Abstract
Class switch DNA recombination (CSR) is central to the maturation of the antibody response, as it diversifies antibody effector functions. Like somatic hypermutation, CSR requires AID, whose expression is restricted to B cells, as induced by CD40 engagement or dual TLR-BCR engagement (primary CSR-inducing stimuli). By constructing conditional knockout Igh+/Cγ1-creRab7fl/fl mice, we identified a B cell-intrinsic role for Rab7, a small GTPase involved in intracellular membrane functions, in mediating AID induction and CSR. Igh+/Cγ1-creRab7fl/fl mice displayed normal B and T cell development, and were deficient in Rab7 only in B cells undergoing IghCγ1-cre Iγ1-Sγ1-Cγ1-cre transcription, as induced – like Igh germline Iγ1-Sγ1-Cγ1 and Iε-Sε-Cε transcription – by IL-4 in conjunction with a primary CSR-inducing stimulus. These mice could not mount T-independent or T-dependent class-switched IgG1 or IgE responses, while maintaining normal IgM levels. Igh+/Cγ1-creRab7fl/fl B cells showed, in vivo and in vitro, normal proliferation and survival, normal Blimp-1 expression and plasma cell differentiation, as well as intact activation of the non-canonical NF-κB, p38 kinase and ERK1/2 kinase pathways. They, however, were defective in AID expression and CSR in vivo and in vitro, as induced by CD40 engagement or dual TLR1/2-, TLR4-, TLR7- or TLR9-BCR engagement. In Igh+/Cγ1-creRab7fl/fl B cells, CSR was rescued by enforced AID expression. These findings, together with our demonstration that Rab7 mediated canonical NF-κB activation, as critical to AID induction, outline a novel role of Rab7 in signaling pathways that lead to AID expression and CSR, likely by promoting assembly of signaling complexes along intracellular membranes.
Introduction
The maturation of the antibody response is critical to effective host defense against microbial infections and tumors. It depends on two B lymphocyte differentiation processes: immunoglobulin (Ig) class switch DNA recombination (CSR) and somatic hypermutation (SHM) (1). CSR replaces an Ig heavy chain (IgH) constant (CH) region, e.g., Cμ, with a downstream CH region (Cγ, Cα or Cε), thereby diversifying the biological effector functions of an antibody without changing its specificity for antigen (2). SHM inserts mainly point-mutations in the Ig V(D)J DNA, thereby providing the structural substrate for the positive selection by antigen for higher affinity antibody mutants (1). CSR and SHM require deamination of deoxycytosines in IgH switch (S) region and V(D)J region DNA, respectively, by activation-induced cytidine deaminase (AID, encoded by AICDA/Aicda) (3). In CSR, AID is targeted to an upstream (donor) S and a downstream (acceptor) S regions by 14-3-3 adaptors, which display high affinities for 5′-AGCT-3′ repeats, as recurring in all S regions, as well as for H3K9acS10ph histone posttranslational modifications, as induced specifically in the donor and acceptor S regions – these S regions also undergo germline IH-S-CH transcription initiated upon activation of their respective upstream intervening IH promoter (4, 5). High densities of deoxyuracils, as generated through AID-mediated DNA deamination, are processed by uracil DNA glycosylase (Ung) recruited by Rev1 (6-8), leading to the generation of S region (DSBs), which are obligatory CSR intermediates. Repair of such DSBs results in looping out of the intervening DNA as an S circle, formation of a S-S junction and juxtaposition of the expressed VHDJH DNA to the downstream CH DNA. Transient transcription of the S circle gives rise to circle transcripts, such as Iγ-Cμ, Iα-Cμ or Iε-Cμ, which are hallmarks of ongoing CSR from IgM to IgG, IgA or IgE; transcription of the IgH locus gives rise to post-recombination Iμ-Cγ, Iμ-Cα or Iμ-Cε transcripts. After undergoing CSR and SHM, B cells differentiate into plasma cells and memory B cells for mature antibody production and generation of long-term immunity, respectively (9-12).
In T-independent antibody responses, B cells are induced to express AID and undergo CSR upon engagement of their B cell receptors (BCRs) by repetitive antigenic ligands together with engagement of their Toll-like receptors (TLRs) and other innate immune receptors by microbe-associated molecular patterns (MAMPs) (13). TLR-signaling also plays an important role in T-dependent responses, e.g., by activating B cells before the emergence of specific TH cells (14). Once specific T helper (TH) cells emerge, they will bear surface trimeric CD154 for engagement of CD40, which is constitutively expressed on B cells. CD154:CD40 engagement leads to further B cell differentiation, including increased AID expression and CSR, which can be enhanced by BAFF/BlyS and APRIL (15-18). The primary CSR-inducing stimuli, e.g., dual TLR/BCR engagement and CD40 engagement, enable secondary stimuli, i.e., cytokine IL-4 and TGF-β (as well as IFN-γ in the mouse), to induce IgH germline IH-S-CH transcription and histone posttranslational modifications in the donor and acceptor S regions (19-23), thereby directing CSR to specific Ig isotypes. IL-4 and TGF-β can also enhance AID expression, as induced by primary CSR-inducing stimuli, through activation of Stat6 and Smad transcription factors, respectively, which are recruited to the Aicda promoter and enhancers (24, 25). T-independent and T-dependent primary CSR-inducing stimuli activate NF-κB through both the canonical and non-canonical pathways, leading to recruitment of NF-κB to the Aicda promoter for induction of AID expression, which is restricted to activated B cells (2, 24-27).
In B cells, signals from TLRs, BCR or CD40 are transduced by multiple pathways, including those involving TRAF6 or PI(3)K (13, 28, 29). These pathways mediate NF-κB activation, thereby linking receptor signals with AID induction. Genetic, biochemical and structural studies have furthered our understanding of the recruitment of signal adaptors through “signalosomes” along plasma membrane lipid rafts (30-32). Nevertheless, the preservation of selected signals in B cells that are virtually ablated in plasma membrane signalosomes, e.g., the intact ERK activation in PLCγ2-deficient B cells (33), indicates that a B cell can use signaling pathways mediated by intracellular membranes. These would include the ER membrane, which could mediate NF-κB activation by different surface receptors, such as CD40 (BL41 B cells), TNF receptor (HEK 293T cells) and T cell receptor (Jurkat T cells) (34). In addition, autophagy-related double-membrane structures, which originate from ER or mitochondria membranes (35), play a role in MAPK p38 activation triggered by BCR and TLR9 (36). Finally, a role of intracellular membranes in B cell signal transduction is suggested by the regulation of CD40 and BCR signaling as well as immunity and inflammation by autophagy-related (Atg) factors (37-40), including Atg5 (41, 42).
The Rab7 small GTPase mediates the maturation of endosomes by replacing Rab5 through a “GTPase switch” process. It also promotes the conversion of endosomes to lysosomes as well as fusion of endosomes with autophagosomes to form amphisomes in different cell types (43). In stressed cells, such as those having phagocytosed large extracellular particles or engulfed a portion of the cytoplasm in response to unfavorable metabolic conditions (e.g., serum starvation), Rab7 mediates the fusion of autophagosomes or amphisomes with lysosomes to form autolysosomes, in which the cargo is degraded. Rab7 also promotes cell death induced by growth factor withdrawal and clearance of apoptotic bodies (44-46). Here, we reasoned that in proliferating or differentiating immune cells, which are not deprived of nutrients or growth factors, Rab7 would play additional and specific roles. This was prompted by the putative role of intracellular membranes in NF-κB activation and the association of Rab7 with those membranes (43).
Rab7 has been shown to regulate T cell functions (47), but its role in B cells is unknown. To address the B cell-intrinsic role of Rab7 in the antibody response, we constructed conditional Igh+/Cγ1-creRab7fl/fl mice, in which Rab7 expression is abrogated only in B cells undergoing IghCγ1-cre Iγ1-Sγ1-Cγ1-cre transcription, as induced by IL-4 in conjunction with a primary stimulus. We stimulated Igh+/Cγ1-creRab7fl/fl B cells with CD154 to activate CD40 signaling and T-independent stimuli to activate both TLR and BCR signaling, including LPS (engaging TLR4 and BCR through its lipid A and polysaccharidic moieties, respectively) or CpG ODN plus anti–δ mAb/dex (engaging TLR9 and BCR, respectively) to address the role of Rab7 in NF-κB activation, AID expression and CSR. We also used Igh+/Cγ1-creRab7fl/fl mice and B cells to analyze the contribution of Rab7 to other B cell functions and differentiation processes, such as plasma cell differentiation, in vivo and in vitro. Our findings suggest an important role of Rab7 in induction of AID expression and CSR for the maturation of T-dependent and T-independent antibody responses.
Materials and Methods
Mice and immunization
Igh+/Cγ1-creRab7fl/fl mice and their Igh+/Cγ1-creRab7+/fl littermate controls (Fig. 1, 2A) were generated by the breeding of Igh+/Cγ1-cre mice ((48), JAX stock number 010611) with Rab7fl/fl mice ((47), JAX stock number 021589), both of which had been backcrossed onto the C57BL/6 background for at least 9 generations. Mice were maintained in pathogen-free vivaria, and were used for experiments at 8-12 weeks of age and without any apparent infection or disease. For immunization, mice were injected intraperitoneally (i.p.) with 100 μg of NP-CGG (in average 16 molecules of 4-hydroxy-3-nitrophenyl acetyl coupled to 1 molecule of chicken γ-globulin; Biosearch Technologies) in 100 μl of alum (Imject® Alum, Pierce), 100 μg of ovalbumin (OVA) in 100 μl of alum, or 25 μg of NP-conjugated LPS (NP-LPS; in average 0.2 NP molecule conjugated with one LPS molecule; Biosearch Technologies) in 100 μl of PBS. All protocols were in accordance with the rules and regulations of the Institutional Animal Care and Use Committee (IACUC) of the University of Texas Health Science Center at San Antonio and those of IACUC of the University of California, Irvine.
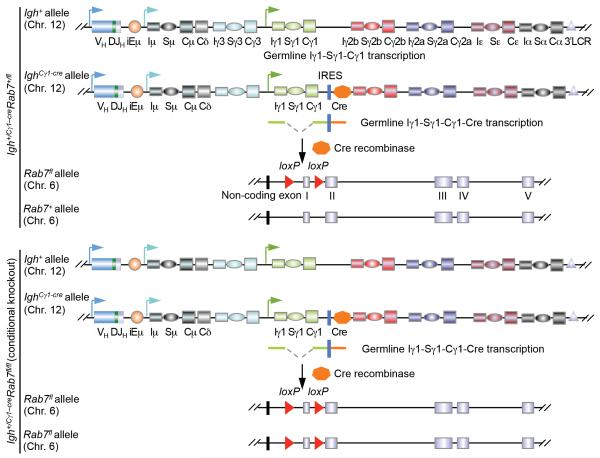
FIGURE 1. Generation of Igh+/Cγ1-creRab7fl/fl mice.
Schematics of expression of the Cre recombinase in “induced” Igh+/Cγ1-creRab7fl/fl B cells (bottom panel) through germline Iγ1-Sγ1-Cγ1-cre transcription in the IghCγ1-cre allele and IRES-dependent translation (germline Iγ1-Sγ1-Cγ1 transcription in the Igh+ allele is also indicated), as well as the consequent Cre-mediated deletion of the first coding exon of the two Rab7 alleles (47) – one Rab7 allele will remain intact in induced Igh+/Cγ1-creRab7+/fl B cells (top panel). This Rab7fl allele is different from a recently reported Rab7 mutant allele in which the second and third exons are floxed (51). In all of our experiments, Igh+/Cγ1-creRab7fl/fl mice were used as conditional knockout mice and Igh+/Cγ1-creRab7+/fl mice were used as their “wildtype” littermate controls, as these mice did not display hypomorphism in the Rab7 protein expression. Igh+/+Rab7fl/fl mice were not chosen as “wildtype” controls because they have two Igh+ alleles capable of encoding IgG1, while Igh+/Cγ1-creRab7fl/fl mice have only one.
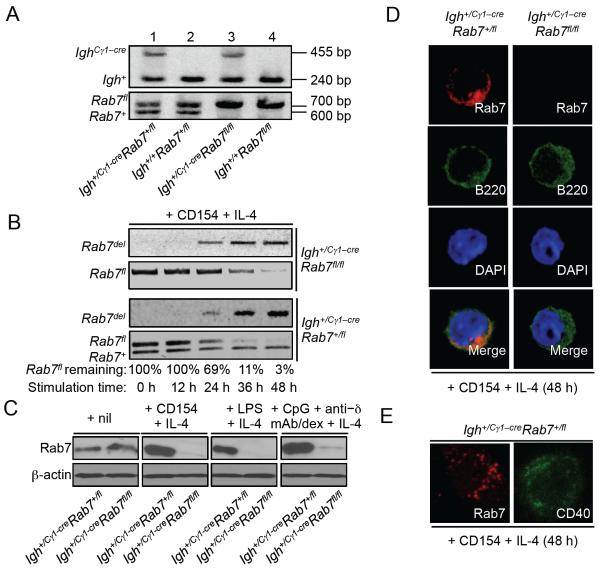
FIGURE 2. Abrogation of Rab7 protein expression in Igh+/Cγ1-creRab7fl/fl B cells stimulated by a primary stimulus plus IL-4.
(A) DNA electrophoresis of PCR products from the genotyping of four littermates born from breeding of an Igh+/Cγ1-creRab7+/fl mouse with an Igh+/+Rab7fl/fl mouse. (B) PCR analysis of the kinetics of Cre-mediated genomic DNA deletion in the Rab7fl allele and appearance of post-deletion Rab7del allele in Igh+/Cγ1-creRab7fl/fl B cells (top panels) and their Igh+/Cγ1-creRab7+/fl B cell counterparts (bottom panels) after stimulation with CD154 plus IL-4 for 0, 12, 24, 36 and 48 h – in Igh+/Cγ1-creRab7+/fl B cells, the Rab7+ allele should remain constant. The proportion of remaining Rab7fl allele DNA was analyzed by the ratio of band intensity of Rab7fl allele DNA (quantified by the ImageJ software) to that of Rab7+ allele DNA (used as the internal loading control) in Igh+/Cγ1-creRab7+/fl B cells, and was expressed as percentages of the value at 0 h (set as 100%). (C) Immunoblotting analysis of Rab7 expression in Igh+/Cγ1-creRab7fl/fl B cells stimulated with CD154, LPS, or CpG plus anti–δ/dex (as indicated) plus IL-4 for 48 h and their Igh+/Cγ1-creRab7+/fl B cell counterparts. (D) Immunofluorescence staining and confocal microscopy analysis of Rab7 and B220 expression (pseudo-colored) in Igh+/Cγ1-creRab7fl/fl B cells stimulated with CD154 plus IL-4 for 48 h and their Igh+/Cγ1-creRab7+/fl B cell counterparts. (E) Stimulated emission depletion (STED) confocal microscopy analysis of Rab7 and CD40 distribution (pseudo-colored) in Igh+/Cγ1-creRab7+/fl B cells stimulated with CD154 plus IL-4 for 48 h (two different cells are shown). Data are representative of three independent experiments.
B cells
Single cell suspensions were prepared from spleens or pooled axillary, inguinal and cervical lymph nodes using a 70-μm cell strainer. Lymph node cells were directly resuspended in RPMI-1640 medium (Invitrogen) supplemented with FBS (10% v/v, Hyclone), penicillin-streptomycin (1% v/v, Invitrogen) and amphotericin B (1% v/v, Invitrogen) (RPMI-FBS), before purification of B cells. Spleen cells were resuspended in ACK Lysis Buffer (Lonza) to lyse red blood cells and, after quenching with RPMI-FBS, were resuspended in RPMI-FBS before B cell purification. B cells were purified by depletion of cells expressing CD43, CD4, CD8, CD11b, CD 49b, CD90.2, Gr-1 or Ter-119 using the EasySep™ Mouse B cell Isolation kit (StemCell Technologies) following the manufacturer’s protocol. For carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen) labeling, B cells were resuspended in PBS at 107 cell/ml and incubated with 4 μM CFSE at 37°C for 5 m and immediately quenched with RPMI-FBS. After pelleting, B cells were resuspended and cultured at 3 × 105 cell/ml in RPMI-FBS supplemented with 50 μM β-mercaptoethanol.
CSR induction and analysis
For CSR induction, B cells were stimulated with the following primary stimuli: CD154 (3 U/ml, mouse CD154-containing membrane fragments of baculovirus-infected Sf21 insect cells (5, 49)), LPS (3 μg/ml, from E. coli, serotype 055:B5, deproteinized by chloroform extraction, Sigma-Aldrich), TLR1/2 ligand Pam3CSK4 (100 ng/ml, Invivogen), TLR4 ligand monophophoryl lipid A (lipid A, 1 μg/ml, Sigma-Aldrich), TLR7 ligand R-848 (30 ng/ml, Invivogen) or TLR9 ligand CpG ODN1826 with a phosphorothioate backbone (CpG, 1 μM, sequence 5′–TCCATGACGTTCCTGACGTT–3′; Operon). Anti–Igδ mAb (clone 11-26c) conjugated to dextran (anti–δ mAb/dex; Fina Biosolutions) was added, as indicated, to crosslink BCRs. Recombinant IL-4 (3 ng/ml) was added for CSR to IgG1 and IgE, IFN-γ (50 ng/ml) for CSR to IgG2a, and TGF-β (2 ng/ml) for CSR to IgG2b (all from R&D Systems). In selected experiments, Igh+/Cγ1-creRab7fl/fl B cells and their Igh+/Cγ1-creRab7+/fl B cell counterparts were stimulated with CD154 or LPS plus IL-4 for 48 h. After washing for three times with RPMI-FBS to remove residual stimuli, the B cells were recultured in LPS plus TGF-β and anti–δ mAb/dex for induction of CSR to IgA.
After stimulation, B cells were harvested, stained with fluorochrome-conjugated mAbs (Supplemental Table S1) in Hank’s Buffered Salt Solution (HBSS) for 15 m and analyzed by flow cytometry for Igγ3, Igγ1, Igγ2b and Igγ2a expression (CSR to IgG3, IgG1, IgG2b and IgG2a) and other B cell surface molecules. For intracellular Igε analysis, B cells (2 × 106) stimulated by CD154 or LPS plus IL-4 were harvested and treated with 200 μl of trypsin for 2 m to cleave off FcεRI, followed by quenching with 1 ml RMPI-FBS, as described (50). Cells were then fixed in 200 μl of formaldehyde (3.7% v/v) at 25°C for 10 m and, after quenching with 22 μl 1 M glycine (pH 7.0), were permeabilized in 500 μl 90% cold methanol on ice for 30 m before staining with fluorochrome-conjugated Abs and flow cytometry analysis. FACS data were analyzed by the FlowJo® software (Tree Star).
B cell proliferation and survival
For analysis of B cell proliferation in vivo in mice immunized with NP-CGG, mice were injected i.p. with bromodeoxyuridine (BrdU) twice, 2 mg (in 200 μl PBS) each time, within a 20-h interval and sacrificed 4 h after the second injection. B cells were analyzed by flow cytometry for BrdU incorporation and DNA contents in spleen B cells using a BrdU Flow Kit (BD Biosciences), following the manufacturer’s instructions. Briefly, spleen cells were stained with fluorochrome-conjugated anti–B220 mAb and, after washing, resuspended in the BD Cytofix/Cytoperm™ buffer for 15 m on ice. After washing, cells were incubated with 7–AAD and APC-conjugated anti–BrdU mAb for staining of DNA and incorporated BrdU, respectively, and then analyzed by flow cytometry. For analysis of B cell proliferation in vitro, purified CFSE-labeled naïve B cells were cultured for 96 h in the presence of appropriate stimuli and then analyzed by flow cytometry for CFSE intensity (CFSE distributes equally into the two daughter cells when a cell divides). The number of B cell divisions was determined by the “Proliferation Platform” of the FlowJo® software. CSR as a function of the cell division number was the ratio of class-switched B cells in each division over the total number of B cells in that division. For analysis of B cell viability, cells were stained with 7–AAD and/or fluorochrome-conjugated Annexin V in HBSS and analyzed by flow cytometry.
Immunofluorescence
To visualize Rab7 expression, B cells were spun onto cover slips pre-coated with poly-D-lysine (10 μg/ml, Sigma-Aldrich) and fixed with 2% paraformaldehyde for 10 m. After blocking with 1% BSA, cells were stained with FITC-conjugated anti–B220 mAb. After washing with HBSS and permeabilization with 0.5% Triton X-100 for 10 m, cells were stained with a rabbit anti–Rab7 mAb (Supplemental Table S1) at 25°C for 1 h, followed by staining with Alexa Fluor647®-conjugated goat anti–rabbit Ab. After washing, cells were mounted using ProLong® Gold with DAPI (Invitrogen) for confocal microscopic analysis.
To analyze Rab7 and CD40 distributions at high resolution, B cells were activated by CD154 and IL-4 for 48 h and then fixed with paraformaldehyde on the poly-D-lysine-coated cover slips. After permeabilization with 1% Triton X-100 and 10% normal donkey serum, B cells were stained with the rabbit anti–Rab7 mAb or a rat anti–CD40 mAb, followed by staining with an Alexa Fluor 532®-conjugated goat anti–rabbit Ab or an Alexa Fluor 488®-conjugated goat anti–rat Ab. A Leica TCS SP8 confocal laser scanning microscope equipped with a time-gated stimulated emission depletion (STED) module was used to achieve super-resolution of Rab7 and CD40 staining by 592 nm depletion laser.
To visualize germinal centers, spleens from immunized mice were embedded in OCT (Tissue-tek) and snap-frozen on dry ice. Cryostat sections (7 μm) were fixed in pre-chilled acetone at −80°C for 30 m and then air dried at 25°C for 30 m. Sections were stained with PE-conjugated anti–B220 mAb (Supplemental Table S1) and FITC-conjugated peanut agglutinin (PNA; EY Laboratories) at 25°C for 1 h in a moist chamber. After washing with HBSS, sections were mounted using ProLong® Gold Antifade Reagent (Invitrogen) and examined under an Olympus FluoView 1000 confocal microscope.
Retroviral transduction
The pTAC and pTAC-AID retroviral vectors were as we described (13). For the generation of retrovirus, retroviral vectors were used to transfect along with the pCL–Eco retrovirus–packaging vector (Imgenex) HEK293T cells using the ProFection Mammalian Transfection System® (Promega). Transfected cells were cultured in FBS-RPMI in the presence of chloroquine (25 μM) for 8 h. After the removal of chloroquine, retrovirus-containing culture supernatants were harvested every 12 h. For transduction and CSR analysis, mouse B cells were activated with CD154 plus IL-4 for 48 h and then incubated with viral particles that were pre-mixed with 6 μg/ml DEAE-dextran at 25°C for 30 m (Sigma-Aldrich). After incubation at 37°C for 5 h with gentle mixing every hour, cells were centrifuged at 500 g for 1 h and then 1,000 g for 4 m. Transduced B cells were cultured in virus-free FBS-RPMI in the presence of CD154 plus IL-4 for 72 h and then harvested for flow cytometry analysis of viability (7–AAD−) and surface IgG1 and CD19 expression. Dead (7–AAD+) cells were excluded from IgG1 and CD19 analysis.
Secreted Ig
To determine titers of total IgM, IgG1, IgG2a, IgG2b, IgG3, IgA or IgE, sera and culture supernatants were first diluted 4 – 100 folds and 4 –10 folds, respectively, with PBS (pH 7.4) plus 0.05% (v/v) Tween-20 (PBST). Two-fold serially diluted samples and standards for each Ig isotypes were incubated in 96-well plates coated with pre-adsorbed goat anti–IgM, –IgG (to capture IgG1, IgG2a, IgG2b and IgG3), –IgA or – IgE Abs (all 1 mg/ml; Supplemental Table S1). After washing with PBST, captured Igs were detected with biotinylated anti–IgM, –IgG1, –IgG2a, –IgG2b, –IgG3, –IgA or –IgE Abs (Supplemental Table S1), followed by reaction with horseradish peroxidase (HRP)-labeled streptavidin (Sigma-Aldrich), development with o-phenylenediamine and measurement of absorbance at 492 nm. Ig concentrations were determined using Prism® (GraphPad Software) or Excel® (Microsoft) software.
To analyze titers of high-affinity NP-specific Abs, sera were diluted 1,000-fold in PBST. Two-fold serially diluted samples were incubated in a 96-well plate pre-blocked with BSA and coated with NP3-BSA (in average 3 NP molecules on one BSA molecule). Captured Igs were detected with biotinylated Ab to IgM, IgG1, IgG2a, IgG2b, IgG3 or IgA. To analyze titers of OVA-specific IgG1 and IgE, sera were diluted 100- (for IgG1) or 10-fold (for IgE) and incubated in plates coated with anti–IgG1 or anti–IgE. Captured Igs were detected with biotinylated OVA. Data are relative values based on end-point dilution factors.
ELISPOT
MultiScreen® filter plates (Millipore) were activated with 35% ethanol, washed with PBS and coated with with NP3-BSA. Single cell suspensions were cultured at 50,000 cells/ml in FBS-RPMI supplemented with 50 μM β-mercaptoethanol at 37°C for 16 h. After supernatants were removed, plates were incubated with biotinylated goat anti–mouse IgM or anti–IgG1 Ab for 2 h and, after washing, incubated with HRP-conjugated streptavidin. After washing, plates were developed using the Vectastain AEC peroxidase substrate kit (Vector Laboratories) to stain antibody forming cells (AFCs). The stained area in each well was quantified using the CTL Immunospot software (Cellular Technology) and depicted as the percentage of total area of each well. This was used to quantify the production of AFCs.
RNA isolation and qRT–PCR analysis of transcripts
Total RNA was extracted from 5 × 106 B cells using the RNeasy Mini Kit (Qiagen). First-strand cDNA was synthesized from 2 μg RNAs using the SuperScript™ III System (Invitrogen) and analyzed by qPCR using SYBR Green (Dynamo HS kit; New England Biolabs) and appropriate primers (Supplemental Table S2). PCR was performed in a MyiQ Real-Time PCR System (Bio-Rad Laboratories) according to the following protocol: 95°C for 30 s, 40 cycles of 95°C for 10 s, 60°C for 30 s, 72°C for 30 s. Melting curve analysis was performed at 72°C–95°C. The ΔΔCt method was used to analyze levels of transcripts and data were normalized to the level of Cd79b, which encodes the BCR Igβ chain constitutively expressed in B cells.
Immunoblotting
B cells (1 × 107) were harvested by centrifugation at 500 g for 5 m, resuspended in 0.5 ml of lysis buffer (20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 1% (v/v) NP-40) supplemented with phosphatase inhibitors, including sodium pyrophosphate (1 mM), NaF (10 mM) and NaVO3 (1 mM), and a cocktail of protease inhibitors (Sigma-Aldrich). Cell lysates were separated by SDS–PAGE and transferred onto nitrocellulose membranes for immunoblotting involving specific Abs (Supplemental Table S1). Membranes were then stripped with 200 mM glycine (pH 2.5) for re-immunoblotting with an anti–β-actin mAb.
Results
Igh+/Cγ1-creRab7fl/fl mice show normal B and T lymphocyte development and survival
Unlike mice with Rab7 deletion in germ cells (47, 51), Igh+/Cγ1-creRab7fl/fl mice were born at Mendelian ratio due to their intact Rab7 locus throughout embryogenesis. They were indistinguishable from their Igh+/+Rab7+/fl, Igh+/+Rab7fl/fl and Igh+/Cγ1-creRab7+/fl littermates in the size, fertility and organ morphology during development and maturation (Fig. 2A and data not shown). This was expected, as in Igh+/Cγ1-creRab7fl/fl mice, efficient Cre recombinase expression and Cre-mediated Rab7 KO occurred exclusively in B cells activated to undergo germline Iγ1-Sγ1-Cγ1-cre transcription in the IghCγ1-cre allele (Fig. 1). Such transcription process, like IgH germline Iγ1-Sγ1-Cγ1 transcription, was induced by IL-4 in the presence of a primary stimulus, such as CD154, LPS or CpG ODN plus anti–δ mAb/dex, leading to DNA deletion in the Rab7fl allele in 89% and 97% of cells after 36 h and 48 h, respectively (Fig. 2B), as well as a virtual abrogation of Rab7 protein expression in Igh+/Cγ1-creRab7fl/fl B cells after 48 h of stimulation (Fig. 2C, 2D). Notably, Rab7, which localized in the cytoplasm (Fig. 2D), formed distinct “foci” structures, as revealed by super-resolution microscopy analysis, in contrast to the more “diffused” pattern of CD40 (Fig. 2E).
Consistent with the lack of IghCγ1-cre germline Iγ1-Sγ1-Cγ1-cre transcription in B cells and T cells during their development and maturation, Igh+/Cγ1-creRab7fl/fl mice displayed normal frequencies, cellularity and viability of B and T cells at different developmental stages in central and peripheral lymphoid organs, including B1 cells in the peritoneal cavity (Fig. 3 and not shown). Thus, Igh+/Cγ1-creRab7fl/fl mice show normal B and T cell development and maturation; Igh+/Cγ1-creRab7fl/fl B cells show abrogated Rab7 expression upon stimulation by a primary CSR-inducing stimulus plus IL-4.
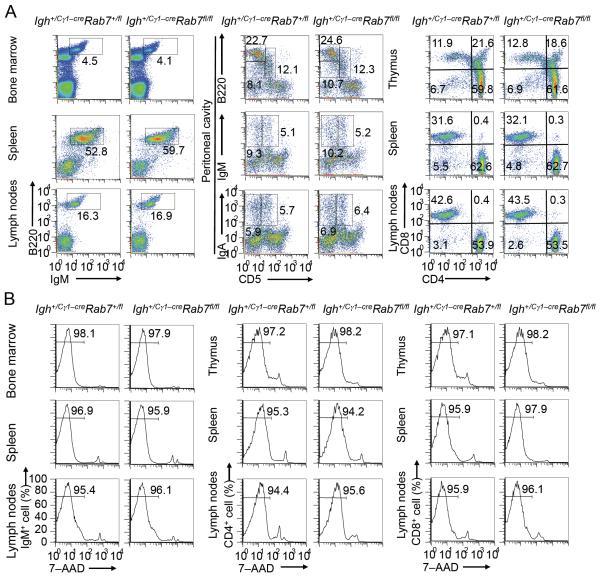
FIGURE 3. Igh+/Cγ1-creRab7fl/fl mice show normal B and T lymphocyte development and survival.
(A) Flow cytometry analysis of expression of B220 and IgM (B cell markers) in bone marrow, spleen and lymph node cells (left panels), expression of CD5, a B1 cell marker in humans and mice (83), and B220 in peritoneal cavity cells (B2 cells were CD5−B220hi, B1a cells were CD5+B220+ and B1b cells were CD5− B220+) as well as that of CD5 and IgM or IgA (middle panels), and expression of CD4 and CD8 in CD3+ T cells from the thymus, spleen and lymph nodes (right panels) in Igh+/Cγ1-creRab7fl/fl and Igh+/Cγ1-creRab7+/fl littermate mice. (B) Flow cytometry analysis of the viability (7–AAD−) of IgM+ B cells in the bone marrow, spleen and lymph nodes (left panels), as well as that of CD4+ (middle panels) and CD8+ (right panels) T (CD3+) cells in the thymus, spleen and lymph nodes in Igh+/Cγ1-creRab7fl/fl and Igh+/Cγ1-creRab7+/fl mice. Data are representative of three independent experiments.
Igh+/Cγ1-creRab7fl/fl mice show decreased IgG1+ B cells, IgG1-AFCs, serum IgG1 and antigen-specific IgG1
In Igh+/Cγ1-creRab7fl/fl mice and their Igh+/Cγ1-creRab7+/fl littermate controls (which showed phenotypes comparable to those of Igh+/Cγ1-creRab7+/+, data not shown), IgG1 expression was driven solely by the Igh+ allele that had undergone a productive VHDJH recombination and then CSR from Igμ to Igγ1, as the Cre sequence would interfere with the expression of VHDJH–Cγ1 in the IghCγ1-cre allele (48). As compared to their Igh+/Cγ1-creRab7+/fl littermates, Igh+/Cγ1-creRab7fl/fl mice showed normal titers of serum IgM, but much reduced (by 85%) titers of IgG1 (Fig. 4A). They had normal titers of other switched IgG isotypes (IgG2a, IgG2b, IgG3) and IgA, consistent with our prediction that B cells that were set to switch to these Ig isotypes maintained an intact Rab7 locus due to their germline Iγ2a-Sγ2a-Cγ2a, Iγ2b-Sγ2b-Cγ2b or Iγ3-Sγ3-Cγ3 transcription and lack of Iγ1-Sγ1-Cγ1-cre transcription.
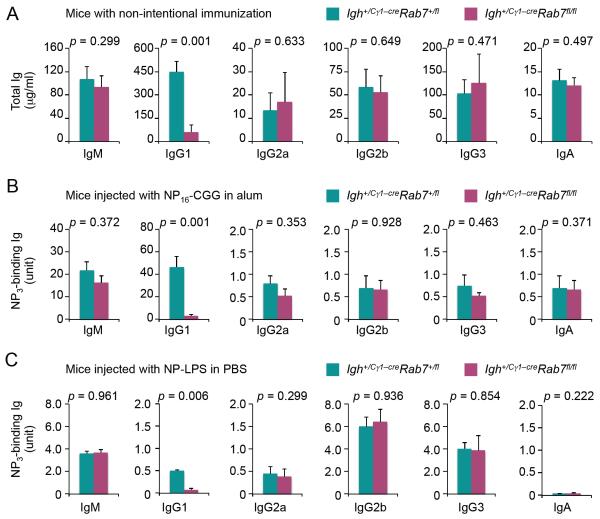
FIGURE 4. Igh+/Cγ1-creRab7fl/fl mice display reduced titers of overall IgG1 and antigen-specific IgG1.
(A) ELISA of serum titers of IgM, IgG1, IgG2a, IgG2b, IgG3 and IgA in Igh+/Cγ1-creRab7fl/fl mice (plum) and their Igh+/Cγ1-creRab7+/fl littermates (teal) (mean and s.e.m. of data from five pairs of mice). (B) ELISA of serum titers of high-affinity NP-specific (NP3-binding) IgG1, IgG2a, IgG2b, IgG3 and IgA as well as NP3-binding IgM (due to the high-avidity) in Igh+/Cγ1-creRab7fl/fl (plum) and their Igh+/Cγ1-creRab7+/fl littermate mice (teal) 9 d after injection with NP-CGG (mean and s.e.m. of data from five pairs of mice). (C) ELISA of serum titers of NP3-binding IgM, IgG1, IgG2a, IgG2b, IgG3 and IgA in Igh+/Cγ1-creRab7fl/fl (plum) and their Igh+/Cγ1-creRab7+/fl littermate mice (teal) 9 d after injection with NP-LPS (mean and s.e.m. of data from three pairs of mice). p values, as calculated by paired student t test, less than 0.05 were considered significant.
We next addressed the role of Rab7 in a specific T-dependent response by injecting mice with NP-CGG, which elicits predominantly NP-binding IgM and IgG1. Igh+/Cγ1-creRab7fl/fl mice showed a severe defect (over 96% reduction) in generation of (high-affinity) NP-specific IgG1, but normal levels of NP-specific IgM and other IgG isotypes (IgG2a, IgG2b, IgG3) and IgA (Fig. 4B). Likewise, Igh+/Cγ1-creRab7fl/fl mice could not produce T-independent NP-specific IgG1 response to NP-LPS, but were competent in generating NP-specific IgM, IgG2a, IgG2b and IgG3 (Fig. 4C) – CSR to IgG2b and IgG3 are characteristically induced by LPS.
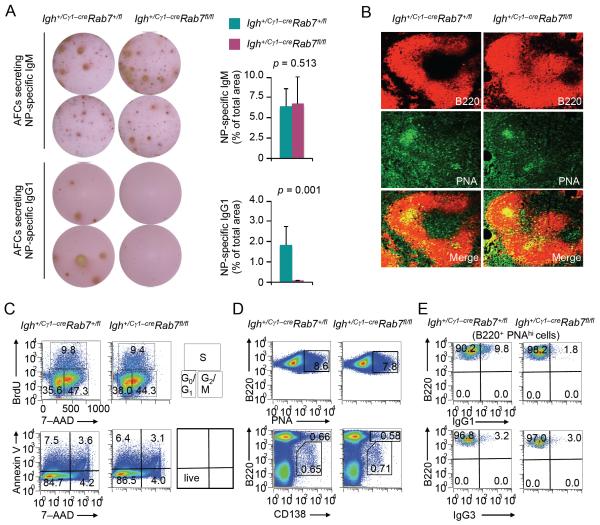
Consistent with an important role of B cell Rab7 in class-switched antibody responses, Igh+/Cγ1-creRab7fl/fl mice injected with NP-CGG were defective in generation of AFCs that secreted NP-specific IgG1, but could generate AFCs that secreted NP-binding IgM (Fig. 5A). These mice showed normal germinal center development, normal B cell proliferation and survival as well as normal differentiation of B cells into germinal center (PNAhi) B cells and plasma cells (Fig. 5B-5D). They, however, failed to generate IgG1+ B cells (Fig. 5E). By contrast, they displayed normal frequencies of IgG3+ B cells in the spleen and IgA+ B cells in the peritoneal cavity and Peyer’s patches (Fig. 3A, 5E and data not shown), consistent with specific ablation of Rab7 in B cells that were to undergo CSR to IgG1. Thus, Rab7 mediates B cell class-switching and generation of AFCs that secrete class-switched antibodies in T-independent and T-dependent responses. It does not play a major role in B cell proliferation, survival or differentiation into plasma cells.
FIGURE 5. Igh+/Cγ1-creRab7fl/fl mice show decreased IgG1+ B cells and IgG1-secreting AFCs.
(A) ELISPOT analysis and quantification ((mean and s.e.m. of data from three pairs of mice) of AFCs that produced NP-specific IgM or IgG1 in spleens of Igh+/Cγ1-creRab7fl/fl mice (plum) and their Igh+/Cγ1-creRab7+/fl littermates (teal) 9 d after injection with NP-CGG. p values, as calculated by paired student t test, less than 0.05 were considered significant. (B) Immunofluorescence staining and confocal microscopy analysis of germinal centers (PNAhi) within B220+ follicles in spleens of Igh+/Cγ1-creRab7+/fl and Igh+/Cγ1-creRab7fl/fl littermate mice 9 d after injection with NP-CGG. (C) Cell cycle analysis (top panels; the quadrant corresponding to the G0/G1, S or G2/M phase of the cell cycle was also depicted) and viability analysis (bottom panels; live cells were Annexin V−7–AAD−) of spleen B cells isolated ex vivo from Igh+/Cγ1-creRab7fl/fl mice and their Igh+/Cγ1-creRab7+/fl littermates 9 d after injection with NP-CGG. (D) Proportion of germinal centers (PNAhi) cells among (B220+) B cells (top panels) and proportion of (CD138+B220hi) plasmablasts and (CD138+B220lo) plasma cells in spleens of Igh+/Cγ1-creRab7fl/fl mice and their Igh+/Cγ1-creRab7+/fl littermates 9 d after injection with NP-CGG. (E) Proportion of IgG1+ (top panels) and IgG3+ (bottom panels) cells among (B220+PNAhi) germinal center B cells in spleens of Igh+/Cγ1-creRab7fl/fl mice and their Igh+/Cγ1-creRab7+/fl littermates 9 d after injection with NP-CGG. Data are representative of three independent experiments.
Igh+/Cγ1-creRab7fl/fl B cells are defective in CSR to IgG1 and IgE, as induced by a primary stimulus plus IL-4
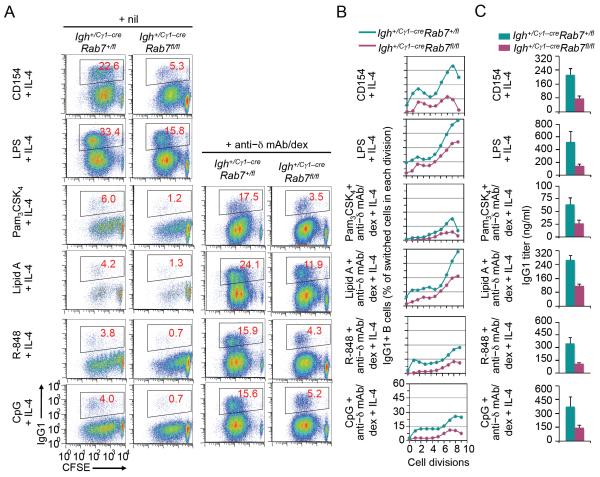
The notion that B cell Rab7 plays an important role in CSR was emphasized by the induction of Rab7 in B cells activated to undergo CSR (Fig. 2C). To address the intrinsic role of Rab7 in CSR, we isolated naïve B cells from Igh+/Cγ1-creRab7fl/fl mice (which expressed Rab7) and cultured them in the presence of a T-dependent or T-independent primary stimulus plus IL-4 to ablate Rab7. Upon stimulation by CD154, LPS alone, or a TLR ligand combined with anti–δ mAb/dex (for dual engagement of TLR1/2, TLR4, TLR7 or TLR9 and BCR), Igh+/Cγ1-creRab7fl/fl B cells gave rise to much fewer IgG1+ cells (up to 80% reduction, overall and across all B cell divisions) and secreted less IgG1, as compared to their Igh+/Cγ1-creRab7+/fl and Igh+/+Rab7fl/fl B cell counterparts (Fig. 6 and Fig. 7A). Consistent with what we reported in C57BL/6 B cells (13), CSR to IgG1 occurred at marginal levels in Igh+/Cγ1-creRab7+/fl B cells stimulated by a TLR ligand alone plus IL-4. It, however, was virtually abolished in Igh+/Cγ1-creRab7fl/fl B cells exposed to the same stimuli (Fig. 6A). Upon stimulation by a primary CSR-inducing stimulus plus a cytokine that direct CSR to IgG3, IgG2a and IgG2b, Igh+/Cγ1-creRab7+/fl B cells showed normal CSR, as expected due to their intact Rab7 locus (Fig. 7B and data not shown). They, however, became Rab7-deficient after stimulation by CD154 or LPS plus IL-4 for 48 h (Fig. 2B, 2C) and defective in CSR to IgA, as subsequently induced by LPS plus TGF-β and anti–δ mAb/dex (Fig. 7C). Despite their deficiency in CSR, Igh+/Cγ1-creRab7fl/fl B cells showed normal IgM expression and plasma cell differentiation (Fig. 7D, 7E).
FIGURE 6. Rab7 deficiency in B cells results in defective CSR to IgG1 in vitro.
(A) Flow cytometry analysis of proliferation and CSR to IgG1 in CFSE-labeled Igh+/Cγ1-creRab7fl/fl B cells after stimulation by a primary stimulus (CD154, LPS, a TLR ligand, alone or together with anti–Igδ mAb/dex, as indicated) plus IL-4 for 4 d and their Igh+/Cγ1-creRab7+/fl B cell counterparts. (B) Depiction of the proportion of switched IgG1+ cells in CFSE-labeled Igh+/Cγ1-creRab7fl/fl B cells and their Igh+/Cγ1-creRab7+/fl B cell counterparts that had completed each cell division after stimulation by a primary stimulus (CD154, LPS, a TLR ligand or together with anti–Igδ mAb/dex, as indicated) plus IL-4 for 4 d. Data are representative of three independent experiments. (C) ELISA of titers of IgG1 secreted into culture supernatants by Igh+/Cγ1-creRab7fl/fl B cells and their Igh+/Cγ1-creRab7+/fl B cell counterparts after stimulation by the same stimuli as in (B) for 7 d (mean and s.e.m. of data from three independent experiments).
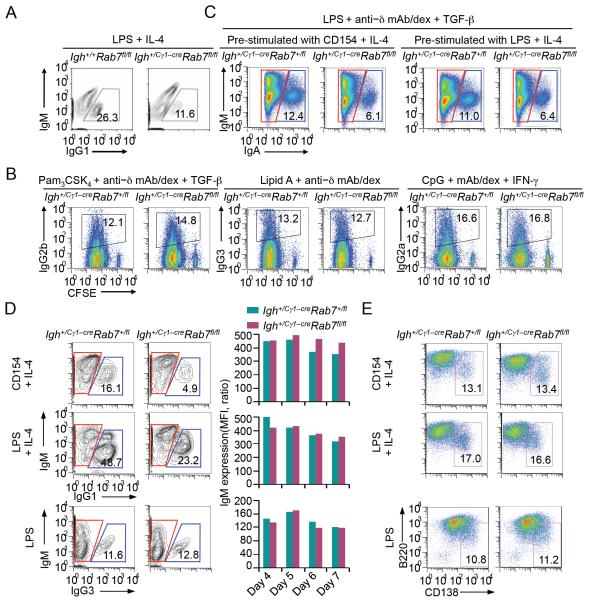
FIGURE 7. Igh+/Cγ1-creRab7fl/fl B cells are normal in IgM expression, class-switching to other IgG isotypes and plasma cell differentiation.
(A) Flow cytometry analysis of CSR to IgG1 in B cells from Igh+/+Rab7fl/fl and Igh+/Cγ1-creRab7fl/fl littermate mice stimulated by LPS plus IL-4 for 96 h. (B) Flow cytometry analysis of proliferation and CSR to IgG2b, IgG3 or IgG2a in CFSE-labeled Igh+/Cγ1-creRab7fl/fl B cells and their counterparts from Igh+/Cγ1-creRab7+/fl littermates stimulated by a selected primary stimulus, as indicated, plus a cytokine (TGF-β, nil or IFN-γ, as indicated) for 4 d (left panels), and depiction of the proportion of switched (IgG2b+, IgG3+ or IgG2a+) cells in B cells that had completed each cell division (right panels). (C) Flow cytometry analysis of CSR to IgA in Igh+/Cγ1-creRab7fl/fl and Igh+/Cγ1-creRab7+/fl B cells pre-stimulated by CD154 (left panels) or LPS (right panels) plus IL-4 for 48 h and then stimulated by LPS plus TGF-β and anti–Igδ mAb/dex for 72 h to undergo CSR to IgA. (D) Flow cytometry analysis of CSR to IgG1 in Igh+/Cγ1-creRab7fl/fl B cells stimulated by CD154 or LPS plus IL-4 for 5 d and their Igh+/Cγ1-creRab7+/fl B cell counterparts and analysis of CSR to IgG3 after those B cells were stimulated by LPS for 4 d (left panels; cells within red gates were unswitched IgM+IgG1− or IgM+IgG3− cells, and those within blue gates were switched IgG1+IgM− or IgG3+IgM− cells), quantification of IgM expression levels in unswitched IgM+IgG1− or IgM+IgG3− cells (middle panels). (E) Proportion of (CD138+B220lo) plasma cells after Igh+/Cγ1-creRab7fl/fl B cells and their counterparts from Igh+/Cγ1-creRab7+/fl littermates were stimulated by CD154 or LPS plus IL-4 or by LPS alone for 4 d. Data are representative of three independent experiments.
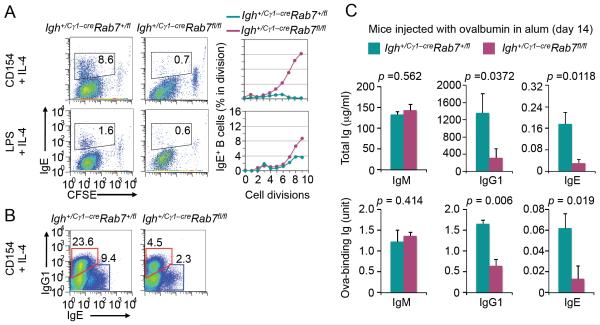
In addition to inducing Igh+ germline Iγ1-Sγ1-Cγ1 transcription and direct CSR to IgG1, IL-4 in conjunction with a primary stimulus (particularly CD154) induces Igh+ Iε-Sε-Cε transcription and CSR to IgE. The abrogation of Rab7 expression in Igh+/Cγ1-creRab7fl/fl B cells stimulated with CD154 or LPS plus IL-4 (Fig. 2B) prompted us to analyze the impact of Rab7 deficiency on CSR to IgE. As expected (5, 50), Igh+/Cγ1-creRab7+/fl B cells underwent CSR to IgE upon stimulation by CD154 plus IL-4, and less efficiently upon stimulation by LPS plus IL-4 (Fig. 8A). By contrast, Igh+/Cγ1-creRab7fl/fl B cells failed to undergo CSR to IgE, in spite of largely normal B cell dividing rates (Fig. 8A). Their defect in CSR to IgE was comparable in magnitude to their defect in CSR to IgG1 (Fig. 8B), reflecting an Ig isotype-independent role of Rab7 in CSR. Accordingly, in Igh+/Cγ1-creRab7fl/fl mice injected with OVA, the levels of OVA-specific IgE and IgG1 as well as total IgE and IgG1 were significantly reduced, as compared to those in Igh+/Cγ1-creRab7+/fl littermates, while IgM levels were normal (Fig. 8C). These in vivo and in vitro experiments further show that Rab7 plays an important role in mediating CSR independently of Ig isotypes. Such a role is central to the CSR machinery, as induced by all the T-dependent and T-independent primary stimuli we tested.
FIGURE 8. Igh+/Cγ1-creRab7fl/fl mice/B cells display impairment in CSR to IgE in vivo and in vitro.
(A) Intracellular staining and flow cytometry analysis of CSR to IgE in CFSE-labeled Igh+/Cγ1-creRab7fl/fl B cells and their counterparts from Igh+/Cγ1-creRab7+/fl littermates stimulated by CD154 or LPS plus IL-4 for 5 d (left panels) and depiction of the proportion of switched IgE+ cells in B cells that had completed each cell division (right panels). (B) Intracellular staining and flow cytometry analysis of CSR to IgE and IgG1 in Igh+/Cγ1-creRab7fl/fl B cells stimulated by CD154 plus IL-4 for 5 d and their Igh+/Cγ1-creRab7+/fl B cell counterparts (cells within red gates were switched IgE+IgG1− cells, and those within blue gates were switched IgG1+IgE− cells). Data are representative of three independent experiments. (C) ELISA of titers of total (top panels) and ovalbumin-binding (bottom panels) IgM, IgG1 and IgE in Igh+/Cγ1-creRab7fl/fl mice (plum) and their Igh+/Cγ1-creRab7+/fl littermates (teal) 14 d after injection with ovalbumin (mean and s.e.m. of data from three pairs of mice). p values, as calculated by paired student t test, less than 0.05 were considered significant.
Rab7-deficient B cells display normal IgH germline IH-S-CH transcription but impaired Aicda induction
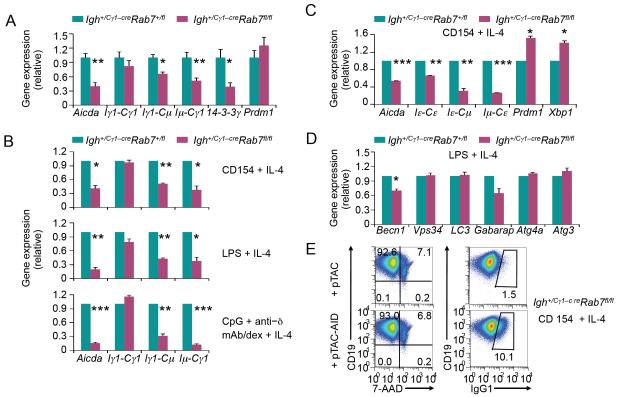
Our studies strongly suggested that Rab7 mediated processes that were induced mainly by primary CSR-inducing stimuli, but not secondary stimuli (which specify the isotype that a B cell will switch to). Indeed, Igh+/Cγ1-creRab7fl/fl B cells underwent normal germline Iγ1-Sγ1-Cγ1 and Iε-SεCε transcription, when induced in vivo and in vitro by IL-4 plus a primary stimulus (Fig. 9A-9C). They, however, showed significantly reduced expression of the Aicda transcripts and protein (Fig. 9A-9C, 10A) as well as reduced levels of circle Iγ1-Cμ and Iε-Cμ transcripts and post-recombination Iμ-Cγ1 and Iμ-Cε transcripts. Rab7 deficiency also decreased the expression of 14-3-3γ adaptor molecules, which stabilize AID and other CSR factors on the donor and receptor S region DNA for CSR to unfold (4, 52). By contrast, it did not decrease (or even increased) expression of Blimp-1 (encoded by Prdm1) and Xbp-1 (encoded by Xbp1), which drive plasma cell differentiation and/or functions (Fig. 9A-9C, 10B), consistent with the normal differentiation of Igh+/Cγ1-creRab7fl/fl B cells into plasma cells – expression of autophagy-related genes was also normal with the exception of Bcln1 (Fig. 9D). Enforced expression of AID in Igh+/Cγ1-creRab7fl/fl B cells that were pre-stimulated with CD154 plus IL-4 for 48 h (to abrogate Rab7) rescued CSR, resulting in over 10% of cells being IgG1+ after 72 h of stimulation with CD154 plus IL-4 (Fig. 9E) – the proportion (10%) of IgG1+ B cells was within the range of CSR levels in normal B cells stimulated for 72 h. Thus, Rab7 plays an important role in mediating AID induction and, therefore, CSR.
FIGURE 9. Igh+/Cγ1-creRab7fl/fl B cells are defective in the induction of AID in vivo and in vitro.
(A) qRT-PCR analysis of levels of Aicda, IgH germline Iγ1-Sγ1-Cγ1 transcripts, circle Iγ1-Cμ transcript, post-recombination Iμ-Cγ1 transcripts, 14-3-3γ transcripts and Pdrm1 transcripts in B cells isolated ex vivo from Igh+/Cγ1-creRab7fl/fl mice (plum) and their Igh+/Cγ1-creRab7+/fl littermates (teal) 9 d after injection with NP-CGG. Data were normalized to the level of Cd79b and are expressed as ratios of values in Igh+/Cγ1-creRab7+/fl B cells to those in Igh+/Cγ1-creRab7fl/fl B cells (mean and s.e.m. of data from three pairs of mice). (B) qRT-PCR analysis of levels of Aicda transcripts, IgH germline Iγ1-Cγ1 transcripts, circle Iγ1-Cμ transcripts and post-recombination Iμ-Cγ1 transcripts in Igh+/Cγ1-creRab7fl/fl B cells (plum) stimulated by CD154, LPS or CpG plus anti–δ mAb/dex in the presence of IL-4 for 48 h and in their Igh+/Cγ1-creRab7+/fl B cell counterparts (teal). (C) qRT-PCR analysis of levels of Aicda transcripts, IgH germline Iε-Cε transcripts, circle Iε-Cμ transcripts and post-recombination Iμ-Cε transcripts as well as those of Prdm1 and Xbp1 transcripts in Igh+/Cγ1-creRab7fl/fl B cells (plum) stimulated by CD154 plus IL-4 for 48 h and in their Igh+/Cγ1-creRab7+/fl B cell counterparts (teal). (D) qRT-PCR analysis of transcript levels of genes involved in autophagy, as indicated, in Igh+/Cγ1-creRab7fl/fl B cells (plum) stimulated by LPS plus IL-4 for 48 h and in their Igh+/Cγ1-creRab7+/fl B cell counterparts (teal). Data were normalized to the level of Cd79b transcripts and are expressed as ratios of values in Igh+/Cγ1-creRab7+/fl B cells to those in Igh+/Cγ1-creRab7fl/fl B cells (mean and s.e.m. of data from three independent experiments). ***, p < 0.005; **, p < 0.01; *, p < 0.05 (the p values were calculated by paired student t test). (E) Flow cytometry analysis of (CD19+) B cell viability (7-AAD−, left panels) and CSR (IgG1+, right panels) in Igh+/Cγ1-creRab7fl/fl B cells pre-stimulated by CD154 plus IL-4 for 48 h, transduced by pTAC or pTAC-AID retrovirus, and then stimulated by CD154 plus IL-4 for 72 h.
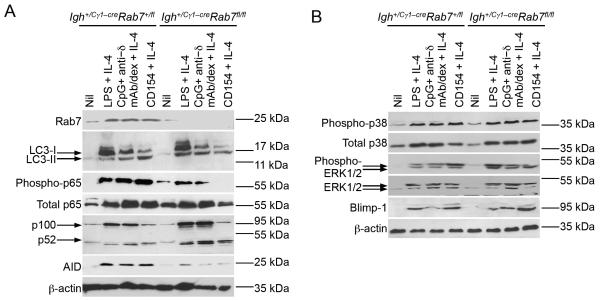
FIGURE 10. Rab7 deficiency in B cells results in defective canonical NF-κB signaling in vitro.
(A) Immunoblotting analysis of levels of Rab7, LC3-I (no processing) and LC3-II (processed and conjugated to phosphatidylethanolamine), phosphorylated and total p65 in the canonical NF-κB pathway, p100 and p52 in the non-canonical NF-κB pathway, AID and β-actin in Igh+/Cγ1-creRab7fl/fl B cells and their Igh+/Cγ1-creRab7+/fl control B cells stimulated by CD154, LPS or CpG plus anti–δ mAb/dex, as indicated, with IL-4 for 48 h. (B) Immunoblotting analysis of levels of phosphorylated and total p38 as well as phosphorylated and total ERK1/2 in MAPK pathways, Blimp-1 and β-actin in Igh+/Cγ1-creRab7fl/fl B cells and their Igh+/Cγ1-creRab7+/fl control B cells stimulated by CD154, LPS or CpG plus anti–δ mAb/dex, as indicated, with IL-4 for 48 h.
Rab7 deficiency results in specific impairment of canonical NF-κB activation
As we and others have shown (13, 24, 25), NF-κB activation is required for AID induction. This together with the role of Rab7 in the induction of AID expression (as well as 14-3-3γ expression, which also depends on NF-κB (49)) prompted us to analyze NF-κB activation in Igh+/Cγ1-creRab7fl/fl B cells, as stimulated by CD40, LPS or CpG plus anti-δ mAb/dex in the presence of IL-4. These B cells were defective in activation of p65 of the canonical NF-κB pathway (Fig. 10A), which, as we have shown, is induced by CD40 or TLR signaling, but not BCR signaling (13). By contrast, they were normal in the activation of the non-canonical NF-κB pathway (i.e., upregulation of p100 expression and processing of p100 to p52), which is induced by CD40 and BCR signaling, but not TLR signaling, and also plays an important role in Aicda induction (13). Stimulated Igh+/Cγ1-creRab7fl/fl B cells were normal in activation of p38 and ERK1/2 MAPK pathways, which are associated with cell metabolisms and induction of Prdm1 expression (53, 54), consistent with their normal proliferation/survival and plasma cell differentiation (Fig. 10B). Finally, they were normal in induction of LC3 (LC3-I) and conversion of LC3-I to LC3-II (Fig. 10A). Thus, Rab7 plays a specific role in the activation of the canonical NF-κB pathway, which is indispensable for AID induction.
Discussion
In this study, we have unveiled an important (B cell-intrinsic) role of Rab7 in the antibody response by constructing conditional KO mice that lack Rab7 only in B cells induced to undergo Iγ1-Sγ1-Cγ1 transcription. This design has circumvented any complications potentially arising from possible roles of Rab7 in B cell and T cell development. It has also allowed us to outline an important role of Rab7 in CSR, through activation of the canonical NF-κB pathway and induction of AID expression. The role of Rab7 in the generation of class-switched B cells and AFCs, but not plasma cells, contrasts the role of Atg5 in mediating the development of peritoneal B1 cells and generation/function of plasma cells, but not CSR (55-57). Rab7 may play a role in the bone marrow homing of plasma cells originating in secondary lymphoid organs, as suggested by our (unpublished) observations in Igh+/Cγ1-creRab7fl/fl mice; it might also mediate the differentiation of IgM+ and residual class-switched B cell into memory B cells and the maintenance/function of memory B cells – the Atg7 autophagic protein has been shown to play such a role (58). Addressing these possibilities would entail the generation of new conditional Rab7 KO mice, including those with tamoxifeninducible Cre expression, long after primary immunization (59) to ablate Rab7 in plasma cells and memory B cells – use of Tg(Prdm1-cre) mice would not be advisable due to the critical role of Rab7 in embryogenesis (47, 51) and possibly high Cre expression (together with Blimp-1 expression) during this process (60, 61). Finally, the normal IgM expression, proliferation and survival of Igh+/Cγ1-creRab7fl/fl B cells suggests that the role of Rab7 is redundant with that of other small GTPases in mediating basal membrane functions and maintaining B cell homeostasis.
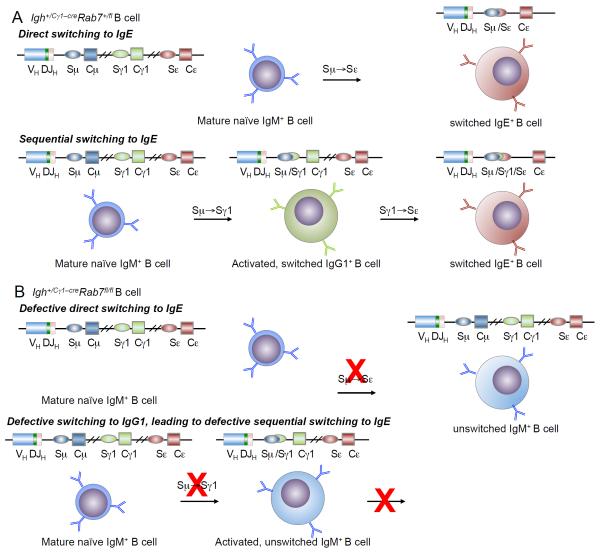
The role of Rab7 in CSR induction was intrinsic to B cells, as shown by the failure of Igh+/Cγ1-creRab7fl/fl B cells to undergo CSR in vivo in a T-independent antibody response and in vitro. In T-dependent antibody responses, Rab7 might mediate, in addition to CSR, autophagy in B cells for antigen presentation (62-65), thereby activating TH cells, which, in turn, induce further B cell activation and differentiation. In Igh+/Cγ1-creRab7fl/fl mice injected with (T-dependent) NP-CGG, however, B cells displayed normal proliferation, survival, germinal center reaction, IgM production and plasma cell differentiation, suggesting that Rab7-deficient B cells were competent in activating TH cells, possibly by using a different Rab GTPase (e.g., Rab9) for autophagy-mediated antigen-presentation, thereby receiving TH cell help. Igh+/Cγ1-creRab7fl/fl B cells showed impaired CSR to not only IgG1, but also IgE in vivo and in vitro despite normal Igh+ germline Iε-SεCε transcription, further suggesting that mediating induction of AID expression is the mechanism by which Rab7 plays a role in CSR. Decreased AID expression in these B cells would impair direct Sμ→Sε (IgM→IgE) switching and sequential Sμ→Sγ1→Sε (IgM→IgG1 and then →IgE) switching (Fig. 11), although the latter would be a minor contributor to the overall CSR to IgE in vitro, as suggested by the intact or even increased IgE production in B cells lacking the Iγ1 promoter or Sγ1 DNA (66, 67). Also, the normal IgG3, IgG2a, IgG2b or IgA levels in Igh+/Cγ1-creRab7fl/fl mice indicated that CSR to IgG2a, IgG2b or IgA is not dependent on sequential switching Sμ→Sγ1→Sγ2a, Sγ2b or Sα. As, to best of our knowledge, no other Igγ or Igα sublocus-specific cre knockin mice exist, verification of Rab7 role in CSR to non-IgG1 or IgE Ig isotypes in vitro would entail Rab7 gene ablation, through retroviral transduction to express Cre in Igh+/+Rab7fl/fl B cells or, as shown here, through stimulation of Igh+/Cγ1-creRab7fl/fl B cells with IL-4 in the presence of a primary CSR-inducing stimulus for 36 – 48 h, followed by culturing of B cells with stimuli that induce CSR to those Ig isotypes. The in vivo verification would entail generation of conditional Rab7 KO mice using Aicda+/cre (68) or BacTg(Aicda-cre)1Rcas allele (69). In these mice, Cre is induced in B cells activated to transcribe Aicda, in a fashion likely dependent on the initially available Rab7 (once Rab7 is ablated, Cre expression would be extinguished).
FIGURE 11. Depiction of defective CSR to IgE in Igh+/Cγ1–creRab7fl/fl B cells.
(A) Depiction of direct switching from IgM to IgE (top) and sequential switching from IgM to IgG1 and then IgE in Igh+/Cγ1–creRab7fl/fl B cells. (B) Depiction of defective direct switching from IgM to IgE in Igh+/Cγ1–creRab7fl/fl B cells (top). As the generation of IgG1+ B cells is also impaired (bottom) due to defective CSR from IgM to IgG1, sequential switching from IgM to IgG1 and then IgE is blocked in these B cells.
The role of Rab7 in CSR induction is tightly associated with its role in AID induction, as enforced expression of AID in Igh+/Cγ1-creRab7fl/fl B cells, which showed impaired AID expression, rescued CSR. The CSR rescue was perhaps incomplete, as expression of selected other important CSR factors, such as 14-3-3 adaptor proteins, whose induction also dependent on the canonical NF-κB activation (49), likely remained suboptimal. As we and others have shown, AID expression is tightly regulated, at several levels, including transcription, post-transcription, targeting and enzymatic activity (27, 70). Primary inducing stimuli induced expression or activation of upstream factors, including Rab7 and NF-κB, resulting in high levels of Aicda expression. The function of the canonical NF-κB pathway in AID induction can be potentiated, but not replaced, by the non-canonical NF-κB pathway, a non-redundancy emphasized by the inability of BCR crosslinking alone to induce AID or CSR despite its efficient activation of the non-canonical NF-κB pathway (13). Accordingly, the defective AID induction in Igh+/Cγ1-creRab7fl/fl B cells would at least partially result from impaired canonical NF-κB activation. By contrast, the non-canonical NF-κB pathway in these B cells was normal and would have mediated – perhaps in a compensatory manner – germline Iγ1-Sγ1-Cγ1 transcription. Consistent with this notion, this transcription process is critically regulated by the κB sites in the Iγ1 promoter and the 3′ locus control region (71, 72), and can be induced by BCR crosslinking (13, 73). The normal non-canonical NF-κB pathway would also support homeostasis of Igh+/Cγ1-creRab7fl/fl B cells. These B cells showed lower levels of CSR than Igh+/+Rab7fl/fl B cells, suggesting that their defective AID induction does not result from modulation of Aicda transcription by the loxP sites inserted in the Rab7 locus, which lies 30 Mbp 5′ of Aicda locus, including a distal regulatory region (24, 25).
As suggested by our findings here, NF-κB activation in B cells would be mediated by Rab7-dependent transduction of signals triggered by surface or intracellular receptors. The predominant localization of this small GTPase in mature endosomes and endosome-derived membranes suggests an important role of intracellular membranes in signal transduction in B cells. Such a role has been underappreciated for CD40 or surface TLRs, which are also likely endocytosed into endosomes upon ligand engagement. CD40 can be internalized by B cells stimulated by an anti-CD40 Ab, and the TLR4-LPS complex can be endocytosed by non-B cells (perhaps together with the TLR4 co-receptor CD14) to regulate IRF3 activation through Rab11a small GTPase and Ca2+ signaling (74-78). Rab7-containing endosomes, which may be the source of Rab7+ foci-like structures we detected by super-resolution microscopy, would house internalized receptors and recruit signaling adaptors necessary for specific activation of the canonical NF-κB pathway, such as the E3 ubiquitin ligase TRAF6. Thus, Rab7 would nucleate the formation of macromolecular complexes, perhaps within the foci structures, to further stabilize interactions of immune receptors and adaptors, thereby enhancing signaling strength and specificity. In B cells stimulated through CD40 or surface TLRs (e.g., TLR1/2 and TLR4), canonical NF-κB activation mediated by intracellular membranes would complement and strengthen that mediated by signals triggered at the plasma membrane. The normal induction of autophagy-related genes and normal conversion of LC3-I to LC3-II in Igh+/Cγ1-creRab7fl/fl B cells suggest that induction of autophagosome formation is independent of Rab7 – lack of significant increase of LC3-II in these B cells suggests that the overall degree of Rab7-mediated generation of autolysosomes (by fusion of autophagosomes with lysosomes) in B cells is low or another Rab protein plays a compensatory role in the fusion process to complete the autophagic flux. Our observation that ERK1/2 activation was normal Rab7-deficient B cells suggests that autophagosomes may recruit ERK1/2 and localize them onto their cytosolic/extra-luminal side to mediate ERK phosphorylation (79), likely in an LC3-dependent but Rab7-independent manner. Rab7-dependency of CSR and Atg5-dependency of plasma cell differentiation reflect putative roles of different intracellular membranes in transducing different signals from some of the same receptors. While internalized BCR might not trigger signals and merely be processed in lysosomes for antigen presentation (80), it could collaborate with TLR9-CpG complexes in double-membrane structures to activate NF-κB and MAPK (36). This raises the possibility that intracellular membranes provide a platform for relay of signals from individual receptors, as well as integration of signals from different receptors.
Our findings outline an important role of Rab7 in antibody responses, i.e., by regulating expression of genes critical to B cell differentiation, namely Aicda, possibly through biogenesis of endosomes, trafficking of immune receptors (CD40 or TLRs) along these membranes, and stabilization of receptor co-localization with specific signaling molecules (e.g., TRAF6). This would reflect a complex role of Rab7, a highly conserved molecule in phylogeny (81). In plants and early animals, engulfment and subsequent degradation of microbes by Rab7-dependent xenophagy would be an important component of immunity. In later animals that use the TLR-NF-κB and CD40-NF-κB signaling modules (82), Rab7 gained a new function, i.e., in signal transduction and gene regulation. Such functions would endow some non-mammals and all mammals with a sophisticated adaptive immunity, as centered on induction of AID for CSR and/or SHM in T-dependent and T-dependent and antibody responses.
Supplementary Material
Acknowledgements
We thank Christie-Lynn Mortales for help with ELISPOT analysis, Dr. Peilin Zheng for expert help with stimulated emission depletion imaging analysis and Dr. Andrew Lees (Fina Biosolutions) for anti-δmAb/dextran.
This work was supported by NIH grants AI 105813 and AI 079705 to P.C., and AI GM 089919 to A.L.E. P.C. was also supported by the Zachry Foundation Distinguished Chair and the Alliance for Lupus Research Target Identification in Lupus Grant ALR 295955. D. L. was supported by Texas Children’s Hospital Pediatric Pilot Research Fund and the Lymphoma SPORE Developmental Research Program from Baylor College of Medicine and the Methodist Research Institute.
Abbreviations used in this article
- AFCs
antibody forming cells
- AID
activation-induced cytidine deaminase
- Atg
autophagy-related
- BCR
B cell receptor
- CSR
class switch DNA recombination
- IgH
immunoglobulin heavy chain
- LPS
lipopolysaccharides
- MAMP
microbe-associated molecular pattern
- NP
4-hydroxy-3-nitrophenyl acetyl
- NP-CGG
NP-conjugated chicken γ-globulin
- NP-LPS
NP-conjugated lipopolysaccharides
- OVA
ovalbumin
- SHM
somatic hypermutation
- TLR
toll like-receptor
Footnotes
Disclosures
The authors declare no competing financial interests.
References
- 1.Casali P. Somatic recombination and hypermutation in the immune system. In: Krebs JE, Goldstein ES, Kilpatrick ST, editors. Lewin's Genes XI. Jones & Bartlett; Sudbury, MA: 2014. pp. 459–507. [Google Scholar]
- 2.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kato L, Stanlie A, Begum NA, Kobayashi M, Aida M, Honjo T. An evolutionary view of the mechanism for immune and genome diversity. J. Immunol. 2012;188:3559–3566. doi: 10.4049/jimmunol.1102397. [DOI] [PubMed] [Google Scholar]
- 4.Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CW, Park SR, Steinacker P, Li Z, Yates JRI, Herron B, Otto M, Zan H, Fu H, Casali P. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, White CA, Lam T, Pone EJ, Tran DC, Hayama KL, Zan H, Xu Z, Casali P. Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination. Cell Rep. 2013;5:702–714. doi: 10.1016/j.celrep.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, Gearhart PJ. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat. Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zan H, White CA, Thomas LM, Mai T, Li G, Xu Z, Zhang J, Casali P. Rev1 recruits Ung to switch regions and enhances du glycosylation for immunoglobulin class switch DNA recombination. Cell Rep. 2012;2:1220–1232. doi: 10.1016/j.celrep.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yousif AS, Stanlie A, Mondal S, Honjo T, Begum NA. Differential regulation of S-region hypermutation and class-switch recombination by noncanonical functions of uracil DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1016–1024. doi: 10.1073/pnas.1402391111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat. Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 10.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin Immunol. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 11.McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. Molecular programming of B cell memory. Nat. Rev. Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Victora GD, Nussenzweig MC. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 13.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, Patel PJ, Al-Qahtani A, Zan H, Xu Z, Casali P. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-κB pathway. Nat. Commun. 2012;3(767):1, 12. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pone EJ, Xu Z, White CA, Zan H, Casali P. B cell TLRs and induction of immunoglobulin class-switch DNA recombination. Front. Biosci. 2012;17:2594–2615. doi: 10.2741/4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerutti A, Cols M, Puga I. Activation of B cells by non-canonical helper signals. EMBO Rep. 2012;13:798–810. doi: 10.1038/embor.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goenka R, Matthews AH, Zhang B, O'Neill PJ, Scholz JL, Migone TS, Leonard WJ, Stohl W, Hershberg U, Cancro MP. Local BLyS production by T follicular cells mediates retention of high affinity B cells during affinity maturation. J. Exp. Med. 2014;211:45–56. doi: 10.1084/jem.20130505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goenka R, Scholz JL, Sindhava VJ, Cancro MP. New roles for the BLyS/BAFF family in antigen-experienced B cell niches. Cytokine Growth Factor Rev. 2014 doi: 10.1016/j.cytogfr.2014.01.001. DOI: 10.1016/j.cytogfr.2014.1001.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magri G, Miyajima M, Bascones S, Mortha A, Puga I, Cassis L, Barra CM, Comerma L, Chudnovskiy A, Gentile M, Llige D, Cols M, Serrano S, Arostegui JI, Juan M, Yague J, Merad M, Fagarasan S, Cerutti A. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat. Immunol. 2014;15:354–364. doi: 10.1038/ni.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J. Exp. Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuang FL, Luo Z, Scharff MD. H3 trimethyl K9 and H3 acetyl K9 chromatin modifications are associated with class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5288–5293. doi: 10.1073/pnas.0901368106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, Cho YW, Sun HW, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22190–22195. doi: 10.1073/pnas.1016923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Zan H, Xu Z, Casali P. Epigenetics of the antibody response. Trends Immunol. 2013;34:460–470. doi: 10.1016/j.it.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat. Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tran TH, Nakata M, Suzuki K, Begum NA, Shinkura R, Fagarasan S, Honjo T, Nagaoka H. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat. Immunol. 2010;11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- 26.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat. Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop GA. The multifaceted roles of TRAFs in the regulation of B-cell function. Nat. Rev. Immunol. 2004;4:775–786. doi: 10.1038/nri1462. [DOI] [PubMed] [Google Scholar]
- 29.Bishop GA. The many faces of CD40: multiple roles in normal immunity and disease. Semin Immunol. 2009;21:255–256. doi: 10.1016/j.smim.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Fruman DA, Satterthwaite AB, Witte ON. Xid-like phenotypes: a B cell signalosome takes shape. Immunity. 2000;13:1–3. doi: 10.1016/s1074-7613(00)00002-9. [DOI] [PubMed] [Google Scholar]
- 31.Pham LV, Tamayo AT, Yoshimura LC, Lo P, Terry N, Reid PS, Ford RJ. A CD40 Signalosome anchored in lipid rafts leads to constitutive activation of NF-kappaB and autonomous cell growth in B cell lymphomas. Immunity. 2002;16:37–50. doi: 10.1016/s1074-7613(01)00258-8. [DOI] [PubMed] [Google Scholar]
- 32.Rawlings DJ, Schwartz MA, Jackson SW, Meyer-Bahlburg A. Integration of B cell responses through Toll-like receptors and antigen receptors. Nat. Rev. Immunol. 2012;12:282–294. doi: 10.1038/nri3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Feng J, Wen R, Marine JC, Sangster MY, Parganas E, Hoffmeyer A, Jackson CW, Cleveland JL, Murray PJ, Ihle JN. Phospholipase Cgamma2 is essential in the functions of B cell and several Fc receptors. Immunity. 2000;13:25–35. doi: 10.1016/s1074-7613(00)00005-4. [DOI] [PubMed] [Google Scholar]
- 34.Alexia C, Poalas K, Carvalho G, Zemirli N, Dwyer J, Dubois SM, Hatchi EM, Cordeiro N, Smith SS, Castanier C, Le Guelte A, Wan L, Kang Y, Vazquez A, Gavard J, Arnoult D, Bidere N. The endoplasmic reticulum acts as a platform for ubiquitylated components of nuclear factor kappaB signaling. Sci. Signal. 2013;6:ra79. doi: 10.1126/scisignal.2004496. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 36.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity. 2008;28:799–809. doi: 10.1016/j.immuni.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virgin HW, Levine B. Autophagy genes in immunity. Nat. Immunol. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLeod IX, Jia W, He YW. The contribution of autophagy to lymphocyte survival and homeostasis. Immunol Rev. 2012;249:195–204. doi: 10.1111/j.1600-065X.2012.01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K, Ichinose S, Hayashizaki K, Tsubata T. Induction of autophagy by B cell antigen receptor stimulation and its inhibition by costimulation. Biochem. Biophys. Res. Commun. 2008;374:274–281. doi: 10.1016/j.bbrc.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe K, Tsubata T. Autophagy connects antigen receptor signaling to costimulatory signaling in B lymphocytes. Autophagy. 2009;5:108–110. doi: 10.4161/auto.5.1.7278. [DOI] [PubMed] [Google Scholar]
- 43.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edinger AL, Cinalli RM, Thompson CB. Rab7 prevents growth factor-independent survival by inhibiting cell-autonomous nutrient transporter expression. Dev. Cell. 2003;5:571–582. doi: 10.1016/s1534-5807(03)00291-0. [DOI] [PubMed] [Google Scholar]
- 45.Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero Rosales K, Peralta ER, Guenther GG, Wong SY, Edinger AL. Rab7 activation by growth factor withdrawal contributes to the induction of apoptosis. Mol. Biol. Cell. 2009;20:2831–2840. doi: 10.1091/mbc.E08-09-0911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy SG, Stevens MW, So L, Edinger AL. Reciprocal effects of rab7 deletion in activated and neglected T cells. Autophagy. 2013;9:1009–1023. doi: 10.4161/auto.24468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mai T, Pone EJ, Li G, Lam TS, Moehlman J, Xu Z, Casali P. Induction of AID-targeting adaptor 14-3-3γ is mediated by NF-κB-dependent recruitment of CFP1 to the 5′-CpG-3′ - rich 14-3-3γ promoter and is sustained by E2A. J. Immunol. 2013;191:1895–1906. doi: 10.4049/jimmunol.1300922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wesemann DR, Magee JM, Boboila C, Calado DP, Gallagher MP, Portuguese AJ, Manis JP, Zhou X, Recher M, Rajewsky K, Notarangelo LD, Alt FW. Immature B cells preferentially switch to IgE with increased direct Sμ to Sε recombination. J. Exp. Med. 2011;208:2733–2746. doi: 10.1084/jem.20111155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawamura N, Sun-Wada G, Aoyama M, Harada A, Takasuga S, Sasaki T, Wada Y. Delivery of endosomes to lysosomes via microautophagy in the visceral endoderm of mouse embryos. Nat. Commun. 2012;3(1071):1–10. doi: 10.1038/ncomms2069. [DOI] [PubMed] [Google Scholar]
- 52.Lam T, Thomas LM, White CA, Li G, Pone EJ, Xu Z, Casali P. Scaffold functions of 14-3-3 adaptors in B cell immunoglobulin class switch DNA recombination. PLoS One. 2013;8:e80414. doi: 10.1371/journal.pone.0080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasuda T, Kometani K, Takahashi N, Imai Y, Aiba Y, Kurosaki T. ERKs induce expression of the transcriptional repressor Blimp-1 and subsequent plasma cell differentiation. Sci. Signal. 2011;4:ra25. doi: 10.1126/scisignal.2001592. [DOI] [PubMed] [Google Scholar]
- 54.Allman DM, Cancro MP. pERKing up the BLIMP in plasma cell differentiation. Sci. Signal. 2011;4:pe21. doi: 10.1126/scisignal.2001987. [DOI] [PubMed] [Google Scholar]
- 55.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, Virgin H. W. t. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 56.Conway KL, Kuballa P, Khor B, Zhang M, Shi HN, Virgin HW, Xavier RJ. ATG5 regulates plasma cell differentiation. Autophagy. 2013;9 doi: 10.4161/auto.23484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, Orfanelli U, Ponzoni M, Sitia R, Casola S, Cenci S. Plasma cells require autophagy for sustainable immunoglobulin production. Nat. Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 58.Chen M, Hong MJ, Sun H, Wang L, Shi X, Gilbert BE, Corry DB, Kheradmand F, Wang J. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat. Med. 2014;20:503–510. doi: 10.1038/nm.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang NS, McHeyzer-Williams LJ, Okitsu SL, Burris TP, Reiner SL, McHeyzer-Williams MG. Divergent transcriptional programming of class-specific B cell memory by T-bet and RORalpha. Nat. Immunol. 2012;13:604–611. doi: 10.1038/ni.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 61.Ohinata Y, Payer B, O'Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 62.Menendez-Benito V, Neefjes J. Autophagy in MHC class II presentation: sampling from within. Immunity. 2007;26:1–3. doi: 10.1016/j.immuni.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In Vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J. Exp. Med. 2011;208:2625–2632. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu. Rev. Immunol. 2012;30:611–646. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 66.Jung S, Siebenkotten G, Radbruch A. Frequency of immunoglobulin E class switching is autonomously determined and independent of prior switching to other classes. J. Exp. Med. 1994;179:2023–2026. doi: 10.1084/jem.179.6.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Misaghi S, Garris CS, Sun Y, Nguyen A, Zhang J, Sebrell A, Senger K, Yan D, Lorenzo MN, Heldens S, Lee WP, Xu M, Wu J, DeForge L, Sai T, Dixit VM, Zarrin AA. Increased targeting of donor switch region and IgE in Sgamma1-deficient B cells. J. Immunol. 2010;185:166–173. doi: 10.4049/jimmunol.1000515. [DOI] [PubMed] [Google Scholar]
- 68.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin B, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montano C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R. Regulation of AID expression in the immune response. J. Exp. Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stavnezer J, Schrader CE. IgH chain class switch recombination: mechanisms and regulaton. J. Immunol. 2014;193:5370–5378. doi: 10.4049/jimmunol.1401849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warren WD, Roberts KL, Linehan LA, Berton MT. Regulation of the germline immunoglobulin Cgamma1 promoter by CD40 ligand and IL-4: dual role for tandem NF-kappaB binding sites. Mol. Immunol. 1999;36:31–44. doi: 10.1016/s0161-5890(98)00114-x. [DOI] [PubMed] [Google Scholar]
- 72.Dunnick WA, Shi J, Graves KA, Collins JT. The 3′ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four gamma genes. J. Exp. Med. 2005;201:1459–1466. doi: 10.1084/jem.20041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turner ML, Corcoran LM, Brink R, Hodgkin PD. High-affinity B cell receptor ligation by cognate antigen induces cytokine-independent isotype switching. J. Immunol. 2010;184:6592–6599. doi: 10.4049/jimmunol.0903437. [DOI] [PubMed] [Google Scholar]
- 74.Hostager BS, Catlett IM, Bishop GA. Recruitment of CD40 and tumor necrosis factor receptor-associated factors 2 and 3 to membrane microdomains during CD40 signaling. J. Biol. Chem. 2000;275:15392–15398. doi: 10.1074/jbc.M909520199. [DOI] [PubMed] [Google Scholar]
- 75.Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, Halaas O, Nilsen NJ, Stenmark H, Latz E, Lien E, Mollnes TE, Bakke O, Espevik T. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147:868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chiang CY, Veckman V, Limmer K, David M. Phospholipase Cγ-2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J. Biol. Chem. 2012;287:3704–3709. doi: 10.1074/jbc.C111.328559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aksoy E, Taboubi S, Torres D, Delbauve S, Hachani A, Whitehead MA, Pearce WP, Berenjeno-Martin I, Nock G, Filloux A, Beyaert R, Flamand V, Vanhaesebroeck B. The p110δ isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat. Immunol. 2012;13:1045–1054. doi: 10.1038/ni.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hou P, Araujo E, Zhao T, Zhang M, Massenburg D, Veselits M, Doyle C, Dinner AR, Clark MR. B cell antigen receptor signaling and internalization are mutually exclusive events. PLoS Biol. 2006;4:e200. doi: 10.1371/journal.pbio.0040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mackiewicz P, Wyroba E. Phylogeny and evolution of Rab7 and Rab9 proteins. BMC Evol. Biol. 2009;9:101. doi: 10.1186/1471-2148-9-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leulier F, Lemaitre B. Toll-like receptors--taking an evolutionary approach. Nat. Rev. Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 83.Kasaian MT, Ikematsu H, Casali P. Identification and analysis of a novel human surface CD5- B lymphocyte subset producing natural antibodies. J. Immunol. 1992;148:2690–2702. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.