Mutations in SPAST, which encodes the microtubule-severing protein spastin, are the commonest cause of hereditary spastic paraplegia type 4. However, increasing evidence suggests that diminished microtubule severing cannot fully explain the disease. Solowska and Baas review these data and discuss various alternative pathological mechanisms, many involving the spastin isoform M1.
Keywords: hereditary spastic paraplegia, SPAST, spastin, microtubule endoplasmic reticulum, endosome
Mutations in SPAST, which encodes the microtubule-severing protein spastin, are the commonest cause of hereditary spastic paraplegia type 4. However, increasing evidence suggests that diminished microtubule severing cannot fully explain the disease. Solowska and Baas review these data and discuss various alternative pathological mechanisms, many involving the spastin isoform M1.
Abstract
Mutations in more than 70 distinct loci and more than 50 mutated gene products have been identified in patients with hereditary spastic paraplegias, a diverse group of neurological disorders characterized predominantly, but not exclusively, by progressive lower limb spasticity and weakness resulting from distal degeneration of corticospinal tract axons. Mutations in the SPAST (previously known as SPG4) gene that encodes the microtubule-severing protein called spastin, are the most common cause of the disease. The aetiology of the disease is poorly understood, but partial loss of microtubule-severing activity resulting from inactivating mutations in one SPAST allele is the most postulated explanation. Microtubule severing is important for regulating various aspects of the microtubule array, including microtubule number, length, and mobility. In addition, higher numbers of dynamic plus-ends of microtubules, resulting from microtubule-severing events, may play a role in endosomal tubulation and fission. Even so, there is growing evidence that decreased severing of microtubules does not fully explain HSP-SPG4. The presence of two translation initiation codons in SPAST allows synthesis of two spastin isoforms: a full-length isoform called M1 and a slightly shorter isoform called M87. M87 is more abundant in both neuronal and non-neuronal tissues. Studies on rodents suggest that M1 is only readily detected in adult spinal cord, which is where nerve degeneration mainly occurs in humans with HSP-SPG4. M1, due to its hydrophobic N-terminal domain not shared by M87, may insert into endoplasmic reticulum membrane, and together with reticulons, atlastin and REEP1, may play a role in the morphogenesis of this organelle. Some mutated spastins may act in dominant-negative fashion to lower microtubule-severing activity, but others have detrimental effects on neurons without further lowering microtubule severing. The observed adverse effects on microtubule dynamics, axonal transport, endoplasmic reticulum, and endosomal trafficking are likely caused not only by diminished severing of microtubules, but also by neurotoxicity of mutant spastin proteins, chiefly M1. Some large deletions in SPAST might also affect the function of adjacent genes, further complicating the aetiology of the disease.
Introduction
Hereditary spastic paraplegia (HSPs) are inherited disorders with a prevalence of 1.8/100 000 in most populations (Ruano et al., 2014). Progressive lower limb spasticity and weakness are the predominant but not exclusive manifestations of the disease. Limited post-mortem studies on HSP patients have consistently revealed distal-end degeneration of ascending sensory fibres and corticospinal tract axons. Corticospinal axons descending from the large pyramidal neurons in layer V of motor cortex can include some of the longest CNS axons and tend to be vulnerable to upper motor neuron diseases. While some corticospinal axons synapse directly with lower motor neurons, most establish synapses with spinal interneurons that form connections with lower motor neurons innervating skeletal muscles. Even late in the progression of HSPs, there is generally very little neuronal cell death, which is consistent with the pathology being one of nerve degeneration. Mutations in more than 70 distinct loci (SPG1–72) and more than 50 mutated gene products have been identified in patients with HSPs (Lo Giudice et al., 2014; Novarino et al., 2014). While they have diverse functions, proteins encoded by HSP genes cluster within a small number of predicted cellular activities such as membrane traffic and organelle shaping, mitochondria regulation, myelination and lipid/sterol modification, axonal path-finding and axonal transport. Whether or not mutations in these various genes produce HSP pathology by a common, similar or different mechanism remains unknown. The clinical presentations and cellular pathways of HSPs have been previously reviewed (Salinas et al., 2008; Blackstone, 2012; Fink, 2013; Lo Giudice et al., 2014).
Mutations in the SPAST gene (located on 2p22.3), which encodes for an enzyme called spastin, are the most common cause of HSP and, depending upon the ethnic background of patients, account for 15–40% of all HSP cases (Hazan et al., 1999; Fonknechten et al., 2000; Shoukier et al., 2009). Autosomal-dominant HSP-SPG4 in most cases is considered a prototypical ‘pure’ or ‘uncomplicated’ HSP with gait impairment due to spasticity and weakness of the lower extremities (each of variable degree and age-of-onset), but without loss of function in the upper limbs or diminished life expectancy. In some cases, HSP-SPG4 patients also manifest cognitive impairment (Orlacchio et al., 2004; Shoukier et al., 2009), cerebellar ataxia (Nielsen et al., 2004), thin corpus callosum (Orlacchio et al., 2004) or lower motor neuron dysfunction (McDermott et al., 2006), and therefore their syndromes are classified clinically as ‘complicated.’ The key characteristics of HSP-SPG4 are summarized in Table 1.
Table 1.
Clinical characteristics of HSP-SPG4
| Affected gene | SPAST (previously known as SPG-4) mapped to 2p22.3 |
|---|---|
| Protein | Spastin, a microtubule-severing ATP-ase |
| Mutations | Missense, nonsense, splice site, insertions, small and large deletions |
| Inheritance | Autosomal dominant |
| Clinical presentation | Uncomplicated HSP, in some cases late onset of cognitive impairment. No genotype–phenotype correlation. |
| Genetic penetrance | Age dependent, as high as 80–90% |
| Age of onset | Infancy through senescence. Average age of onset 29 ± 17 years. |
| Functional impairment | Highly variable from slight stiffness in the legs to wheelchair bound. |
| Progression | Faster in patients with late-onset of the disease |
Structure and function of spastin, the protein encoded by SPAST
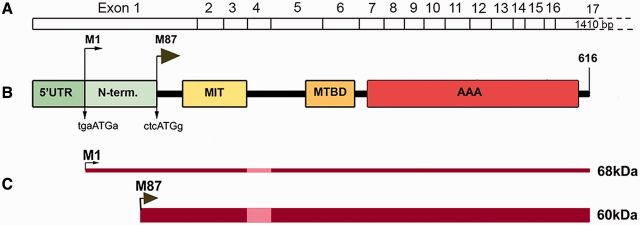
The SPAST gene spans the region of ∼90 kb of genomic DNA and contains 17 exons. Human spastin, encoded by SPAST, is a member of the AAA (ATPase associated with various cellular activities) protein family. The domain organization of spastin is presented in Fig. 1. The spastin AAA cassette contains three conserved ATPase domains, including Walker motif A (amino acids GPPGNGKT in positions 382–389) corresponding to the ATP-binding domain, Walker motif B (amino acids IIFIDE in positions 437–442), and the AAA minimal consensus sequence in position 480–498 (Hazan et al., 1999). Spastin open reading frame (1848 base pairs) flanked by 5’UTR and 3’UTR has two initiation codons (Claudiani et al., 2005). A weak Kozak sequence tgaATGa surrounding the M1 initiation codon deviates considerably from a good consensus sequence g(a)ccATGg and leads to leaky scanning of the first AUG. A better Kozak sequence ctcATGg is present at the M87 initiation codon (Kozak, 2002; Claudiani et al., 2005). As a result, a 616 amino acid (68 kDa) isoform called M1 and a 530 amino acid (60 kDa) isoform called M87 are synthesized simultaneously but at different levels, as depicted in Fig. 1C. In rodents, the shorter isoform is the predominant spastin isoform in all tissues at all stages of development, whereas M1 is only detectably present in adult spinal cord (Solowska et al., 2008). Similarly, analysis of spastin expression in adult human CNS has revealed the presence of M87 both in spinal cord and cerebral cortex, whereas significant levels of M1 were detected only in spinal cord and not in brain (Solowska et al., 2010). Two additional spastin isoforms might be generated as a result of alternative mRNA splicing of exon 4 (Fig. 1C) (Svenson et al., 2001; Claudiani et al., 2005).
Figure 1.
Schematic representation of spastin structure. (A) Spastin exons 1–17. (B) Spastin functional domains: N-term = N-terminal sequence present only in M1 spastin isoform; MIT = microtubule interacting and trafficking domain; MTBD = microtubule-binding domain; AAA = ATPase associated with various cellular activities. The Kozak’s sequence tgaAUGa surrounding M1 start codon deviates considerably from a good consensus sequence g(a)ccAUGg. A better Kozak sequence ctcAUGg is present at the M87 initiation codon. (C) A leaky scanning of the first initiation codon with a poor Kozak’s sequence leads to a preferred initiation of translation at the second AUG. As a result, both 68 kDa M1 and 60 kDa M87 spastin isoforms are expressed simultaneously but at different levels. A thin dark red line represents low levels of M1 expression and a dark red bar represents considerably higher levels of M87. Light red represents the M1 and M87 regions that are not present in spastin isoforms encoded by alternatively spliced mRNA lacking exon 4.
Spastin is a microtubule-severing ATPase that breaks longer microtubules into shorter ones and thereby regulates the number and mobility of microtubules and the distribution of their dynamic plus-ends (Errico et al., 2002; Evans et al., 2005; Roll-Mecak and Vale, 2005; Baas et al., 2006). To sever microtubules, spastin assembles into hexamers that dock on microtubules and break them by tugging the negatively charged C-terminal of tubulin through the central pore of the hexamer (White et al., 2007; Roll-Mecak and Vale, 2008). Polyglutamylation of tubulin stimulates spastin-mediated severing most likely because of an increase in negative charge of the microtubule tails (Lacroix et al., 2010). Because modifications such as polyglutamylation tend to correspond to microtubule stability, spastin has a stronger proclivity for severing in the stable region of axonal microtubules as opposed to the more labile/dynamic region situated toward the microtubule’s plus-end. Consistent with this, knockdown of spastin tends to shift the microtubule mass of the axon toward a higher proportion of the more stable regions (Riano et al., 2009).
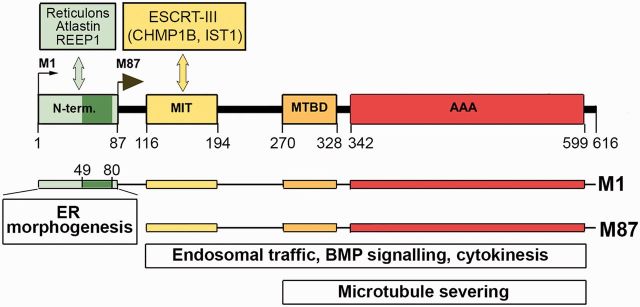
Figure 2 illustrates schematically the interactions of spastin functional domains with other proteins involved in different cellular activities. Functional analyses of truncated spastin cDNAs have revealed that the microtubule-binding domain (MTBD) situated between residues 270–328, and ATPase AAA domain, spanning residues 342–599, are sufficient for hexamerization and microtubule severing (White et al., 2007). The microtubule interacting and trafficking (MIT) domain, spanning residues 116–194, is required for interaction with two endosomal sorting complex required for transport III (ESCRT-III) proteins, charged multivesicular body protein 1B (CHMP1B) and increased sodium tolerance (IST1) (Reid et al., 2005; Yang et al., 2008; Connell et al., 2009; Blackstone et al., 2011; Guizetti et al., 2011; Allison et al., 2013).
Figure 2.
M1 and M87 interacting proteins and involvement of spastin isoforms in cellular functions. Insertion of hydrophobic region (dark green, amino acids 49–80) of the M1-specific N-terminal domain (amino acids 1–87) into endoplasmic reticulum (ER) membrane and interaction with reticulons, atlastin and REEP1 is responsible for endoplasmic reticulum morphogenesis. Interaction of MIT domain (amino acids 116–194) with ESCRT-III complex proteins is required for endosomal trafficking and cytokinesis. Microtubule-binding domain (MTBD) (amino acids 270–328) and AAA domain (amino acids 342–599) are essential for microtubule severing as well as endosomal trafficking and cytokinesis. BMP = bone morphogenic protein.
ESCRT-III proteins are required for completion of abscission at the end stage of cytokinesis, when the two daughter cells are physically severed from one another. CHMP1B recruits spastin to the midbody, presumably to sever microtubules and enable full constriction at the abscission site. Depletion of spastin from HeLa cells results in delayed abscission because the microtubule disruption that normally accompanies abscission does not occur (Yang et al., 2008; Connell et al., 2009; Guizetti et al., 2011). However, this function may be redundant with another protein or pathway, as spastin depletion in vivo does not result in any defects in cell division.
Recruitment of spastin into the ESCRT-III complex facilitated by IST1 might promote fission of recycling tubules from the endosome, one of the steps in a process that controls the balance between receptor degradation and recycling (Blackstone et al., 2011; Allison et al., 2013). Depletion of spastin by siRNA from HeLa cells leads to increased tubulation of the early endosomal compartment and results in defective receptor sorting through endosomal tubular recycling compartments. An increase in complex tubular structures has also been observed in axonal growth cones of motor neurons cultured from spastin-depleted zebrafish embryos (Allison et al., 2013).
Analyses of endogenous spastin distribution in HeLa cells and rat cortical neurons have revealed co-localization of M87 spastin with the centrosomal protein NA14, suggesting that NA14 might act as a molecular adaptor involved in targeting spastin to the centrosome and midbody, to facilitate microtubule severing in these specific locations (Errico et al., 2004; Goyal et al., 2014). Spastin activity is also implicated in downregulation of bone morphogenic protein (BMP) signalling (Tsang et al., 2009), which regulates axonal growth, guidance and differentiation during mammalian development (Charron and Tessier-Lavigne, 2007).
As shown in Fig. 2, the MIT, MTBD and AAA domains are present in both M1 and M87 spastin isoforms, whereas the 86-amino acid N-terminal domain is present only in M1 spastin. The M1 hydrophobic region spanning amino acids 49–80 has been suggested to form a hairpin that can partially insert into endoplasmic reticulum membrane. Spastin hydrophobic hairpins can interact with hydrophobic hairpins of atlastin 1 (encoded by ATL1) and REEP1 (Evans et al., 2006; Sanderson et al., 2006; Park et al., 2010; Blackstone et al., 2011). Atlastin 1 is a large integral- membrane GTPase that mediates homotypic fusion of endoplasmic reticulum tubules (Hu et al., 2009). Mutations of atlastin 1 are the cause of HSP-SPG3A, the second most common HSP (Zhao et al., 2001). REEP1 (receptor expression enhancing protein 1) is an endoplasmic reticulum morphogen that links endoplasmic reticulum membranes to microtubules. Mutations in REEP1 have been found in patients with HSP-SPG31 (Beetz et al., 2008). Interaction of spastin with the endoplasmic reticulum-shaping protein reticulon 1 (RTN1) has also been reported (Mannan et al., 2006). These findings suggest that coordinated interaction of atlastin 1, M1 spastin and REEP1 might be important for shaping of the endoplasmic reticulum tubules through hydrophobic wedging and for endoplasmic reticulum–microtubule interactions that build the tubular endoplasmic reticulum network (Park et al., 2010; Blackstone et al., 2011, 2012; Lumb et al., 2012). It is unclear whether M1 inserted into endoplasmic reticulum membrane would at the same time form hexamers with M87 to sever microtubules.
Loss-of-function model of HSP-SPG4
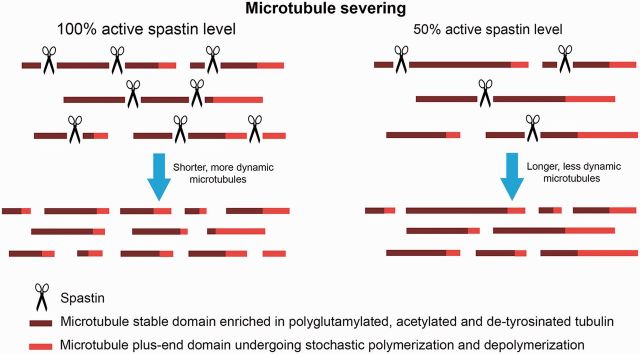
Over 200 different mutations have been found in SPAST (Hazan et al., 1999; Fonknechten et al., 2000; Lindsey et al., 2000; Shoukier et al., 2009). Missense mutations are clustered mainly in the AAA domain, while nonsense, splice site mutations and insertions/deletions have been found in all locations in SPAST. Interestingly, no mutations have been detected in SPAST exon 4, which might be alternatively spliced (Svenson et al., 2001; Shoukier et al., 2009). It has been also demonstrated that 18–20% of patients carry large deletions in the spastin gene (Beetz et al., 2006; Depienne et al., 2007; Boone et al., 2014). The spectrum of mutations found in HSP-SPG4 has prompted a loss-of-function explanation for the disease. As most pathogenic mutations in SPAST affect spastin domains necessary for its hexamerization, microtubule binding, or ATP hydrolysis, inadequate microtubule severing resulting from inactivation of one spastin allele (haploinsufficiency) has become a postulated explanation for the symptoms of HSP-SPG4 (Hazan et al., 1999; Burger et al., 2000; Fonknechten et al., 2000). Figure 3 schematically depicts the hypothetical consequences of the partial loss of spastin activity on microtubule severing and dynamics. This model posits that spastin expression levels are the limiting factor in microtubule severing. Loss-of-function is a plausible mechanism for HSP-SPG4 if indeed the number of severing events is directly proportional to the concentration of spastin and if a decrease in the number of severing events is detrimental specifically to a subset of corticospinal axons.
Figure 3.
Hypothetical effects of spastin depletion on microtubule severing and dynamics. The model posits that the levels of spastin control the extent of microtubule severing and that a lower concentration of spastin leads directly to a proportionally lower number of severing events.
In neurons, severing of long microtubules into shorter ones is critical for efficient microtubule transport because only short microtubules are able to move in a rapid and concerted fashion within the axon (Wang and Brown, 2002; Baas et al., 2006). Microtubule severing also creates greater numbers of free, dynamic microtubule plus-ends that can interact with plus-end associated proteins and cellular cortical structures (Roll-Mecak and Vale, 2006). An abundance of short mobile microtubules and free microtubule ends, as well as a greater fraction of labile/dynamic microtubule mass, is important for axonal growth and for the formation of new axonal branches, all of which are compromised in cultured rodent neurons depleted of spastin (Yu et al., 2008; Riano et al., 2009; Qiang et al., 2010).
Drosophila model of HSP-SPG4
Negative effects of spastin depletion on axon outgrowth and branching in cultured rodent neurons suggest the importance of microtubule severing during development. Indeed, morpholino-based knockdown of spastin from developing zebrafish embryos results in impaired outgrowth of axons from spinal and branchiomotor neurons (Wood et al., 2006). In Drosophila, however, loss of spastin function does not result in such developmental deficits of axonal outgrowth. Instead, ubiquitous depletion of spastin (encoded by spas) with RNAi or deletion of the entire SPAST gene (homozygous null spastin mutation) resulted in low eclosion rates of ∼20% and decreased neuromuscular junction synaptic area with an increased number of synaptic buttons in third instar larvae. The effects of lack of spastin on neuromuscular junction synaptic area were likely due to inhibition of microtubule severing, as the levels of acetylated tubulin at the neuromuscular junction presynaptic terminals become elevated while the total tubulin levels are reduced, indicating a shift toward a greater fraction of stable microtubules (Sherwood et al., 2004; Trotta et al., 2004; Orso et al., 2005). Spas-null flies that survived to adulthood were short-lived and could not fly or jump. Interestingly, partial or complete deletion of the first exon of Drosophila SPAST, which should abolish protein synthesis due to a removal of translation initiation codon(s), resulted in a milder phenotype than removal of the entire gene. Homozygous flies lacking the first spastin exon eclosed at normal frequencies and exhibited less severe motor defects than flies with the entire spastin gene deleted (Sherwood et al., 2004).
When depletion of spastin with RNAi was limited to the CNS, newly eclosed flies exhibited normal motor functions and their brains had normal anatomical and histological organization. Over time, however, locomotor performance declined more rapidly in experimental than in control flies, and signs of neurodegeneration began to appear in their brains (Orso et al., 2005). It has been also reported that loss of one copy of the spastin gene severely disrupted the capacity of Drosophila axons to regenerate (Stone et al., 2012).
Mouse model of HSP-SPG4
For the purpose of evaluating the significance of spastin loss in mammals, two spastin knockout mouse models have been generated and analysed (Tarrade et al., 2006; Kasher et al., 2009). In the first model, spastin exons 5–7 were deleted via homologous recombination. As a result of the accompanying frame-shift, a termination codon was created 29 base pairs downstream from the new exon 4–8 junction. Interestingly, truncated spastin transcripts lacking exons 5–7 were readily detected in various tissues including brain, indicating that nonsense-mediated mRNA decay did not completely eliminate the mRNA with a premature termination codon. Truncated spastin protein, however, was not detected in brain (Tarrade et al., 2006). In the second mouse model, exon 7 of spastin was deleted as a result of a point mutation in the splice donor site. Sequencing revealed that the mutated transcription product would encode truncated spastin with 50 novel C-terminal amino acids followed by a termination codon. Again, truncated mRNA was detected but truncated protein was not detected in mouse brain (Kasher et al., 2009). Neither of these spastin mutations generated in mice led to developmental abnormalities, and compared to many human HSP-SPG4 patients carrying just one copy of mutated Spast, homozygous Spast knockout mice had only mild motor defects and heterozygous mice did not exhibit any detectable gait abnormalities.
Human neuronal model of HSP-SPG4
To generate human models of HSP-SPG4, fibroblasts from one patient with G>T substitution located in SPAST exon 4 (Denton et al., 2014) and from two patients carrying SPAST R562X mutation (Havlicek et al., 2014) were infected with lentiviruses carrying pluripotency genes SOX2, POU5F1 (also known as OCT3/4), KLF4 and MYC (also known as c-MYC). Subsequently, the induced pluripotent stem cells were grown either in neural differentiation medium and differentiated into telencephalic glutamatergic neurons expressing TBR1+ (Denton et al., 2014) or underwent spontaneous undirected differentiation into mostly glutamatergic neurons, expressing CTIP2 (encoded by BCL11B) protein found in vivo in layer 5/6 neurons (Havlicek et al., 2014). Identically treated fibroblasts from healthy individuals were used as controls. SPG4 patient-derived neurons show decreased expression of wild-type spastin and axonal swellings filled with loosely arranged, fragmented microtubules and accumulated mitochondria. The number of swellings in heterozygous SPG4 patient-derived neurons was significantly higher than the number of swellings in cortical neuron cultures from homozygous Spast knockout mice. One of the two studies found that the levels of acetylated tubulin in SPG4 patient-derived neurons were no higher than in control neurons with the authors suggesting that microtubule dynamics were relatively normal due to the increase in expression of katanin (another microtubule-severing protein) (Havlicek et al., 2014). The other study, however, reported a dramatic increase in acetylated tubulin levels in SPG4-derived neurons (Denton et al., 2014). In addition, SPG4 patient-derived neurons had lower numbers of shorter and less branched primary neurites, which is similar to the phenotype observed when rat hippocampal neurons in culture were depleted of spastin by siRNA (Qiang et al., 2010). How these observations apply to the nerve degeneration in HSP remains unclear, as such deficits in neurite outgrowth have not been reported in HSP-SPG4 patients. As mutated spastin proteins were not detected, the phenotypes observed in SPG4 patient-derived neurons were attributed to the loss of spastin microtubule-severing activity that cannot be fully substituted by other severing enzymes.
Perturbation in axonal transport in HSP-SPG4
The most prominent phenomenon observed in HSP-SPG4 mammalian models has been the presence of focal axonal swellings. In homozygous mice, such swellings present in both descending and ascending tracts of the spinal cord are filled with organelles and filaments. Axonal swellings were also found in SPG4 patient-derived neurons and sections of spinal cord from two HSP-SPG4 patients (Kasher et al., 2009). These findings might be best explained by perturbations of axonal transport. In cultured neurons derived from one of the mouse models of HSP-SPG4, selective reduction of anterograde transport was found (Kasher et al., 2009). On this basis, it was proposed that spastin depletion results in misregulation of microtubule dynamics, leading to increased cargo stalling, possibly by disturbing cargo loading on microtubules (Fassier et al., 2013). This explanation, however, is not entirely satisfactory for various reasons. First, axonal swellings have not always been observed in cultured neurons depleted of spastin by siRNA (Yu et al., 2008; Qiang et al., 2010) and were not reported in Drosophila models (Sherwood et al., 2004; Trotta et al., 2004). Second, it has been shown that kinesin 1, the motor protein responsible for most anterograde organelle transport in the axon, preferentially moves on stable microtubules rich in acetylated and detyrosinated α-tubulin (Cai et al., 2009), and such modifications are not less but rather more numerous on the microtubules of spastin-depleted cells (Sherwood et al., 2004; Trotta et al., 2004; Orso et al., 2005; Riano et al., 2009; Fassier et al., 2013). It has also been shown that endoplasmic reticulum extension (sliding) along microtubules is facilitated by tubulin acetylation (Friedman et al., 2010). In SPG4 patient-derived neurons, retrograde transport was decreased and anterograde transport seemed to be only marginally affected (Denton et al., 2014; Havlicek et al., 2014).
Finally, while severing increases the number of microtubule plus-ends that could then undergo polymerization and depolymerization, there is no indication that the rate of microtubule polymerization or depolymerization, within itself, is affected by wild-type spastin. Indeed, no decrease in microtubule polymerization has been found in spastin-depleted cultured neurons (Qiang et al., 2010; Fassier et al., 2013). It is, therefore, unclear why the use of vinblastine at nanomolar concentrations resulted in improved motor function in Drosophila (Orso et al., 2005) as well as decreased axonal swellings in spastin-depleted neuronal cells (Fassier et al., 2013) or SPG4 patient-derived neurons (Denton et al., 2014). At this concentration vinblastine is a ‘kinetic stabilizer’ of microtubules that inhibits both the frequency and the rate of microtubule polymerization and depolymerization (Yang et al., 2010; Baas and Ahmad, 2013), not a destabilizer that would increase microtubule dynamics as has been proposed in these studies. As microtubule-modifying drugs have been suggested as a potential therapy for HSP-SPG4, further studies and clarification on these issues is of great practical importance. Interestingly, sub-stoichiometric concentrations of either vinblastine or taxol restored neurite outgrowth in SPG3A patient-derived forebrain neurons expressing mutated atlastin 1 protein (Zhu et al., 2014), suggesting that microtubules might be a therapeutic target in various HSPs.
Loss of spastin activity in HSP-SPG4 patients
Most (but not all) human patients carrying a single mutated SPAST allele exhibit a phenotype characteristic of HSP, indicating high but not complete, age-dependent genetic penetrance of 50% at age 27 and 80% at 50 years. Approximately 6% of individuals with SPAST mutations are completely asymptomatic (Dürr et al., 2012). The mean age at onset, reported to be 29 ± 17 years, encompasses a broad range of ages from early childhood to late adulthood onset (1 to 74 years). The disability levels vary from patients being able to walk but not run or walk with aid, to patients being wheelchair-bound. Typically, the level of disability notably increases with the duration of the disease. Despite that different SPAST mutations presumably decrease spastin activity to different degrees, no clear correlation has been identified between the type of mutation and the severity of the phenotype (Fonknechten et al., 2000; Yip et al., 2003; McDermott et al., 2006; Shoukier et al., 2009).
Theoretically, full-length spastins carrying missense mutations could have dominant-negative activity and thereby further decrease activity of normal spastin encoded by the wild-type allele. As a result, patients carrying such mutations would have less than half of normal spastin activity. On the other end of the spectrum, some SPAST splice site mutations result in only slight reduction in wild-type spastin mRNA expression (Svenson et al., 2001), and there are patients with typical HSP-SPG4 symptoms who carry missense mutations that are localized outside the MTBD and AAA domains and do not affect spastin microtubule-severing activity (Sauter et al., 2002; Patrono et al., 2005; Crippa et al., 2006; Solowska et al., 2010). Some of these mutations localized in the MIT domain might, however, encode spastins unable to interact with ESCRT-III proteins and hence unable to participate in endosomal dynamics.
The compound heterozygosity for the missense S44L and P361L or D470V SPAST mutations causes a rare severe infantile HSP (Chinnery et al., 2004; Svenson et al., 2004). P361L or D470V mutations are localized in the AAA domain and most likely inactivate spastin. The S44L mutation is located outside of the DNA region encoding the M87 isoform and does not affect the severing activity of M87 (Solowska et al., 2010). The S44L mutation found in ∼0.6–3% of the population is asymptomatic in the overwhelming majority of carriers and therefore is considered a polymorphism that might act as a phenotypic modifier (Svenson et al., 2004; Erichsen et al., 2007). It has been suggested, however, that the S44L mutation might affect a cryptic M87 promoter in exon 1 and thereby downregulate the expression of the M87 isoform (Mancuso and Rugarli, 2008), or it might increase the stability of the M1 isoform (Schickel et al., 2007). Infantile onset of HSP was also diagnosed in cases of heterozygous missense SPAST mutations such as G471D (Blair et al., 2007) and D613H (but not D613A) (Aulitzky et al., 2014), and there are cases in which the compound heterozygosity (e.g. S44L and R503W) did not result in infantile onset of HSP-SPG4 (Shoukier et al., 2009), indicating once again that the severity of the HSP-SPG4 cannot be predicted on the basis of SPAST mutation type. Significant clinical variability is also typically observed in families where all individuals have the identical mutation in the SPAST gene, and therefore presumably experience the same loss of spastin function (Meijer et al., 2002). Such apparent lack of correlation between the severity of HSP-SPG4 phenotype and the degree to which spastin activity is lost due to a particular SPAST mutation suggests that factors other than loss of active spastin contribute to the aetiology of the disease.
Mutated spastin proteins in HSP-SPG4
The overwhelming majority of mutations found in HSP-SPG4 patients would abolish microtubule-severing activity of spastin encoded by the mutated SPAST allele and theoretically result in microtubules that are lower in number but more stable (Fig. 3). In human patients, such loss of microtubule severing does not, however, lead to abnormalities during nervous system development, which is when greater microtubule mobility and a higher fraction of labile/dynamic microtubule mass are particularly important to accommodate the growth of axons (Baas et al., 2006). One possibility is that reducing active spastin levels has no functional impact because the total levels of spastin and other microtubule-severing proteins such as katanin are higher during development than in the adult (Solowska et al., 2008). Another possibility, however, is that insufficient microtubule severing, within itself, does not adequately explain the disease and that a toxic gain-of-function mechanism contributes to HSP-SPG4 pathology. A comparison of the phenotypes observed in the spastin knockout mouse with that of cattle with bovine spinal dysmyelination strongly suggests that both mutated spastin proteins and the loss of spastin function might play a role in the aetiology of HSP-SPG4. Motor defects observed in homozygous knockout mice are generally mild compared to those suffered by many human patients carrying just one mutated SPAST allele (Tarrade et al., 2006; Kasher et al., 2009). In cattle with bovine spinal dysmyelination, a naturally occurring neurodegenerative disease caused by the missense inactivating R560Q spastin mutation, heterozygous carriers are asymptomatic (Thomsen et al., 2010). As the genetic penetrance of HSP-SPG4 is highly age-dependent in humans, it is possible that the animals carrying heterozygous R560Q mutation simply did not live long enough to develop symptoms. Bovine natural lifespan is ∼25 years, but in modern dairy farming, lifespans usually do not exceed 5–7 years. Interestingly, in humans heterozygous SPAST mutations R562Q (Meijer et al., 2002), R562G (Svenson et al., 2001) or R562X (Fonknechten et al., 2000; Meijer et al., 2002) that correspond to bovine R560Q result in HSP.
Calves homozygous for the R560Q spastin mutation, unlike knockout mice, manifest clinical signs immediately after birth with complete penetrance of the phenotype. Newborns are alert to their surroundings but cannot raise or move their limbs. Microscopic examination indicated bilateral symmetrical dysmyelination of axons in the cervical and thoracic spinal cords of affected animals. Such myelination defects were not observed in any other parts of the nervous system. Ultrastructural examination showed the presence of slightly swollen axons with accumulation of disoriented microtubules and intermediate filaments as well as the presence of highly swollen axons filled with organelles such as mitochondria and lysosomal bodies (Thomsen et al., 2010). Interestingly, some loss of myelin has also been observed in spinal cords of heterozygous HSP-SPG4 patients and accompanying decreases in phosphorylated epitopes of neurofilament and tubulin proteins has suggested that the dysmyelination occurred in the context of axonal degeneration (Wharton et al., 2003).
The question arises as to why phenotypes observed in the absence of active spastin are so profoundly different in two mammalian species. One possibility is that mice are particularly resistant to spastin inactivation. That, however, would make mice an unsuitable model for human HSP-SPG4. The other and more likely explanation lies in different expression of mutated spastin proteins in those two models. In mouse models carrying truncating mutations, despite the presence of corresponding truncated transcripts, proteins have not been detected, suggesting that such proteins are efficiently degraded. The R560Q missense mutation, however, would produce full-length inactive spastin, and experimental results indicate that such spastin is metabolically as stable as wild-type spastin (Solowska et al., 2010, 2014). Therefore the major difference between mice and bovine, each equally depleted of spastin microtubule-severing activity but exhibiting significantly different phenotypes, likely lies in the presence or absence of mutated spastin proteins (Table 2).
Table 2.
Animal models for spastin mutations
| Organism | Mutation | Predicted spastin inactivation | Predicted level of mutated spastin | Onset/severity |
|---|---|---|---|---|
| Mouse | Homozygous deletion exons 5–7 | 100% | Not detected | Late onset, mild |
| Mouse | Homozygous splice site mutation | 100% | Not detected | Late onset, mild |
| Bovine | Homozygous R560Q mutation | 100% | Wild-type level | Early onset, very severe |
Missense SPAST mutations
Neurotoxic proteins are a central feature of many neurodegenerative diseases, but toxicity of spastins has gone relatively unstudied mainly because it is often argued that most mutated spastins are expressed at levels that are too low to be toxic (Burger et al., 2000). There are, however, compelling reasons why neurotoxicity of mutated spastins should not be dismissed. About 30% of SPAST mutations are missense mutations, which are expected to produce stable mutated spastin mRNAs and proteins. Full-length spastins with missense mutations could act in dominant-negative fashion to lower the activity of spastin encoded by the wild-type allele. In studies on Drosophila, RNAi knockdown or expression of spastin carrying the missense K467R mutation resulted in similar phenotypes, suggesting that K468R mutated protein acting through a dominant-negative mechanism interferes with the function of endogenous spastin (Orso et al., 2005). In human patients, however, missense SPAST mutations do not lead to more severe symptoms that would be expected if wild-type spastin activity were further lowered by dominant-negative inhibition of function. In vitro experiments have shown that while mutated spastin may increase the length of the so-called pre-severing phase (time from the addition of spastin and ATP to microtubules immobilized on microscopic coverslips and the start of severing) the actual severing of microtubules by wild-type spastin was not affected by the presence of mutated spastin (Eckert et al., 2012). Also, the M1 or M87 isoform carrying the pathogenic C448Y mutation did not diminish severing activity of wild-type M87 when the two were co-expressed in fibroblasts (Solowska et al., 2014). Therefore, not all SPAST missense mutations generate proteins with dominant-negative properties.
Truncating SPAST mutations
Many of the splice site mutations and insertions/deletions in SPAST (with the exception of in-frame exon deletions) could generate premature termination codons. Premature termination codons are also created by missense point mutations. While the majority of mRNAs with premature termination codons do undergo a rapid decay, some escape degradation and are stable. In fact, truncated mRNAs have been readily detected in mouse models with premature termination codons generated in their SPAST gene (Tarrade et al., 2006; Kasher et al., 2009). During mRNA splicing, a multi-subunit protein complex termed the exon junction complex is deposited 20–24 nucleotides upstream of the exon–exon junction and then removed from the mRNA by the elongating ribosome. Generally, premature termination codons located more than 50–55 nucleotides upstream of the exon-exon junction prevent the removal of exon junction complex and such leftover exon junction complex(es) serve as an anchoring point for the assembly of the nonsense-mediated decay complex and subsequent degradation of mRNA. As the removal of exon junction complexes requires a pioneer round of translation, the truncated proteins are synthesized, but inefficiently (Matsuda et al., 2008; Rebbapragada and Lykke-Andersen, 2009; Schweingruber et al., 2013). There are also some premature stop codons in SPAST located <50–55 nucleotides upstream of the 3’-most exon–exon junction, and therefore unlikely to produce mRNAs undergoing degradation (Beetz et al., 2006; Aridon et al., 2007; Shoukier et al., 2009).
As discussed earlier, the lack of detectable levels of truncated spastins in the brain of mouse models of HSP-SPG4 (Tarrade et al., 2006; Kasher et al., 2009; Fassier et al., 2013), in lymphoblastoid cell lines derived from human HSP-SPG4 patients (Riano et al., 2009) and in neuronal cells derived from pluripotent stem cells generated from fibroblast of HSP-SPG4 patients (Denton et al., 2014; Havlicek et al., 2014) has led to the view that a partial loss of spastin microtubule-severing activity, not the presence of neurotoxic spastin proteins, best explains the observed HSP pathologies. However, none of these models truly mirrors the human disease, as HSP-SPG4 in most cases develops over many years and affects mainly a subset of long upper motor neuron axons. The absence of mutated spastins in short-term cultured cells does not preclude the possibility that mutated spastins are present in the affected corticospinal axons of HSP-SPG4 patients. Moreover, as both mRNAs carrying premature termination codons and truncated proteins could be unstable, detection of such proteins may require immunoprecipitation and stringent extraction, and these methods were not used to detect mutated spastins in HSP-SPG4 patient-derived lymphoblastoid or neuronal cell lines. Studies of mutant forms of Cu, Zn-superoxide dismutase (SOD1) that contribute to the cases of familial amyotrophic lateral sclerosis (ALS) revealed, for example, that very minute levels of a C-terminally truncated version of SOD1 expressed in transgenic mice, induce late-onset ALS with rapid progression to death. Analyses of spinal cord and brain of a patient with ALS carrying the same mutation showed that the mutated protein was expressed below 0.5% of the SOD1 levels in controls (Jonsson et al., 2004). In another study, pathogenic SOD1 was detected only after immunoprecipitation that allowed for concentration of small amounts of mutated protein expressed in transgenic mice (Watanabe et al., 2005). To the best of our knowledge, the spinal cord of only one patient carrying a SPAST splice mutation resulting in in-frame skipping of exon 11 (Svenson et al., 2004) has been examined for the presence of mutant spastins using western blotting with an anti-human spastin antibody, a method that allows M1 to be distinguished from M87. In this case, a prominent band corresponding to the truncated mutant M1 was detected in thoracic but not in cervical spinal cord (Solowska et al., 2010). Interestingly, post-mortem studies in HSP patients consistently report the most severe axon degeneration in the thoracic spinal cord (Fink, 2013).
Large deletions of SPAST
Another category of mutations in spastin gene are large deletions varying from 1.3 kb to 1283.9 Such deletions are most likely facilitated by Alu-rich genomic architecture of SPAST that render the locus susceptible to a variety of genomic Alu-mediated rearrangements (Beetz et al., 2006; Depienne et al., 2007; Boone et al., 2014). Of 50 different combinations of SPAST deleted exons, none has been a deletion of the entire spastin gene (Boone et al., 2014), indicating that in many cases synthesis of truncated spastins cannot be ruled out, particularly because correspondingly shortened transcripts have been found in patient lymphocytes (Beetz et al., 2006). Furthermore, almost half of the deletions extended beyond the boundaries of SPAST and some of them interrupted adjacent genes, potentially affecting their expression. Twelve deletions extending the 3’end of SPAST might form chimeric genes that yield fusion transcripts with possible phenotypic consequences. Chimeras between SPAST and SLC30A6 (solute carrier Family 30, member 6) encoding zinc transporter ZnT6 might be of particular interest, as alterations in zinc and zinc transporters dynamics have been found in the brain of humans with Alzheimer’s disease (Smith et al., 2006). In rare HSP-SPG4 cases, deletions of the entire SPAST gene that would preclude synthesis of any mutated spastin protein were reported (Depienne et al., 2007). It is unclear, however, how such large DNA deletions of at least 90 kb affected the integrity of SPAST neighbouring genes (or if any other HSP genes were mutated in these patients), and therefore it is difficult to judge whether HSP in these cases developed solely as the result of the inactivation of one SPAST allele.
Pathogenic M1 and M87 spastin isoforms
Determining the role of the spastin isoforms M1 and M87 in HSP-SPG4 is likely to be crucial for understanding the aetiology of the disease. Analysis of spastin expression pattern in rodents revealed the presence of appreciable levels of M1 spastin only in adult spinal cord (Solowska et al., 2008). Also in adult human CNS, levels of M1 in spinal cord are significantly higher than in cerebral cortex (Solowska et al., 2010). M1 expression level seems to be strictly controlled by GC-rich 5’UTR, the upstream AUG sequence and a weak Kozak consensus sequence at the first initiation codon (Fig. 1). Such translation-reducing measures are often used to prevent harmful overproduction of toxic proteins (Kozak, 2002). As a result, wild-type M87 isoform is always more abundant in both neuronal and non-neuronal tissues. Experimental expression of truncated spastins revealed, however, that the levels of M87 were often significantly lower than the levels of the simultaneously expressed M1, despite a weak Kozak sequence at the M1 initiation codon (our unpublished observations), indicating that truncating mutations might significantly decrease the stability of M87 and at least to some extent explain why truncated M87 spastins were not detected in lymphocytes of HSP-SPG4 patients (Riano et al., 2009) or in human induced pluripotent stem cell-derived neuronal cells generated from fibroblasts of HSP-SPG4 patients (Denton et al., 2014; Havlicek et al., 2014).
Removal of 5’UTR from spastin cDNA and creation of a good Kozak sequence at the M1 initiation codon prevented leaky translation from the M87 start codon. Moreover, the synthesis of both isoforms under control of identical promoter and Kozak sequence resulted in significantly higher expression of wild-type or mutated M1 than of the corresponding M87, suggesting that the degradation of M87 is more efficient than that of M1 (Solowska et al., 2010, 2014). Toxicity of individual spastin isoforms carrying missense C448Y mutation found in HSP-SPG4 patients (Hazan et al., 1999; Fonknechten et al., 2000) was tested in rat primary cortical neurons and in transgenic Drosophila (Solowska et al., 2014). The high levels of mutated M1 with pathogenic C448Y mutation might at least partly explain why M1 C448Y was significantly more detrimental to neurite outgrowth in cultured neurons and caused notably more severe motor defects in transgenic Drosophila than M87 C448Y. The flies expressing pathogenic M1 also experienced greater progressive decline of climbing activity than flies expressing pathogenic M87 (Solowska et al., 2014). Thus, the specificity of the disease for adult corticospinal tracts might relate directly to the elevated levels of mutated M1 not M87, particularly given that truncating mutations significantly decrease stability of many mutated M87 but not M1. The motor defects observed in Drosophila and reduced neurite outgrowth in cultured cortical neurons expressing spastins with the C488Y mutation did not result from dominant-negative activity, suggesting that at least some SPAST mutations produce neurotoxic proteins (Solowska et al., 2014). Interestingly, experiments in which cultured primary cortical neurons were transfected with truncated, GFP-tagged mouse spastins showed that only M1 but not M87 isoform had detrimental effects on neurite outgrowth, and even low (1–10 nM) concentrations of truncated M1 significantly inhibited fast axonal transport in squid axoplasm (Solowska et al., 2008).
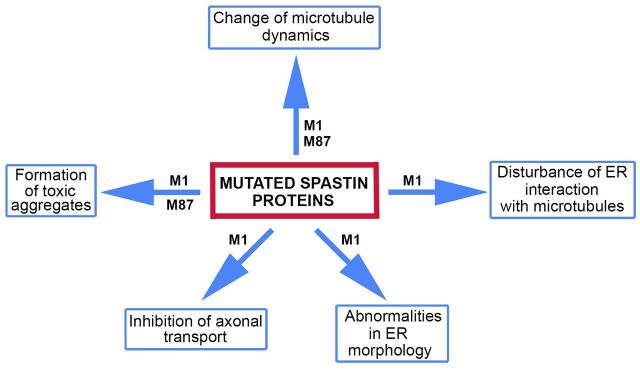
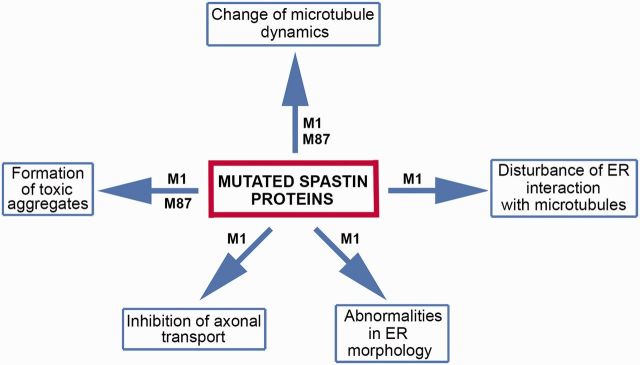
Why mutated spastin proteins are toxic is unknown. Hypothetically, as shown in Fig. 4, mutated spastin proteins might affect various cellular activities. It has been shown that M1 with missense mutation C448Y decorates a subset of microtubules in cultured fibroblast and neuronal cells and decreases the rates of microtubule polymerization and depolymerization (Solowska et al., 2014). As microtubule decoration has been reported for a number of spastins carrying inactivating missense mutations in the AAA domain (Errico et al., 2002; McDermott et al., 2003; Evans et al., 2005), it is possible that many mutated spastin proteins decrease microtubule dynamics via a gain-of-function mechanism, and exacerbate the effects of the lost microtubule-severing activity. Interestingly, in the case of the pathogenic C448Y mutation, the M87 protein that does not bind to microtubules causes the inverse effect on microtubules, rendering them more dynamic rather than less dynamic (Solowska et al., 2014).
Figure 4.
Cellular activities hypothetically affected by mutated spastin proteins. Mutated M1 protein might affect endoplasmic reticulum (ER) morphology and interaction with microtubules, as well as axonal transport. Mutated M1 and M87 might affect microtubule dynamics and form toxic aggregates.
A more permanent association of mutated M1 with microtubules might also cause a decrease of endoplasmic reticulum sliding along microtubules while lower rates of polymerization and depolymerization might decrease growth and retraction of endoplasmic reticulum tubules attached to microtubule plus-ends. Truncated M1 isoforms lacking the microtubule-binding domain could theoretically be inserted into endoplasmic reticulum membranes but would not be able to participate in the interaction of endoplasmic reticulum with microtubules. Mutant M1 inserted into endoplasmic reticulum membranes could also adversely affect endoplasmic reticulum morphogenesis.
Mutant M1 bound to microtubules could interfere with a variety of proteins that are involved in axonal transport. As truncated mouse M1 spastin inhibited fast axonal transport in squid axoplasm (Solowska et al., 2008), it has been speculated that mutated M1, similarly to other neuropathogenic peptides including huntingtin, filamentous tau, presenilin 1 and oligomeric amyloid-β, promotes abnormal activation of selected protein kinases. Such activation might lead to aberrant phosphorylation and regulation of molecular motor proteins, kinesin 1 and cytoplasmic dynein, that are responsible for axonal transport (Morfini et al., 2009). Finally, spastin mutations might result in synthesis of misfolded proteins and promote formation of toxic aggregates, particularly in the case of M1 with inappropriately exposed hydrophobic domain.
Concluding remarks
Mutations in the SPAST gene encoding spastin have long been known to exist in the most common form of HSP. A breakthrough in mechanistic understanding of the disease came when it was discovered that spastin is an enzyme that severs long microtubules into shorter ones. Because most pathogenic SPAST mutations destroy the enzymatic activity of spastin, the idea that insufficient microtubule severing is the cause of HSP-SPG4 became popular. However, this explanation, while appealing in its simplicity, cannot fully explain the disease. For example, it is unclear why a partial loss of microtubule-severing activity would be more damaging to adult axons than to developing axons, when the latter rely more than the former on increased microtubule number, mobility and dynamics. It is also unclear why the corticospinal tracts would suffer more than other long axons in the body. Some insights are provided by considering that a leaky scanning of the first initiation codon of SPAST and a preferential translation from the second initiation codon lead to a simultaneous synthesis of two spastin isoforms termed M1 and M87. There is far more M87 than M1, indicating that deficiencies in M87 might be mainly responsible for any aspects of the disease that are caused by insufficient microtubule severing, including deficits in endosomal trafficking. The M1 isoform, owing to the presence of the hydrophobic N-terminal domain, can be inserted into endoplasmic reticulum membranes and involved in endoplasmic reticulum morphogenesis and endoplasmic reticulum–microtubule interactions. M1 carrying inactivating missense mutations can persistently associate with microtubules, potentially causing deficits in axonal transport. Most truncating mutations result in M1 that is not able to interact with microtubules but such mutant spastins could promote activation of selected protein kinases leading to abnormal phosphorylation of motor proteins, and hence cause defects in axonal transport in an entirely different manner. The hydrophobic domain of M1 could also promote aggregation of the mutated spastin, and if this is the case, the capacity of a particular patient to dispose of such misfolded protein might affect the severity of the disease. M1 is only detectably present in adult spinal cord, which is consistent with a mechanism by which either loss of M1 function or toxicity of mutant M1 could be especially relevant to degeneration of the corticospinal tracts. In addition, some deletions of parts or all of SPAST may also include neighbouring genes and affect their function. Finally, it is relevant to note that endosome trafficking, endoplasmic reticulum morphogenesis, bone morphogenic protein signalling, and microtubule dynamics, all of which involve spastin in one way or another, are also affected by mutations in other HSP associated genes, none of which severs microtubules. In conclusion, while identifying spastin as a microtubule-severing protein was an important mechanistic breakthrough, it seems certain that insufficient microtubule severing alone is not an adequate explanation for HSP-SPG4.
Acknowledgements
We are thankful to past and present members of our laboratory who contributed to our work on hereditary spastic paraplegia, as well as our invaluable collaborators, Drs Gerardo Morfini, Daniel Marenda, and Terry Heiman-Patterson. The authors declare no financial or conflicting interests.
Glossary
Abbreviations
- AAA
ATPase associated with various cellular activities
- HSP
hereditary spastic paraplegia
Funding
The relevant work in the Baas Laboratory was supported by National Science Foundation Grant IOS0841245 and National Institutes of Health Grants R01 NS28785, and also by grants from the Philadelphia Institute of Neurodegenerative Diseases, the Pennsylvania Department of Health CURE program to Drexel University College of Medicine, and the Spastic Paraplegia Foundation.
References
- Allison R, Lumb JH, Fassier C, Connell JW, Ten Martin D, Seaman MN, et al. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J Cell Biol 2013; 202: 527–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridon P, Ragonese P, De Fusco M, Lo Coco D, Salemi G, Casari G, et al. Autosomal dominant hereditary spastic paraplegia: report of a large Italian family with R581X spastin mutation. Neurol Sci 2007; 28: 171–4. [DOI] [PubMed] [Google Scholar]
- Aulitzky A, Friedrich K, Gläser D, Gastl R, Kubisch C, Ludolph AC, et al. A complex form of hereditary spastic paraplegia in three siblings due to somatic mosaicism for a novel SPAST mutation in the mother. J Neurol Sci 2014; 347: 352–5. [DOI] [PubMed] [Google Scholar]
- Baas PW, Ahmad FJ. Beyond taxol: microtubule-based treatment of disease and injury of the nervous system [Review]. Brain 2013; 136: 2937–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Vidya Nadar C, Myers KA. Axonal transport of microtubules: the long and short of it [Review]. Traffic 2006; 7: 490–8. [DOI] [PubMed] [Google Scholar]
- Beetz C, Nygren AO, Schickel J, Auer-Grumbach M, Bürk K, Heide G, et al. High frequency of partial SPAST deletions in autosomal dominant hereditary spastic paraplegia. Neurology 2006; 67: 1926–30. [DOI] [PubMed] [Google Scholar]
- Beetz C, Schüle R, Deconinck T, Tran-Viet KN, Zhu H, Kremer BP, et al. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain 2008; 131: 1078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C, O'Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway [Review]. Nat Rev Neurosci 2011; 12: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone C. Cellular pathways of hereditary spastic paraplegia [Review]. Annu Rev Neurosci 2012; 35:25–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair MA, Riddle ME, Wells JF, Breviu BA, Hedera P. Infantile onset of hereditary spastic paraplegia poorly predicts the genotype. Pediatr Neurol 2007; 36: 382–6. [DOI] [PubMed] [Google Scholar]
- Boone PM, Yuan B, Campbell IM, Scull JC, Withers MA, Baggett BC, et al. The Alu-rich genomic architecture of SPAST predisposes to diverse and functionally distinct disease-associated CNV alleles. Am J Hum Genet 2014; 95: 143–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J, Fonknechten N, Hoeltzenbein M, Neumann L, Bratanoff E, Hazan J, et al. Hereditary spastic paraplegia caused by mutations in the SPG4 gene. Eur J Hum Genet 2000; 8: 771–6. [DOI] [PubMed] [Google Scholar]
- Cai D, McEwen DP, Martens JR, Meyhofer E, Verhey KJ. Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS Biol 2009; 7: e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron F, Tessier-Lavigne M. The Hedgehog, TGF-beta/bone morphogenic protein and Wnt families of morphogens in axon guidance [Review]. Adv Exp Med Biol 2007; 621: 116–33. [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Keers SM, Holden MJ, Ramesh V, Dalton A. Infantile hereditary spastic paraparesis due to codominant mutations in the spastin gene. Neurology 2004; 63: 710–12. [DOI] [PubMed] [Google Scholar]
- Claudiani P, Riano E, Errico A, Andolfi G, Rugarli EI. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp Cell Res 2005; 309: 358–69. [DOI] [PubMed] [Google Scholar]
- Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic 2009; 10: 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa F, Panzeri C, Martinuzzi A, Arnoldi A, Redaelli F, Tonelli A, et al. Eight novel mutations in SPG4 in a large sample of patients with hereditary spastic paraplegia. Arch Neurol 2006; 63: 750–5. [DOI] [PubMed] [Google Scholar]
- Denton KR, Lei L, Grenier J, Rodionov V, Blackstone C, Li XJ. Loss of spastin function results in disease-specific axonal defects in human pluripotent stem cell-based models of hereditary spastic paraplegia. Stem Cells 2014; 32: 414–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Fedirko E, Forlani S, Cazeneuve C, Ribaï P, Feki I, et al. Exon deletions of SPG4 are a frequent cause of hereditary spastic paraplegia. J Med Genet 2007; 44: 281–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr A, Tallaksen C, Depienne C. Spastic paraplegia 4. In: Pagon RA, Adam MP, Ardinger HH, Bird TD, Dolan CR, et al., editors. GeneReviews® [Internet] Seattle (WA): University of Washington, Seattle. 1993-2014. 2003 Apr 17 [updated 2012 Aug 16]. [Google Scholar]
- Eckert T, Link S, Le DT, Sobczak JP, Gieseke A, Richter K, et al. Subunit Interactions and cooperativity in the microtubule-severing AAA ATPase spastin. J Biol Chem 2012; 287: 26278–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erichsen AK, Inderhaug E, Mattingsdal M, Eiklid K, Tallaksen CM. Seven novel mutations and four exon deletions in a collection of Norwegian patients with SPG4 hereditary spastic paraplegia. Eur J Neurol 2007; 14: 809–14. [DOI] [PubMed] [Google Scholar]
- Errico A, Ballabio A, Rugarli EI. Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet 2002; 11: 153–63. [DOI] [PubMed] [Google Scholar]
- Errico A, Claudiani P, D'Addio M, Rugarli EI. Spastin interacts with the centrosomal protein NA14, and is enriched in the spindle pole, the midbody and the distal axon. Hum Mol Genet 2004; 13: 2121–32. [DOI] [PubMed] [Google Scholar]
- Evans KJ, Gomes ER, Reisenweber SM, Gundersen GG, Lauring BP. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J Cell Biol 2005; 168: 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K, Keller C, Pavur K, Glasgow K, Conn B, Lauring B. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc Natl Acad Sci USA 2006; 103: 10666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassier C, Tarrade A, Peris L, Courageot S, Mailly P, Dalard C, et al. Microtubule-targeting drugs rescue axonal swellings in cortical neurons from spastin knock-out mice. Dis Model Mech 2013; 6: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink JK. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms [Review]. Acta Neuropathol 2013; 126:307–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonknechten N, Mavel D, Byrne P, Davoine CS, Cruaud C, Bonsch D, et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum Mol Genet 2000; 9: 637–44. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol 2010; 190: 363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal U, Renvoisé B, Chang J, Blackstone C. Spastin-interacting protein NA14/SSNA1 functions in cytokinesis and axon development. PLoS One 2014; 9: e112428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mäntler J, Maar S, Poser I, Leonhardt H, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 2011; 331: 1616–20. [DOI] [PubMed] [Google Scholar]
- Havlicek S, Kohl Z, Mishra HK, Prots I, Eberhardt E, Denguir N, et al. Gene dosage-dependent rescue of HSP neurite defects in SPG4 patients' neurons. Hum Mol Genet 2014; 23: 2527–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Gene 1999; 23: 296–303. [DOI] [PubMed] [Google Scholar]
- Hu, J1, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz WA, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 2009; 138: 549–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson PA, Ernhill K, Andersen PM, Bergemalm D, Brännström T, Gredal O, et al. Minute quantities of misfolded mutant superoxide dismutase-1 cause amyotrophic lateral sclerosis. Brain 2004; 127: 73–88. [DOI] [PubMed] [Google Scholar]
- Kasher PR, De Vos KJ, Wharton SB, Manser C, Bennett EJ, Bingley M, et al. Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients. J Neurochem 2009; 110: 34–44. [DOI] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation [Review]. Gene 2002; 299: 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, et al. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J Cell Biol 2010; 189: 945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey JC, Lusher ME, McDermott CJ, White KD, Reid E, Rubinsztein DC, et al. Mutation analysis of the spastin gene (SPG4) in patients with hereditary spastic paraparesis. J Med Genet 2000; 37: 759–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A. Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. [Review] Exp Neurol 2014; 261: 518–39. [DOI] [PubMed] [Google Scholar]
- Lumb JH, Connell JW, Allison R, Reid E. The AAA ATPase spastin links microtubule severing to membrane modelling [Review]. Biochim Biophys Acta 2012; 1823: 192–7. [DOI] [PubMed] [Google Scholar]
- Mancuso G, Rugarli EI. A cryptic promoter in the first exon of the SPG4 gene directs the synthesis of the 60-kDa spastin isoform. BMC Biol 2008; 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannan AU, Boehm J, Sauter SM, Rauber A, Byrne PC, Neesen J, et al. Spastin, the most commonly mutated protein in hereditary spastic paraplegia interacts with Reticulon 1 an endoplasmic reticulum protein. Neurogenetics 2006; 7: 93–103. [DOI] [PubMed] [Google Scholar]
- Matsuda D, Sato H, Maquat LE. Studying nonsense-mediated mRNA decay in mammalian cells. Methods Enzymol 2008; 449: 177–201. [DOI] [PubMed] [Google Scholar]
- McDermott CJ, Burness CE, Kirby J, Cox LE, Rao DG, Hewamadduma C, et al. Clinical features of hereditary spastic paraplegia due to spastin mutation. Neurology 2006; 67: 45–51. Erratum in: Neurology 2009; 72: 1534. [DOI] [PubMed] [Google Scholar]
- McDermott CJ, Grierson AJ, Wood JD, Bingley M, Wharton SB, Bushby KM, et al. Hereditary spastic paraparesis: disrupted intracellular transport associated with spastin mutation. Ann Neurol 2003; 54: 748–59. [DOI] [PubMed] [Google Scholar]
- Meijer IA, Hand CK, Cossette P, Figlewicz DA, Rouleau GA. Spectrum of SPG4 mutations in a large collection of North American families with hereditary spastic paraplegia. Arch Neurol 2002; 59: 281–6. [DOI] [PubMed] [Google Scholar]
- Morfini GA, Burns M, Binder LI, Kanaan NM, LaPointe N, Bosco DA, et al. Axonal transport defects in neurodegenerative diseases. J Neurosci 2009; 29: 12776–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JE, Johnsen B, Koefoed P, Scheuer KH, Grønbech-Jensen M, Law I, et al. Hereditary spastic paraplegia with cerebellar ataxia: a complex phenotype associated with a new SPG4 gene mutation. Eur J Neurol 2004; 11: 817–24. [DOI] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 2014; 343: 506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlacchio A, Kawarai T, Totaro A, Errico A, St George-Hyslop PH, Rugarli EI, et al. Hereditary spastic paraplegia: clinical genetic study of 15 families. Arch Neurol 2004; 61: 849–55. [DOI] [PubMed] [Google Scholar]
- Orso G, Martinuzzi A, Rossetto MG, Sartori E, Feany M, Daga A. Disease-related phenotypes in a Drosophila model of hereditary spastic paraplegia are ameliorated by treatment with vinblastine. J Clin Invest 2005; 11: 3026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest 2010; 120: 1097–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrono C, Scarano V, Cricchi F, Melone MA, Chiriaco M, Napolitano A, et al. Autosomal dominant hereditary spastic paraplegia: DHPLC-based mutation analysis of SPG4 reveals eleven novel mutations. Hum Mutat 2005; 25: 506. [DOI] [PubMed] [Google Scholar]
- Qiang L, Yu W, Liu M, Solowska JM, Baas PW. Basic fibroblast growth factor elicits formation of interstitial axonal branches via enhanced severing of microtubules. Mol Biol Cell 2010; 21: 334–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? [Review]. Curr Opin Cell Biol 2009; 21: 394–402. [DOI] [PubMed] [Google Scholar]
- Reid E, Connell J, Edwards TL, Duley S, Brown SE, Sanderson CM. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum Mol Genet 2005; 14: 19–38. [DOI] [PubMed] [Google Scholar]
- Riano E, Martignoni M, Mancuso G, Cartelli D, Crippa F, Toldo I, et al. Pleiotropic effects of spastin on neurite growth, depending on expression levels. J Neurochem 2009; 108: 1277–88. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr Biol 2005; 15: 650–5. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J Cell Biol 2006; 175: 849–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 2008; 451: 363–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano L, Melo C, Silva MC, Coutinho P. The global epidemiology of hereditary ataxia and spastic paraplegia: a systematic review of prevalence studies. Neuroepidemiology 2014; 42: 174–83. [DOI] [PubMed] [Google Scholar]
- Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. [Review] Lancet Neurol 2008; 7: 1127–38. [DOI] [PubMed] [Google Scholar]
- Sanderson CM, Connell JW, Edwards TL, Bright NA, Duley S, Thompson A, et al. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet 2006; 15: 307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter S, Miterski B, Klimpe S, Bönsch D, Schöls L, Visbeck A, et al. Mutation analysis of the spastin gene (SPG4) in patients in Germany with autosomal dominant hereditary spastic paraplegia. Hum Mutat 2002; 20: 127–32. [DOI] [PubMed] [Google Scholar]
- Schickel J, Pamminger T, Ehrsam A, Münch S, Huang X, Klopstock T, et al. Isoform-specific increase of spastin stability by N-terminal missense variants including intragenic modifiers of SPG4 hereditary spastic paraplegia. Eur J Neurol 2007; 14: 1322–8. [DOI] [PubMed] [Google Scholar]
- Schweingruber C1, Rufener SC, Zünd D, Yamashita A, Mühlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells [Review]. Biochim Biophys Acta 2013; 1829: 612–23 [DOI] [PubMed] [Google Scholar]
- Sherwood NT, Sun Q, Xue M, Zhang B, Zinn K. Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biology 2004; 2: e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoukier M, Neesen J, Sauter SM, Argyriou L, Doerwald N, Pantakani DV, et al. Expansion of mutation spectrum, determination of mutation cluster regions and predictive structural classification of SPAST mutations in hereditary spastic paraplegia. Eur J Hum Genet 2009; 17: 187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Xiong S, Markesbery WR, Lovell MA. Altered expression of zinc transporters-4 and -6 in mild cognitive impairment, early and late Alzheimer's disease brain. Neuroscience 2006; 140: 879–88. [DOI] [PubMed] [Google Scholar]
- Solowska J, Garbern J, Baas PW. Evaluation of loss-of-function as an explanation for SPG4-based hereditary spastic paraplegia. Human Molec Genetics 2010; 19: 2767–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solowska JM, D'Rozario M, Jean DC, Davidson MW, Marenda DR, Baas PW. Pathogenic Mutation of Spastin Has Gain-of-Function Effects on Microtubule Dynamics. J Neurosci 2014; 34: 1856–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solowska JM, Morfini G, Falnikar A, Himes BT, Brady ST, Huang D, et al. Quantitative and functional analyses of spastin in the nervous system: implications for hereditary spastic paraplegia. J Neurosci 2008; 28: 2147–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Rao K, Gheres KW, Kim S, Tao J, La Rochelle C, et al. Normal spastin gene dosage is specifically required for axon regeneration. Cell Rep 2012; 2: 1340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson IK, Ashley-Koch AE, Gaskell PC, Riney TJ, Cumming WJ, Kingston HM, et al. Identification and expression analysis of spastin gene mutations in hereditary spastic paraplegia. Am J Hum Genet 2001; 68: 1077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson IK, Kloos MT, Gaskell PC, Nance MA, Garbern JY, Hisanaga S, et al. Intragenic modifiers of hereditary spastic paraplegia due to spastin gene mutations. Neurogenetics 2004; 5: 157–64. [DOI] [PubMed] [Google Scholar]
- Tarrade A, Fassier C, Courageot S, Charvin D, Vitte J, Peris L, et al. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum Mol Genet 2006; 15: 3544–58. [DOI] [PubMed] [Google Scholar]
- Thomsen B, Nissen PH, Agerholm JS, Bendixen C. Congenital bovine spinal dysmyelination is caused by a missense mutation in the SPAST gene. Neurogenetics 2010; 11: 175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta N, Orso G, Rossetto MG, Daga A, Broadie K. The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr Biol 2004; 14: 1135–47. [DOI] [PubMed] [Google Scholar]
- Tsang HT, Edwards TL, Wang X, Connell JW, Davies RJ, Durrington HJ, et al. The hereditary spastic paraplegia proteins NIPA1, spastin and spartin are inhibitors of mammalian bone morphogenic protein signalling. Hum Mol Genet 2009; 18: 3805–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown A. Rapid movement of microtubules in axons. Curr Biol 2002; 12: 1496–501. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yasui K, Nakano T, Doi K, Fukada Y, Kitayama M, et al. Mouse motor neuron disease caused by truncated SOD1 with or without C-terminal modification. Brain Res Mol Brain Res 2005; 135: 12–20. [DOI] [PubMed] [Google Scholar]
- Wharton SB, McDermott CJ, Grierson AJ, Wood JD, Gelsthorpe C, Ince PG, et al. The cellular and molecular pathology of the motor system in hereditary spastic paraparesis due to mutation of the spastin gene. J Neuropathol Exp Neurol 2003; 62: 1166–77. [DOI] [PubMed] [Google Scholar]
- White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J Cell Biol 2007; 176: 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JD, Landers JA, Bingley M, McDermott CJ, Thomas-McArthur V, Gleadall LJ, et al. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum Mol Genet 2006; 15: 2763–71. [DOI] [PubMed] [Google Scholar]
- Yang D, Rismanchi N, Renvoisé B, Lippincott-Schwartz J, Blackstone C, Hurley JH. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat Struct Mol Biol 2008; 15: 1278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ganguly A, Cabral F. Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J Biol Chem 2010; 285: 32242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip AG, Durr A, Marchuk DA, Ashley-Koch A, Hentati A, Rubinsztein DC, et al. Meta-analysis of age at onset in spastin-associted hereditary spastic paraplegia provides no evidence for a correlation with mutational class. J Med Genet 2003; 40: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Qiang L, Solowska JM, Karabay A, Korulu S, Baas PW. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell 2008; 19: 1081–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Alvarado D, Rainier S, Lemons R, Hedera P, Weber CH, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet 2001; 29: 326–31. [DOI] [PubMed] [Google Scholar]
- Zhu PP, Denton KR, Pierson TM, Li XJ, Blackstone C. Pharmacologic rescue of axon growth defects in a human iPSC model of hereditary spastic paraplegia. Hum Mol Genet 2014; 23: 5638–48. [DOI] [PMC free article] [PubMed] [Google Scholar]