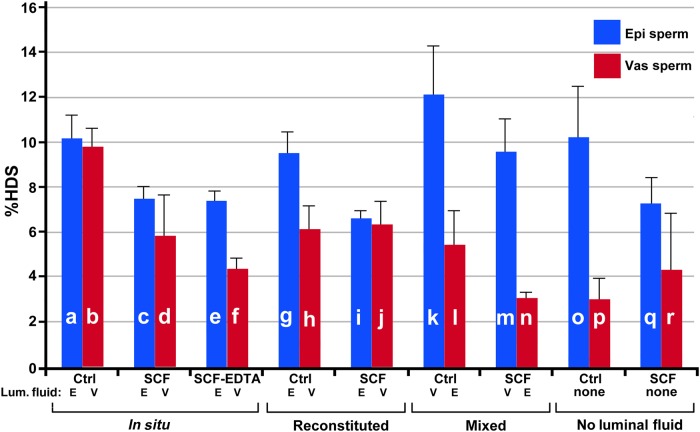
Representative samples for the DFI, the percentage of cells with measureable DNA damage, of the SCSA analysis for both epididymal and vas deferens spermatozoa are shown in Fig. 2. Epididymal sperm show only a slight increase in DFI when induced to undergo SCF (Fig. 2A and B). When vas deferens sperm are induced to undergo SCF, virtually all the sperm accumulate significant DNA damage (Fig. 2C and D). The average DFI values for these conditions are shown in histogram form in Fig. 3a–d. Note that treatment with EDTA did not reverse SCF-induced DNA breaks in either epididymal or vas deferens sperm (Fig. 3e and f). EDTA only reverses some dsDNA breaks (Fig. 1A, lane 3) but not ssDSBs. Since the SCSA test detects both dsDSBs and ssDSBs, it is likely that the amount of ssDSBs is much greater than dsDSBs, and therefore overwhelms the signal. The differences between epididymal and vas deferens sperm SCF were highly significant (Fig. 3c and d, Supplementary data, Table SII). The HDS values, which measure the extent of chromatin decondensation or aberration in packaging, did not vary greatly in these or any subsequent treatments never reaching above 12% (Fig. 4 and Supplementary data, Table SIII). This suggests that SCF does not induce global architectural changes in chromatin structure.
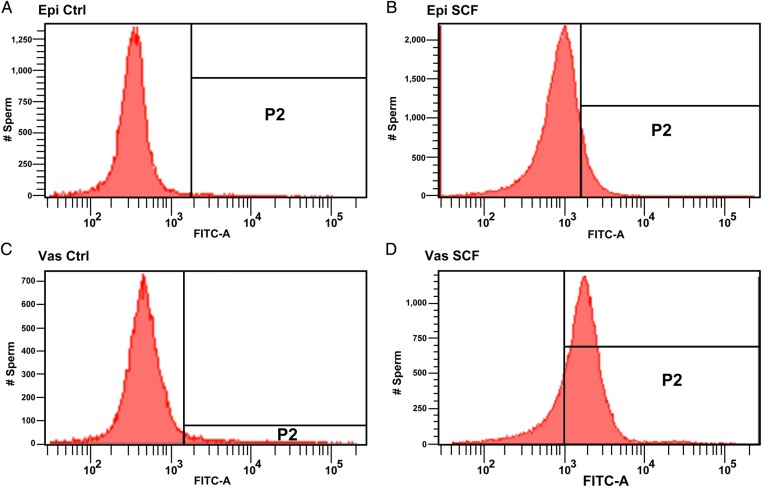
Figure 2.
Examples of DNA fragmentation index (DFI) plots for sperm chromatin structure assay (SCSA). Four examples of epididymal and vas deferens sperm in their luminal fluid without reconstitution. (A) Epididymal sperm control; (B) epididymal sperm induced to undergo sperm chromatin fragmentation (SCF); (C) vas deferens sperm; (D) vas deferens sperm induced to undergo SCF. Data are presented as DFI dot plots (native versus fragmented DNA) and the corresponding frequency histograms demarked as non-fragmented DNA (Box 1), moderate level DNA fragmentation (Box 2) and high level of DNA fragmentation (Box 3).
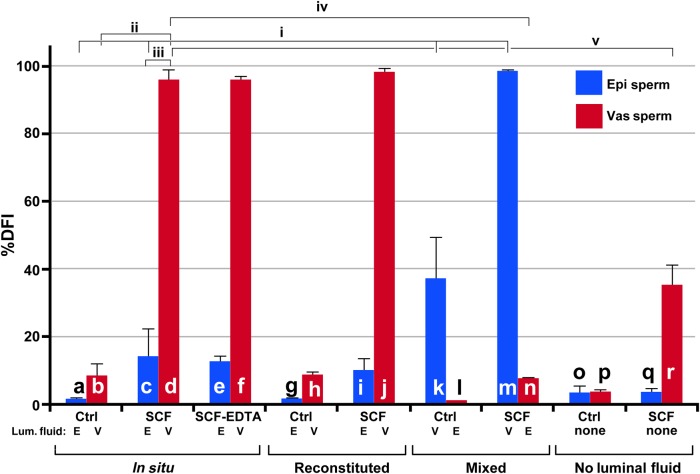
Figure 3.
SCSA analysis of SCF. Percentage DFI of SCF by the SCSA Assay. (a–f) In situ SCF: sperm samples were isolated and diluted with their luminal fluid. Control, untreated epididymal (a) and vas deferens (b) sperm. SCF induced epididymal (c) and vas deferens (d) sperm. SCF reversed with EDTA epididymal (e) and vas deferens (f) sperm; (g–l) reconstituted SCF: sperm were washed and reconstituted in the luminal fluid from the same organ; (g) Control, untreated epididymal sperm; (h) control, untreated vas deferens sperm; (i) SCF induced epididymal sperm; (j) SCF induced vas deferens sperm. (k–n) Mixed SCF: sperm were washed and resuspended in the other organ's luminal fluid: (k) control, epididymal sperm mixed with vas deferens luminal fluid; (l) control, vas deferens sperm mixed with epididymal luminal fluid; (m) epididymal sperm mixed with vas deferens luminal fluid then induced to undergo SCF; (n) vas deferens sperm mixed with epididymal luminal fluid then induced to undergo SCF. (o–r) Sperm induced to undergo SCF in the absence of luminal fluid: sperm were washed and resuspended in TKB; (o) control, epididymal sperm; (p) control, vas deferens sperm; (q) SCF induced epididymal sperm; (r) SCF induced vas deferens sperm. For each bar in the graph, n = 4, and one mouse was used for two experiments. The lines above the bars represent SD. Each experiment was compared with every other experiment in Student's t-test, and these values are shown in Supplementary data, Table SII. The most relevant comparisons are shown as brackets above the bars. These are: (i) SCF in epididymal sperm (c) differs from control (a), and both of these differ from control epididymal sperm incubated with vas deferens fluid (k) and from epididymal sperm induced to undergo SCF with vas deferens fluid (m) (P < 0.05 for all four comparisons); (ii) vas deferens sperm control (b) differs from vas deferens SCF (d) (P < 0.0001); (iii) epididymal sperm induced to undergo SCF (c) has significantly less DNA damage than vas deferens induced to undergo SCF (d) (P > 0.0001); (iv) vas deferens sperm induced to undergo SCF (d) has much more DNA damage than vas deferens sperm induced to undergo SCF in the presence of epididymal fluid (l) (P < 0.0001); (v) vas deferens sperm can undergo SCF without any luminal fluid present (r) but has significantly less DNA damage than vas deferens sperm with luminal fluid (d) (P < 0.0001).
Figure 4.
Chromatin condensation measurements by SCSA. High DNA staining (HDS) by the SCSA for all samples. All samples are exactly as labeled for Fig. 3. For each bar in the graph, n = 4, and one mouse was used for two experiments. The lines above the bars represent SD. None of the values for any experiment was larger than 12%, so HDS is not considered a major factor in SCF. Each experiment was compared with every other experiment in Student's t-test, and these values are shown in Supplementary data, Table SIII. There were several statistically significant differences in these data, although in almost every case, HDS decreased with SCF. The two controls (a and b) were not statistically different (P = 0.5726). In each of the three controls where the sperm were manipulated, epididymal sperm had a slightly higher HDS value than that of vas deferens sperm (g and h, P = 0.0064; k and l, P = 0.0047; o and p, P = 0.0023). In the last two SCF experiments where the sperm were manipulated, epididymal sperm had higher HDS values than vas deferens sperm (m and n, P = 0.0003; q and r, P = 0.0023).
These findings were verified with the TUNEL assay. Our positive control for the TUNEL assay using DNAseI to induce DNA breaks resulted in a strong positive signal, comparable with the strongest SCF signal we obtained (Supplementary data, Fig. S1). Representative examples are shown in Fig. 5. Once again, SCF-induced vas deferens sperm preparations had a higher percentage with significant DNA breaks than SCF-induced epididymal sperm (Fig. 6b and d: P < 0.0001 for comparisons between Fig. 6b and d; Supplementary data, Table SIV). In this case, however, there was less of a difference between SCF-induced epididymal and vas deferens control sperm samples. The TUNEL assay, like the SCSA, quantifies both ssDSB and dsDSB, so no reversal with EDTA was expected, and was not seen.
Figure 5.
Examples of terminal deoxynucleotidyltransferase-mediated dUTP nick-end labeling (TUNEL) Analysis for SCF. Four examples of epididymal and vas deferens sperm in their luminal fluid without reconstitution. (A) Epididymal sperm control; (B) epididymal sperm induced to undergo SCF; (C) vas deferens sperm and (D) vas deferens sperm induced to undergo SCF.
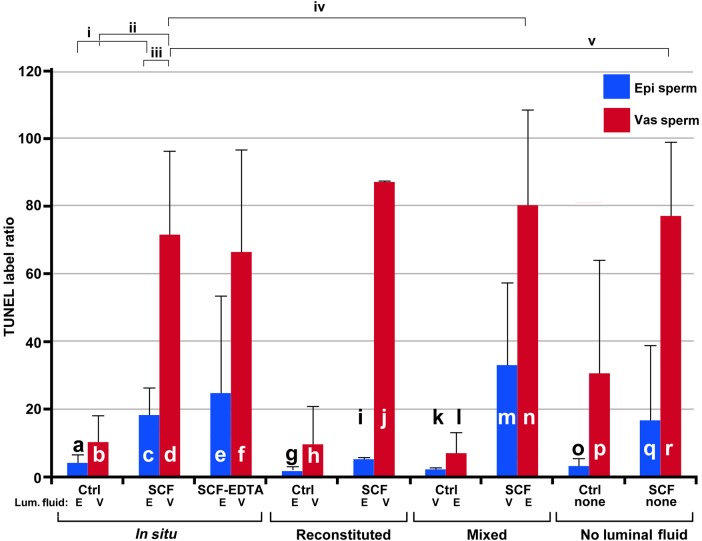
Figure 6.
TUNEL analysis for SCF. The TUNEL label ratio for each sample. Samples are labeled exactly as for Fig. 3. For each bar in the graph, n = 2, and one mouse was used for one experiment. The lines above the bars represent SD. Each experiment was compared with every other experiment in Student's t-test, and these values are shown in Supplementary data, Table SIV. The most relevant comparisons are shown as brackets above the bars. These are: (i) SCF in epididymal sperm (c) differs from control (a) (P < 0.0005); (ii) vas deferens sperm control (b) differs from vas deferens SCF (d) (P < 0.0001); (iii) epididymal sperm induced to undergo SCF (c) has significantly less DNA damage than vas deferens induced to undergo SCF (d) (P > 0.0001); (iv) vas deferens sperm induced to undergo SCF (d) has no statistically different level of DNA damage than vas deferens sperm induced to undergo SCF in the presence of epididymal fluid (l) (P < 0.5943); (v) vas deferens sperm that undergoes SCF without any luminal fluid present (r) has the same level of DNA damage as vas deferens sperm with luminal fluid (d) (P < 0.8).
SCF is largely mediated by the luminal fluid of the reproductive tract
To induce SCF, the luminal fluid that contains the sperm is extracted from the epididymis or the vas deferens and diluted with TKB, and the suspension is then incubated with divalent cations. We have already demonstrated that SCF induction degrades vas deferens sperm DNA much more than epididymal sperm and we tested whether this activity was an intrinsic property of the sperm or mediated by the luminal fluid in which the sperm reside. We centrifuged the sperm from the diluted luminal fluid from each organ, kept the supernatants and washed the sperm at least two more times. We then reconstituted the epididymal sperm with the diluted luminal fluid from either the epididymis or vas deferens, and did the same with vas deferens sperm. We found that the luminal fluid from the vas deferens induced a much greater DNA degradation in epididymal sperm that the epididymal fluid (compare lanes 3 and 5 in Fig. 1B), essentially making it appear identical to SCF of vas deferens sperm, either in situ (Fig. 1A, lane 5) or reconstituted (Fig. 1B, lane 9). Conversely, epididymal luminal fluid retarded the degradation of DNA in vas deferens sperm (compare lanes 7 and 9, Fig. 1B), though epididymal luminal fluid did not reduce SCF in vas deferens sperm to a level comparable with epididymal sperm SCF. This latter point is clear when comparing the reversal with EDTA. The dsDSBs of epididymal sperm induced to undergo SCF whether in situ (Fig. 1A, lane 2) or reconstituted (Fig. 1B, lane 3) completely reverse when treated with EDTA (Fig. 1A, lane 3 and Fig. 1B, lane 4, respectively). However, vas deferens sperm induced to undergo SCF in the presence of epididymal luminal fluid did not reverse completely (Fig. 1B, lane 6).
With SCSA we saw a similar effect. When epididymal sperm are reconstituted with vas deferens luminal fluid then induced to undergo SCF (Fig. 3k), the number of sperm with measureable DNA lesions is much higher than with epididymal luminal fluid (Fig. 3c and i; P < 0.05, for comparisons between Fig. 3k and c, or Fig. 3k and i; Supplementary data, Table SI). Conversely, the vas deferens sperm have much fewer DNA lesions when induced to undergo SCF in the presence of epididymal luminal fluid (Fig. 3n) than with vas deferens luminal fluid (Fig. 3d and j; P < 0.0001 for comparisons between Fig. 3d and k, or Fig. 3j and k; Supplementary data, Table SI). This effect was much greater with the SCSA test than was visible by FIGE, and this was probably because SCSA quantifies both ssDSBs and dsDSBs, while FIGE is only sensitive to dsDSBs. Nevertheless, both assays clearly show that vas deferens luminal fluid induces greater levels of SCF than epididymal fluid, regardless of the source of the sperm.
This effect, however, differed with the TUNEL assay. In this case, the number of epididymal sperm induced to undergo SCF in the presence of vas deferens luminal fluid (Fig. 6m) was greater than with epididymal luminal fluid (Fig. 6c and I; this did not reach statistical significance, Supplementary data, Table SIII), but still less than vas deferens SCF (Fig. 6d and j, P < 0.05; Supplementary data, Table SIII). Thus, while it is clear that in the TUNEL assay vas deferens luminal fluid increased the levels of DNA damage in SCF, there was some activity that was more associated with the source of the sperm. The apparent discrepancy between the source of the luminal fluid and extent of SCF between the FIGE analysis and SCSA test versus the TUNEL assay suggests that these three measures of DNA integrity in sperm detect different types of lesions. This will be addressed in the Discussion section.
Vas deferens sperm acquire the ability to degrade their DNA through SCF
While it is clear that the luminal fluid contributes to the stability of sperm DNA, the TUNEL analysis also suggested that vas deferens sperm are more susceptible to DNA degradation by SCF. We therefore tested the ability of the sperm cells, alone, to cleave their chromatin when incubated with divalent cations. When sperm were isolated and washed five times to remove the luminal fluid then incubated with divalent cations, epididymal sperm DNA was largely intact (Fig. 1C, lane 2) but vas deferens sperm DNA was degraded to a similar extent as epididymal SCF (compare Fig. 1C, lane 5 with Fig. 1A, lane 2). This suggested that vas deferens sperm have acquired the ability to induce DNA degradation even in the absence of the luminal fluid, although to not as great an extent. Note that in this case, the vas deferens SCF was reversible (Fig. 1C, lane 6) suggesting that DNA degradation was limited to the topoisomerase-like degradation previously reported (Shaman et al., 2006). Both the SCSA test (Fig. 3q and r) and the TUNEL assay (Fig. 6q and r) confirmed this finding.
Discussion
SCF occurs in condensed chromatin
The SCSA test allowed us to test a long-held hypothesis of ours that SCF occurs in the fully condensed sperm chromatin. The HDS of SCSA is a measure of the compaction of the sperm chromatin, and this did not change significantly over all the conditions tested (Fig. 4). In many cases, the SCF treated sperm had lower HDS values than the controls (for example, compare Fig. 4a and b with c and d, respectively). Moreover, the vas deferens sperm consistently had lower HDS values than the epididymal sperm, even though the latter digested its DNA less. Thus, there is no correlation between chromatin compaction and the level of degradation by inducing SCF. This is an important concept because of the level of sperm DNA degradation. Mammalian sperm DNA is so tightly packaged that nucleases cannot digest most of the DNA (Hud et al., 1995; Sotolongo et al., 2003; Vilfan et al., 2004). The fact that SCF so completely digests the DNA without decondensing the chromatin suggests that the DNA fragmentation is intimately related to the chromatin structure.
SCF depends on the luminal fluid for complete activity, but the sperm acquire the ability to initiate SCF during it transit through the male reproductive tract
Each of the three tests, we used in this work confirmed that much of the activity in SCF depends on the presence of the luminal fluid. Epididymal sperm that were washed and resuspended in TKB could not be induced to undergo SCF, and vas deferens sperm only initiated the first step of SCF without luminal fluid. Moreover, in all three analyses, incubation of epididymal sperm with vas deferens luminal fluid increased the level of DNA damage by SCF over that seen when the epididymal luminal fluid was used. Conversely, in the FIGE and SCSA analyses, DNA damage in vas deferens sperm SCF was much reduced when the vas deferens luminal fluid was replaced with epididymal luminal fluid. (We believe that the apparent discrepancy of the TUNEL analysis on this point is due to steric hindrance of TdT, as discussed below)
These data suggest three properties of SCF. First, the luminal fluid of both the epididymis and the vas deferens can induce SCF in sperm. Second, the luminal fluid of the vas deferens induces a much stronger SCF reaction in sperm. Third, sperm acquire the ability to initiate the first step of SCF during their transit through the epididymis to the vas deferens. We have previously suggested that the luminal fluid contains a sequestered nuclease, possibly in epididymosome-like structures (Frenette et al., 2006; Schwarz et al., 2013; Akintayo et al., 2015) that are part of an apoptotic-like mechanism that allows for the activation of sperm self-destruction if the sperm cell is damaged in some way.
Three types of lesions in SCF and their relation to sperm chromatin structure
We have identified three types of DNA breaks in SCF: ssDSBs (Ribas-Maynou et al., 2014), reversible dsDSBs, and non-reversible dsDSBs (Sotolongo and Ward, 2000; Shaman et al., 2006). The data suggest that the reversible dsDSBs are also the ‘hidden’ dsDSBs that remain attached to the sperm nuclear matrix and are therefore not detected by conventional neutral Comet Assays (Ribas-Maynou et al., 2014). It is likely that the reversible dsDSBs are caused by topoisomerase II or similar enzyme (Shaman et al., 2006). The irreversible dsDSBs are probably caused by nucleases that enter from the luminal fluid (Shaman et al., 2006; Boaz et al., 2008; Dominguez and Ward, 2009). The ssDSBs may also be caused by the nucleases, but are probably also generated by ROS that are released from the mitochondria in damaged sperm (Aitken and Baker, 2004; Aitken and De Iuliis, 2010; Smith et al., 2013). Because SCF occurs in mature sperm with no treatment other than nonionic detergent and divalent cations, the distribution of these different types of lesions is dependent on, and somewhat limited by, the structure of the sperm chromatin.
Our model for sperm chromatin includes most known elements of sperm DNA compaction. The large majority of sperm DNA, 90–99% depending on the species (Tanphaichitr et al., 1978; Hammoud et al., 2009; Brykczynska et al., 2010), is sequestered by the protamines into crystalline-like toroids that are relatively insensitive to nuclease and other forms of DNA damaging agents (Hud et al., 1995). Each protamine toroid is connected by sections of chromatin that are nuclease sensitive, the toroid linkers (Ward, 2010). These linkers include matrix attachment regions (MARs) by which the DNA is attached to the sperm nuclear matrix. Thus, there are three major fractions of sperm chromatin structure with different accessibilities (Fig. 7). The protamine bound DNA is relatively insensitive to external insults, but would be expected to be somewhat susceptible to attack by ROS, and possibly some nucleases. The toroid linker regions would be the most sensitive to nuclease attack. We suggest that many of the ssDSBs are therefore located in the protamine toroids. We hypothesize that some of the ssDSBs and most of the irreversible dsDSBs are located in this region. The MARs are most likely the site of the reversible dsDSBs of SCF, just as they are in somatic cell apoptosis (Li et al., 1999; Solovyan et al., 2002).
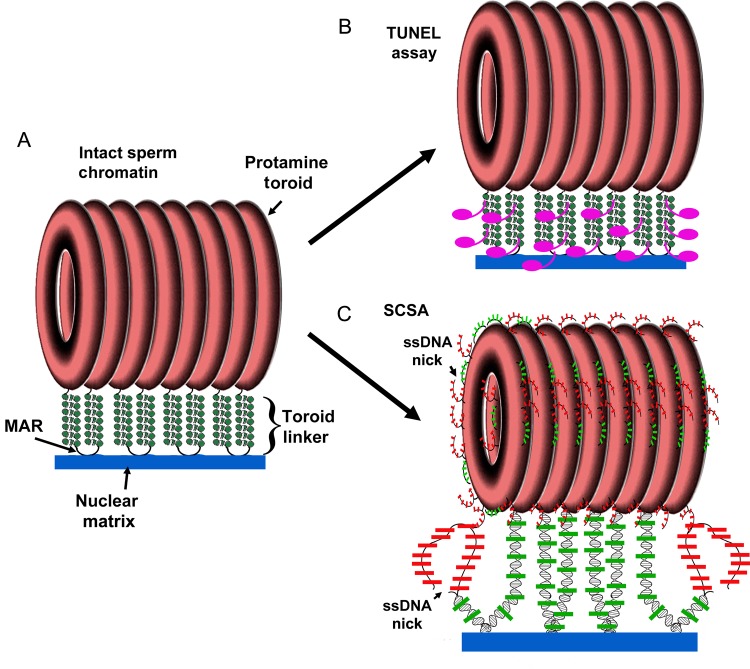
Figure 7.
Model for the abilities of SCSA and TUNEL to detect sperm DNA damage. (A) Sperm DNA is organized into protamine toroids that are linked by nuclease sensitive linkers. (B) The terminal deoxynucleotidyl transferase (TdT) enzyme would be expected to be able to access these linker regions, but not the toroids. (C) The SCSA assay first denatures DNA with acid at sites of existing single and double DNA strand breaks, which also extracts the histones (minimal in mice). Acridine orange then stains the double stranded DNA green and single stranded DNA red. Because it is a very small molecule, it would be expected to access most parts of the sperm chromatin. This model is a modified version of a previously published diagram (Shaman and Ward, 2006).
TUNEL and SCSA identify different DNA lesions in SCF
Gel electrophoresis is able to detect both types of dsDSBs but cannot detect ssDSBs. Unlike the FIGE analysis, both the SCSA and TUNEL assays detect ssDSBs and dsDSBs. But what role, if any, does the chromatin structure play in the ability of these two tests to detect DNA lesions created by SCF? The TUNEL assay requires the enzyme terminal deoxynucleotidyl transferase (TdT) to add dUTP to nicked or broken DNA ends (Gavrieli et al., 1992). However, because of the high degree of compaction of sperm chromatin, its requirement for TdT almost certainly restricts its access to a limited fraction of the in situ DNA. In sharp contrast, the SCSA assay requires only AO, a small planar molecule (MW 265), much smaller than any protein, suggesting that it would detect lesions in a broader fraction of the compact sperm chromatin (Evenson et al., 1999; Evenson, 2013). Both the SCSA and the TUNEL assay are currently used for assessing DNA damage in sperm. The SCSA assay was the pioneering sperm DNA fragmentation assay (Evenson et al., 1980, 1985) followed by the sperm TUNEL assay (Henkel et al., 2003; Sharma et al., 2013). The SCSA has been the most extensively used assay for human (Evenson et al., 1980, 1999; Virro et al., 2004) and animal (Ballachey et al., 1987; Evenson, 1999) fertility studies, as well as human (Rubes et al., 2005) and animal (Evenson et al., 1986; Sailer et al., 1995) toxicology studies. The TUNEL assay has been used mostly for human fertility (Sharma et al., 2013) and chromatin structure studies (Sutovsky et al., 2002; Smith et al., 2013). Both the TUNEL and SCSA would be expected to detect all three types of lesions, albeit with some limitations, discussed below. Based on our model of sperm chromatin structure (Sotolongo et al., 2003; Ward, 2010), we previously suggested that the detection of DNA lesions by both the SCSA and TUNEL assays would be limited to the toroid-linker regions between the protamine bound toroids of sperm chromatin (Shaman and Ward, 2006). In this work, we were able to directly test the potential similarities of these two tests using the same model for sperm DNA damage, and discovered a significant difference prompting us to modify our model for the types of DNA damage they can detect in sperm (Fig. 7).
We found a discrepancy between the SCSA and TUNEL assays when sperm were mixed with luminal fluid from the other organ (compare Fig. 3k–m with Fig. 6k–m). Both assays indicated that vas deferens SCF was more severe than epididymal SCF, and that vas deferens sperm digested their DNA to a greater extent than epididymal sperm in the absence of luminal fluid. However, the SCSA assay showed a much higher DFI in epididymal sperm mixed with vas deferens luminal fluid (Fig. 3m) as compared with vas deferens sperm mixed with epididymal fluid (Fig. 3n). The TUNEL assay, conversely, indicated a higher level of DNA damage in vas deferens sperm regardless of the type of luminal fluid used (Fig. 6m and n). We suggest that this discrepancy is related to the chromatin structure. Because TUNEL requires the enzyme TdT to label the DNA breaks, it will probably have access only to the toroid linker regions (Fig. 7A). Since these toroid linker regions are accessible by nucleases (Sotolongo et al., 2003) they should also be accessible to TdT and certainly to AO. This is the region where SCF is initiated, and the part of the chromatin where SCF-induced sperm induce DNA breaks in the absence of luminal fluid. Because sperm acquire the ability to degrade their own DNA as they migrate through the reproductive tract, vas deferens sperm are more susceptible to SCF activation. Therefore, because the TUNEL assay is focused on the toroid linker because of steric hindrance, the DNA damage it detects will be more related to the source of the sperm than on the luminal fluid. On the other hand, the SCSA assay is not limited by steric hindrance of an enzyme, and could detect DNA damage at least on the outer fibers of the protamine toroid. It is likely that SCF induces ssDSBs created by a combination of nucleases and ROS, so they would probably be dispersed throughout the sperm genome and present in the protamine toroids. SCSA would then detect a greater level of DNA damage. Our data indicate that the full effect of SCF requires the luminal fluid, and our data also suggest that the level of ssDSBs is greater than dsDSBs in SCF. Therefore, when DNA damage is detected outside the protamine linkers, the major factor for SCF is the luminal fluid, not the source of the sperm.
Potential physiological role of SCF
It is important to stress that the SCF that we have induced by treating sperm with divalent cations is an experimental activation of what we believe is a normal physiological process. It is probable that the normal, physiological mechanism for SCF-type DNA degradation in sperm is a discrete molecular pathway that has yet to be unveiled. The work in this manuscript verifies that it does exist, and that it is largely controlled by the luminal fluid of the vas deferens and to some extent that of the epididymis with a contribution from the sperm that is acquired during maturation. After treatment of sperm with the divalent cations used to induce SCF, all sperm were immotile and therefore not capable of normal fertilization. We hypothesize that the SCF that we induce experimentally with cations activates a mechanism for the inactivation of the sperm DNA by degradation that is similar to apoptosis. We suggest that the vas deferens has a mechanism to identify dead or functionally deficient sperm and degrade their DNA to ensure that they do not participate in fertilization. However, much further work will be needed to identify this mechanism.
Conclusions
We used three different assays to assess a model for sperm DNA damage during SCF. We found that sperm acquire the ability to degrade their DNA as they move through the male reproductive tract, and that the luminal fluid plays an important role in the SCF mechanism. SCF is also a useful model to understand the different types of lesions that sperm DNA damage assays detect. We recently demonstrated how SCF could be used to identify a new type of lesion we termed hidden dsDSBs (Ribas-Maynou et al., 2014). Here, we provided evidence that the TUNEL assay is useful for detecting lesions in the toroid linker regions, which may also be the sites of DNA synthesis in the zygote (Shaman et al., 2007), while the SCSA assay is useful for detecting a broader range of DNA damage.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors' roles
J.E.G., S.B., K.K. and H.N. made substantial contributions to design, acquisition of data and data analysis in this work. D.P.E. and W.S.W. both made substantial contributions to design, acquisition of data and data analysis and revising the article critically for important intellectual content. W.S.W. was responsible for the final approval of the version to be published.
Funding
This work was supported by National Institutes of Health, USA (Grant HD060722) to W.S.W. and SCSA Diagnostics, Brookings, SD, USA.
Conflict of interest
D.P.E. is owner of SCSA Diagnostics and holds and the patents for the SCSA test. K.K. works for SCSA Diagnostics.
Supplementary Material
References
- Aitken RJ, Baker MA. Oxidative stress and male reproductive biology. Reprod Fertil Dev 2004;16:581–588. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, De Iuliis GN. On the possible origins of DNA damage in human spermatozoa. Mol Hum Reprod 2010;16:3–13. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Bronson R, Smith TB, De Iuliis GN. The source and significance of DNA damage in human spermatozoa; a commentary on diagnostic strategies and straw man fallacies. Mol Hum Reprod 2013;19:475–485. [DOI] [PubMed] [Google Scholar]
- Akintayo A, Legare C, Sullivan R. Dicarbonyl l-xylulose reductase (DCXR), a “moonlighting protein” in the bovine epididymis. PLoS One 2015;10:e0120869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R. A model for the structure of chromatin in mammalian sperm. J Cell Biol 1982;93:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballachey BE, Hohenboken WD, Evenson DP. Heterogeneity of sperm nuclear chromatin structure and its relationship to bull fertility. Biol Reprod 1987;36:915–925. [DOI] [PubMed] [Google Scholar]
- Boaz SM, Dominguez KM, Shaman JA, Ward WS. Mouse spermatozoa contain a nuclease that is activated by pretreatment with EGTA and subsequent calcium incubation. J Cell Biochem 2008;103:1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 2010;17:679–687. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Traganos F, Sharpless T, Melamed MR. Thermal denaturation of DNA in situ as studied by acridine orange staining and automated cytofluorometry. Exp Cell Res 1975;90:411–428. [DOI] [PubMed] [Google Scholar]
- Dominguez K, Ward WS. A novel nuclease activity that is activated by Ca(2+) chelated to EGTA. Syst Biol Reprod Med 2009;55:193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson DP. Loss of livestock breeding efficiency due to uncompensable sperm nuclear defects. Reprod Fertil Dev 1999;11:1–15. [DOI] [PubMed] [Google Scholar]
- Evenson DP. Sperm chromatin structure assay (SCSA(R)). Methods Mol Biol, 2013;927:147–164. [DOI] [PubMed] [Google Scholar]
- Evenson DP, Darzynkiewicz Z, Melamed MR. Relation of mammalian sperm chromatin heterogeneity to fertility. Science 1980;210:1131–1133. [DOI] [PubMed] [Google Scholar]
- Evenson DP, Higgins PJ, Grueneberg D, Ballachey BE. Flow cytometric analysis of mouse spermatogenic function following exposure to ethylnitrosourea. Cytometry 1985;6:238–253. [DOI] [PubMed] [Google Scholar]
- Evenson DP, Baer RK, Jost LK, Gesch RW. Toxicity of thiotepa on mouse spermatogenesis as determined by dual-parameter flow cytometry. Toxicol Appl Pharmacol 1986;82:151–163. [DOI] [PubMed] [Google Scholar]
- Evenson DP, Jost LK, Marshall D, Zinaman MJ, Clegg E, Purvis K, de Angelis P, Claussen OP. Utility of the sperm chromatin structure assay as a diagnostic and prognostic tool in the human fertility clinic. Hum Reprod 1999;14:1039–1049. [DOI] [PubMed] [Google Scholar]
- Evenson DP, Jost LK, Corzett M, Balhorn R. Characteristics of human sperm chromatin structure following an episode of influenza and high fever: a case study. J Androl 2000;21:739–746. [PubMed] [Google Scholar]
- Frenette G, Girouard J, Sullivan R. Comparison between epididymosomes collected in the intraluminal compartment of the bovine caput and cauda epididymidis. Biol Reprod 2006;75:885–890. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol 1992;119:493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier L, Robaire B, Hales BF. Paternal exposure to cyclophosphamide affects the progression of sperm chromatin decondensation and activates a DNA damage response in the prepronuclear rat zygote. Biol Reprod 2010;83:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald S, Paasch U, Said TM, Sharma RK, Glander HJ, Agarwal A. Caspase activation in human spermatozoa in response to physiological and pathological stimuli. Fertil Steril 2005;83(Suppl 1):1106–1112. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature 2009;460:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel R, Kierspel E, Hajimohammad M, Stalf T, Hoogendijk C, Mehnert C, Menkveld R, Schill WB, Kruger TF. DNA fragmentation of spermatozoa and assisted reproduction technology. Reprod Biomed Online 2003;7:477–484. [DOI] [PubMed] [Google Scholar]
- Hud NV, Downing KH, Balhorn R. A constant radius of curvature model for the organization of DNA in toroidal condensates. Proc Natl Acad Sci USA 1995;92:3581–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Chen AY, Yu C, Mao Y, Wang H, Liu LF. Activation of topoisomerase II-mediated excision of chromosomal DNA loops during oxidative stress. Genes Dev 1999;13:1553–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Crespo M, Pericuesta E, Rey R, Gutierrez-Adan A. OC6 scrotal heat stress in mice affects viability and DNA integrity of sperm, and sex ratio of the offspring. Reprod Domest Anim 2006;41(Suppl 2):104. [Google Scholar]
- Pujianto DA, Curry BJ, Aitken RJ. Prolactin exerts a prosurvival effect on human spermatozoa via mechanisms that involve the stimulation of Akt phosphorylation and suppression of caspase activation and capacitation. Endocrinology 2010;151:1269–1279. [DOI] [PubMed] [Google Scholar]
- Ribas-Maynou J, Gawecka JE, Benet J, Ward WS. Double-stranded DNA breaks hidden in the neutral Comet assay suggest a role of the sperm nuclear matrix in DNA integrity maintenance. Mol Hum Reprod 2014;20:330–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubes J, Selevan SG, Evenson DP, Zudova D, Vozdova M, Zudova Z, Robbins WA, Perreault SD. Episodic air pollution is associated with increased DNA fragmentation in human sperm without other changes in semen quality. Hum Reprod 2005;20:2776–2783. [DOI] [PubMed] [Google Scholar]
- Said TM, Paasch U, Glander HJ, Agarwal A. Role of caspases in male infertility. Hum Reprod Update 2004;10:39–51. [DOI] [PubMed] [Google Scholar]
- Sailer BL, Jost LK, Erickson KR, Tajiran MA, Evenson DP. Effects of X-irradiation on mouse testicular cells and sperm chromatin structure. Environ Mol Mutagen 1995;25:23–30. [DOI] [PubMed] [Google Scholar]
- Sakkas D, Alvarez JG. Sperm DNA fragmentation: mechanisms of origin, impact on reproductive outcome, and analysis. Fertil Steril 2010;93:1027–1036. [DOI] [PubMed] [Google Scholar]
- Schwarz A, Wennemuth G, Post H, Brandenburger T, Aumuller G, Wilhelm B. Vesicular transfer of membrane components to bovine epididymal spermatozoa. Cell Tissue Res 2013;353:549–561. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Ward WS. Sperm Chromatin Stability and Susceptibility to Damage in Relation to its Structure. Cambridge: Cambridge University Press, 2006. [Google Scholar]
- Shaman JA, Prisztoka R, Ward WS. Topoisomerase IIB and an extracellular nuclease interact to digest sperm DNA in an apoptotic-like manner. Biol Reprod 2006;75:741–748. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. The sperm nuclear matrix is required for paternal DNA replication. J Cell Biochem 2007;102:680–688. [DOI] [PubMed] [Google Scholar]
- Sharma R, Masaki J, Agarwal A. Sperm DNA fragmentation analysis using the TUNEL assay. Methods Mol Biol 2013;927:121–136. [DOI] [PubMed] [Google Scholar]
- Smith TB, Dun MD, Smith ND, Curry BJ, Connaughton HS, Aitken RJ. The presence of a truncated base excision repair pathway in human spermatozoa that is mediated by OGG1. J Cell Sci 2013;126:1488–1497. [DOI] [PubMed] [Google Scholar]
- Solovyan VT, Bezvenyuk ZA, Salminen A, Austin CA, Courtney MJ. The role of topoisomerase II in the excision of DNA loop domains during apoptosis. J Biol Chem 2002;277:21458–21467. [DOI] [PubMed] [Google Scholar]
- Sotolongo B, Ward WS. DNA loop domain organization: the three dimensional genomic code. J Cell Biochem 2000;35:23–26. [DOI] [PubMed] [Google Scholar]
- Sotolongo B, Lino E, Ward WS. Ability of hamster spermatozoa to digest their own DNA. Biol Reprod 2003;69:2029–2035. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Neuber E, Schatten G. Ubiquitin-dependent sperm quality control mechanism recognizes spermatozoa with DNA defects as revealed by dual ubiquitin-TUNEL assay. Mol Reprod Dev 2002;61:406–413. [DOI] [PubMed] [Google Scholar]
- Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Exp. Cell Res. 1978;117:347–356. [DOI] [PubMed] [Google Scholar]
- Taylor SL, Weng SL, Fox P, Duran EH, Morshedi MS, Oehninger S, Beebe SJ. Somatic cell apoptosis markers and pathways in human ejaculated sperm: potential utility as indicators of sperm quality. Mol Hum Reprod 2004;10:825–834. [DOI] [PubMed] [Google Scholar]
- Vilfan ID, Conwell CC, Hud NV. Formation of native-like mammalian sperm cell chromatin with folded bull protamine. J Biol Chem 2004;279:20088–20095. [DOI] [PubMed] [Google Scholar]
- Virro MR, Larson-Cook KL, Evenson DP. Sperm chromatin structure assay (SCSA) parameters are related to fertilization, blastocyst development, and ongoing pregnancy in in vitro fertilization and intracytoplasmic sperm injection cycles. Fertil Steril 2004;81:1289–1295. [DOI] [PubMed] [Google Scholar]
- Ward WS. Function of sperm chromatin structural elements in fertilization and development. Mol Hum Reprod 2010;16:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod 1991;44:569–574. [DOI] [PubMed] [Google Scholar]
- Widlak P, Li P, Wang X, Garrard WT. Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J Biol Chem 2000;275:8226–8232. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Boaz SM, Ward WS. Paternal pronuclear DNA degradation is functionally linked to DNA replication in mouse oocytes. Biol Reprod 2007a;77:407–415. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Shaman JA, Ward WS. Topoisomerase II mediated breaks in spermatozoa cause the specific degradation of paternal DNA in fertilized oocytes. Biol Reprod 2007b;76:666–672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.