Abstract
UGT2B enzymes metabolize multiple endogenous and exogenous molecules, including steroid hormones and clinical drugs. However, little is known about the inter-individual variation in gene expression and its determinants. We re-sequenced candidate regulatory regions and the partial coding regions (41.1 kb) of UGT2B genes and identified 332 genetic variants. We measured gene expression in normal breast and liver samples and observed different patterns. The expression levels varied greatly across individuals in both tissues and were significantly correlated with each other in liver. Genotyping of tagging single-nucleotide polymorphisms (SNPs) in the same samples and association tests between genotype and transcript levels identified 62 variants that were associated with at least one UGT2B mRNA levels in either tissue. Most of these cis-regulatory SNPs were not shared between tissues, suggesting that this gene family is regulated in a tissue-specific manner. Our results provide insight into studying the role of UGT2B variation in hormone-dependent cancers and drug response.
Keywords: UGT2B, genetic variation, gene expression, cis-regulatory
Introduction
One of the most important goals of genetics is to identify genetic variation that can explain the observed variation in disease susceptibility and in other phenotypes. However, elucidating the relationship between genetic variation and complex phenotypes at the organismal level is not straightforward. Most polymorphisms that influence complex phenotypes first alter intermediate and molecular phenotypes, including, but not limited to, gene (mRNA, ncRNA, microRNA and so on) expression,1 mRNA secondary structure and stability,2 protein sequence, RNA splicing,3 microRNA interaction4 and codon usage,5 which in turn contribute to diseases and complex phenotypes. Among these molecular phenotypes, variation in expression levels has been proposed as one of the most important types in the human genome.1 To aid in the dissection of the genetic basis of human traits and diseases, numerous association studies of transcript levels have been performed in multiple tissues on a genome-wide scale.1, 6, 7, 8, 9, 10
Glucuronidation is an important clearance pathway for many endogenous and exogenous molecules, including steroid hormones, bile acid, bilirubin, carcinogens and clinical drugs.11, 12 UDP-glucuronosyltransferases (UGTs) transfer glucuronic acid from UDP-glucuronic acid to substrates, thus making them more water soluble than their parent compound and more easily excreted through the biliary and renal systems.11 In humans, there are two major UGT subfamilies, UGT1A and UGT2B.13, 14, 15 UGT2B includes 7 active members (UGT2B4, UGT2B7, UGT2B10, UGT2B11, UGT2B15, UGT2B17 and UGT2B28) located on chromosome 4,13, 14, 15 which are mainly expressed in liver, breast, prostate, colon and kidney.16, 17, 18 The UGT2B17 and UGT2B28 genes are not expressed in some individuals due to whole-gene polymorphic deletions (copy number variation, CNV) that are common in human populations.19 Because UGT2B enzymes are essential in the metabolism of steroid hormones which in turn have a central role in multiple cancers, it has been proposed that variation in the UGT2B genes is involved in breast20 and prostate21, 22, 23, 24, 25 cancer risk.26, 27 Therefore, a systematic survey of genetic variants that affect enzyme activity and gene expression will provide a firm basis for elucidating the role of these genes in hormone-dependent cancers.
Although recent genome-wide association studies of transcript levels have identified multiple potential regulatory elements for this family,6, 7, 10 these studies have several shortcomings. First, most studies used lymphoblastoid cell lines (LCLs)6, 7, 10 that express only a few UGT2B genes. Moreover, the regulation of this gene family is likely to differ among tissues as proposed by a recent study.28 Therefore, results from LCLs cannot easily be extrapolated to other tissues. Second, the current expression arrays cannot detect modest to small changes in expression level; this problem is further aggravated by the cross-hybridization of probes to the highly similar UGT2B genes. Third, only single-nucleotide polymorphisms (SNPs) from the HapMap project were tested, which has low coverage in regions containing duplicated genes like the UGT2B. Indeed, the average SNP density in the UGT2B cluster is 1 per ~1300 bp in the HapMap data compared with the genome-wide average of 1 per ~750 bp.29
In light of the limitations of previous studies, we performed a more detailed survey of sequence and expression variation of the UGT2B gene cluster. Candidate regulatory regions in the UGT2B cluster were re-sequenced in ethnically diverse individuals and tagging SNPs were selected accordingly. Transcript levels for each UGT2B gene were measured by quantitative real-time PCR (qPCR) in normal breast and liver tissue samples and tagging SNPs were genotyped in the same samples. The subsequent association analysis identified multiple potential cis-regulatory variants, which represent strong candidate susceptibility variants for hormone-dependent cancers and for inter-individual variation in drug response.
Materials and methods
Bioinformatics analyses
We first used bioinformatics tools to identify candidate regulatory regions. We used two major approaches: one is the prediction of transcription factor binding sites and the other is the identification of conserved sequences across distantly related species. The binding matrix for transcription factors enriched in the liver or prostate, including HNF1 homeobox A (HNF1A), hepatocyte nuclear factor 4, α (HNF4A), nuclear receptor subfamily 1, group I, member 2 (NR1I2), coxsackie virus and adenovirus receptor (CXADR), POU class 2 homeobox 1 (POU2F1), peroxisome proliferator-activated receptor α (PPARA), retinoid X receptor, γ (RXRG), CCAAT/enhancer binding protein α (CEBPA), forkhead box A2 (FOXA2), nuclear receptor subfamily 2, group F, member 2 (NR2F2) and for nuclear hormone receptors, such as nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor, NR3C1) and estrogen receptor 1 (ESR1), were obtained from the TRANSFAC database (http://www.gene-regulation.com/). Binding site prediction on the human reference sequence was performed by Cluster Buster (http://zlab.bu.edu/cluster-buster/).30 Regions that contain binding site clusters predicted with high probability and that are conserved between humans and at least two other species, as shown on the ECR genome browser (http://ecrbrowser.dcode.org/), were included in the re-sequencing survey (Supplementary Figure S1). Moreover, the promoter regions and partial coding regions were also included. In total, 41.1 kb were chosen for re-sequencing.
Re-sequencing and tagging
SNP selection Fifty-six unrelated HapMap samples (24 YRI, 22 CEU and 10 ASN) were chosen for re-sequencing. cDNA was synthesized using the Super Transcript III kit (Invitrogen, Carlsbad, CA, USA) and utilized for the amplification of UGT2B17 coding regions and UGT2B7 exons 2–6 (that is, the UGT2B7_v4 variant, which is expressed in LCLs, unlike the complete UGT2B7_v1 variant31). All PCR was performed using the primers in Supplementary Table S1. After exonuclease I and Shrimp Alkaline Phosphatase (United States Biochemicals, Cleveland, OH, USA) treatment, sequencing was performed by using PCR and internal primers in Supplementary Table S1 and BigDye Terminator v3.1 (Applied Biosystems, Foster City, CA, USA). Polymorphisms were scored by PolyPhred32 and confirmed through visual inspection. Visual genotype plots were drawn by using the Genome Variation Server (http://gvs.gs.washington.edu/GVS/). The UGT2B15 promoter and exon 1 re-sequencing data in our recent study,28 HapMap genotyping data in UGT2B cluster and UGT2B4 re-sequencing data from Environmental Genome Project (http://egp.gs.washington.edu/welcome.html) were also included in the selection of tagging SNPs. Tagging SNPs were chosen by using ldSelect33 with r2 greater than or equal to 0.8 and minor allele frequency (MAF) greater than or equal to 0.05. All re-sequencing data will be available in the PharmGKB database (http://www.pharmgkb.org).
Tissue collection, RNA and DNA isolation
Eighty-one normal breast (4 European American (CA), 8 African American (AA) and 69 of unknown ethnicity; 78 female, 2 male and 1 unknown gender) and 31 normal liver (3 CA, 1 AA and 27 unknown; 14 female, 16 male and 1 unknown) tissue samples were obtained from the University of Chicago Tissue Core Facility. RNA and DNA were isolated by RNeasy Lipid Tissue Mini Kit and QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA), respectively.
Genotyping of breast and liver samples
CNVs in UGT2B17 and UGT2B28 were genotyped in breast and liver samples by a previously published qPCR assay28 and Taqman assay Hs00852540_s1 (Applied Biosystems) according to manufacturer’s recommendation. Partial UGT2B17 CNV data were reported in our recent study.28
The tagging SNPs were genotyped in all breast and liver samples by iPLEX SNP genotyping assay (Sequenom, San Diego, CA, USA) according to the manufacturer’s protocol. In brief, the PCR and extension primers for each SNP were designed by online tools provided by the manufacturer (https://mysequenom.com/default.aspx) and listed in Supplementary Table S2. Multiplex PCR was performed with HotStarTaq DNA polymerase (Qiagen). After Shrimp Alkaline Phosphatase (Sequenom) treatment, single base extension was performed. The extension product was dispensed onto the SpectroCHIP bioarray (Sequenom) and the mass was determined and distinguished by Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Sequenom). The genotype calls were analyzed by Typer 4.0 software (Sequenom) and confirmed through visual inspection. Fifteen HapMap samples with known genotypes at all sites were included as positive controls.
Quantitative real-time PCR
cDNA for breast and liver samples was synthesized by using the High Capacity Reverse Transcription Kit (Applied Biosystems). Transcript levels for the UGT2B genes in breast and liver tissues were measured by qPCR with power SYBR green (Applied Biosystems) and primers listed in Supplementary information. All qPCR assays had high efficiency (>95%) and all PCR products were sequenced to confirm the primer specificity. The qPCR was performed in triplicate for each gene. β-actin and 18S mRNA level were quantified in breast and liver tissues, respectively, as reference genes due to their relatively conserved expression in these two tissues (data not shown). UGT2B11 and UGT2B28 transcript levels were not quantified because primers specific for these genes could not be designed. All qPCR and Taqman readings in this study were performed on a StepOne Plus Real time PCR System (Applied Biosystems). The details for qPCR are described according to minimum information for publication of qPCR experiment guidelines34 in Supplementary information.
Statistical analyses
First, UGT2B transcript levels in breast or liver were normalized by log2 transformation. For individuals with null expression of a specific gene, we assigned a value of one half the minimum value observed for that gene to avoid taking log transformation on zero. Then, we used linear regression models to assess the relationship between genotypes and gene expression. As the true mode of inheritance was unknown, we used a two degree of freedom linear model to jointly test for differences between the three genotype categories. To gain statistical power, we also fitted an additive linear model to the SNP genotype and gene expression data. To identify SNPs that influence UGT2B17 transcript levels in addition to its CNV, we excluded individuals with homozygous deletions and adjusted for CNV genotype (1 copy or 2 copies) by multiple linear regressions. We also used a permutation method to get multiple test-corrected gene cluster-wide P-values. Similarly, we used linear regression models to examine whether UGT2B gene expression was correlated with sex and age. For genes whose expression was correlated with age or sex, we further adjusted for age and/or sex when we assessed the association between genotype and mRNA levels. Similar adjustments for ancestry could not be made due to incomplete information about the ethnicity of our samples. Finally, we asked whether expression levels of different genes in liver or breast are correlated with each other by using Spearman’s rank correlation coefficients. Analysis of gene cluster-wide genotype expression correlations was conducted using plink v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/).35 Other statistical analysis and data management were conducted using Stata 11.0 (StataCorp LP, College Station, TX, USA).
Results
Re-sequencing survey of the UGT2B gene cluster
Our re-sequencing survey identified 332 genetic variants (see Supplementary Figure S2 and Supplementary Table S3 for details), most (91.8%) of which were not genotyped by the HapMap project. Among all variants, 314 (97.2%) were SNPs while the others were short insertion/deletion polymorphisms. Approximately half of these variants (162 out of 332) showed an MAF >5%. Twenty-seven of them were located in the coding regions and 10 were non-synonymous. We included the HapMap SNP genotype and other available re-sequencing data for the same individuals and used them together with our re-sequencing data (1316 genetic variants total) to select tagging SNPs for the UGT2B cluster. We identified 285 tag bins with MAF greater than or equal to 0.05 for the entire cluster and at least one tagging SNP for each bin was chosen for iPLEX assay design.
SNP and CNV genotyping in breast and liver DNA samples
Two hundred thirty-eight individual tagging SNPs (83.5%) were clustered in seven genotyping assays; the remaining 47 tagging SNPs were rejected by the assay design program and could not be surveyed. Out of the 238 clustered SNPs, 189 SNPs (79.4%) were successfully genotyped (see Supplementary Figure S3 for detail). Given the high sequence similarity across the UGT2B genes, it is not surprising that many SNPs did not yield a high quality assay. For the successfully genotyped SNPs, the comparison between the iPLEX genotype calls and the known genotypes for 15 controls yielded an error rate <1.9%. We also compared the MAF and linkage disequilibrium pattern between the iPLEX genotypes and our re-sequencing data. As shown in Supplementary Figure S4, the MAF in the iPLEX data are highly correlated with those in our re-sequencing data (r2=0.717, P<10−52). The linkage disequilibrium pattern is also similar (result not shown). No deviation from Hardy–Weinberg equilibrium was observed (P>0.05) in most SNPs (85.2% for breast and 95.2% for liver samples). It is worth noting that most (64.3%) of the SNPs with Hardy–Weinberg equilibrium departures in the breast samples showed the same pattern in the HapMap and the re-sequencing data when different populations were pooled into a single sample (not shown), suggesting that the Hardy–Weinberg equilibrium departures are due to high differentiation of these SNPs across populations coupled with the mixed ethnicity of the breast samples. When these SNPs were removed from analysis, only 5.3% SNPs in breast showed deviation from Hardy–Weinberg equilibrium.
The genotype frequencies for UGT2B17 and UGT2B28 CNVs in the breast and liver samples are in good agreement with those for the HapMap CEU and YRI population samples (Supplementary Table S4).19
UGT2B gene expression in breast and liver
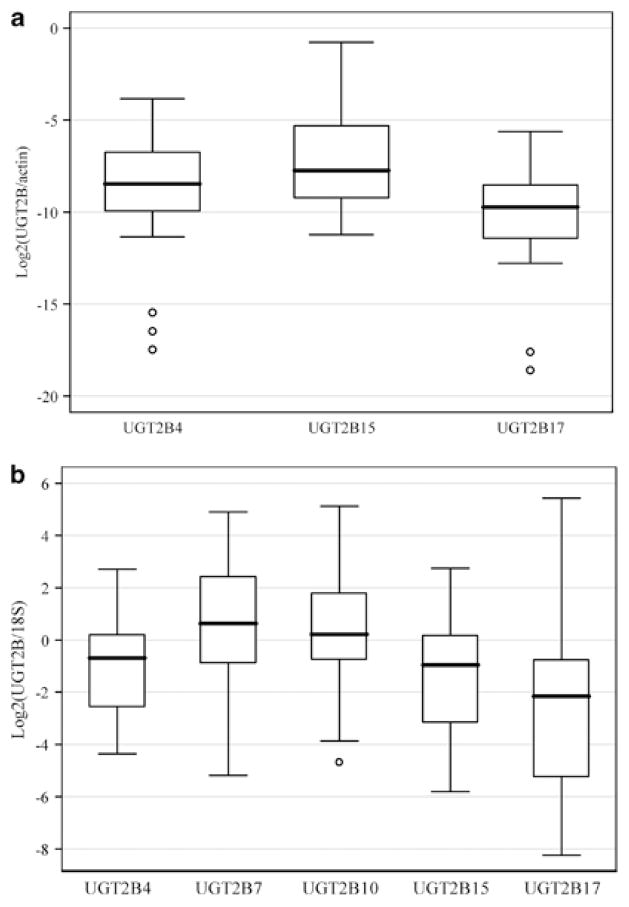
UGT2B transcript levels in breast and liver samples are displayed in Figures 1a and b, respectively. In general, UGT2B15 (quantification cycle (Cq)34 value, mean±s.d., 32.0±2.6) and UGT2B17 (Cq value, mean±s.d., 33.5±3.0) are expressed at intermediate levels in breast while UGT2B4 (Cq value, mean±s.d., 35.3±3.4) is expressed at lower levels. However, the comparison of expression levels across genes should be interpreted with caution; this is because, even though all our PCR assays had high efficiency (>95%), a subtle difference in efficiency across assays may have a non-trivial effect on the estimate of mRNA levels due to the exponential relationship between number of mRNA molecules and number of cycles. In liver, all five genes show high expression levels (Cq value, mean±s.d., for UGT2B4, 29.3±2.0; for UGT2B10, 29.1±2.4; for UGT2B15, 29.5±1.8; for UGT2B17, 33.1±2.1), especially UGT2B7 (Cq value, mean±s.d., 25.3±2.0). These results are consistent with previous reports about the relative expression levels of UGT2B genes.18 UGT2B731 and UGT2B10 transcripts were not detected in breast samples. UGT2B17 transcripts could not be detected in some individuals, which is mostly due to the known common polymorphic deletions of this gene. All genes showed a high degree of inter-individual variation in transcript levels in both breast and liver, with ~133–1400-fold variation between the highest and the lowest non-zero value. This observation also confirmed recent findings in liver.36
Figure 1.
Boxplots of UGT2B gene expression in breast (a) and liver (b) samples. All genes are normalized to β-actin (a) or 18S (b) and log2 transformed.
A recent study36 has suggested a correlation between expression levels of different UGT2B genes in liver. We analyzed published genome-wide expression data in liver samples (not shown)9 and observed a similarly strong correlation between UGT2B genes. To test whether this is the case also in our breast and liver samples, we performed correlation analysis on transcript levels for all pairs of genes. As shown in Table 1a, a significantly positive correlation between UGT2B15 and UGT2B17 was observed (r=0.469, P<10−5) in breast. We also found a negative correlation between UGT2B4 and UGT2B15 (r=−0.308, P=0.005) in breast. In liver, all expressed genes were strongly and positively correlated with each another (see Table 1b). The observation in liver may reflect a shared regulatory mechanism for all UGT2B genes subject to variation in genetic background or environmental exposures.36, 37
Table 1.
Spearman’s correlation coefficients (P-values) among UGT2B gene expression in breast (a) and liver (b) samples
| UGT2B4 | UGT2B15 | UGT2B17 | ||
|---|---|---|---|---|
| (a) | ||||
| UGT2B15 | −0.308 (0.005) | |||
| UGT2B17 | −0.123 (0.27) | 0.469 (<10−5) | ||
| UGT2B4 | UGT2B7 | UGT2B10 | UGT2B15 | |
| (b) | ||||
| UGT2B7 | 0.770 (<10−5) | |||
| UGT2B10 | 0.799 (<10−5) | 0.795 (<10−5) | ||
| UGT2B15 | 0.853 (<10−5) | 0.642 (10−5) | 0.646 (10−5) | |
| UGT2B17 | 0.806 (<10−5) | 0.598 (0.0004) | 0.553 (0.001) | 0.736 (<10−5) |
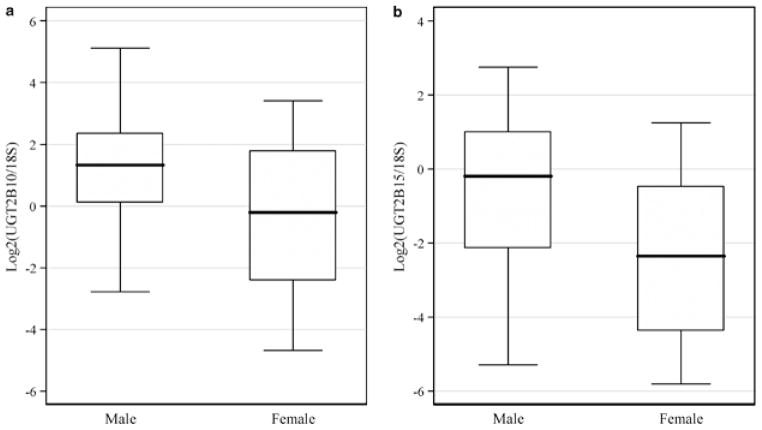
Recent studies have proposed that estrogens can upregulate UGT2B15 in breast cancer cell lines38 while androgens, especially dihydrotestosterone, can downregulate UGT2B15 and UGT2B17 in prostate cancer cell lines.39, 40, 41, 42 These findings raise the possibility that females express some UGT2B members, especially UGT2B15, at higher levels compared with males. In liver, we found that UGT2B10 (P=0.0496; Figure 2a) and UGT2B15 (P=0.039; Figure 2b) had ~3.3-fold higher expression on average in males than females whereas the other UGT2B genes did not show such variation (P>0.13 in other genes, data not shown). This observation is in contrast with the above prediction and it may be due to differences in the regulation of UGT2B genes between liver and breast or prostate, as recently suggested.28 It should also be noted that all previous analyses were performed in cancer cell lines38, 39, 40, 41, 42 rather than in primary cells, as in this study; therefore, this apparent discrepancy could be due, at least in part, to differences in regulation between transformed and primary cells.
Figure 2.
Gender difference in UGT2B10 (a, P=0.0496) and UGT2B15 (b, P=0.039) expression in liver. All genes are normalized to 18S and log2 transformed.
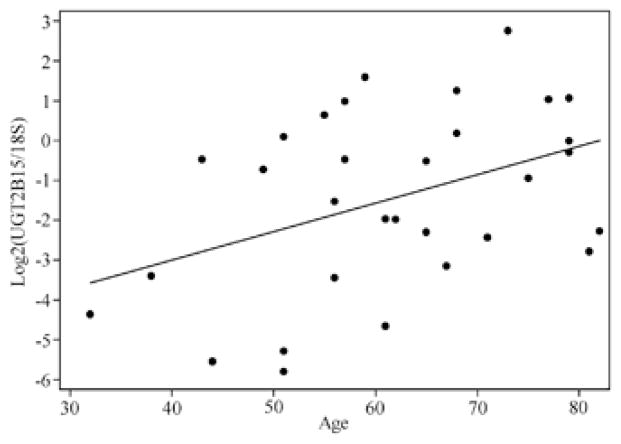
To investigate whether UGT2B expression levels are constant during human life, we examined the correlation between age of the breast and liver donors and UGT2B expression levels. UGT2B15 expression levels in liver are significantly correlated with age, with expression increasing in older individuals (r=0.416, P=0.020; Figure 3). After adjusting for gender in a multiple linear model, we still observed a significant correlation between age and UGT2B15 in liver (P=0.049). This finding might be explained by the known changes in steroid hormone levels during human life.43 No significant correlation was observed for the other genes in liver (P>0.23 in all cases, data not shown) and for all genes in breast (P>0.08 in all genes, data not shown).
Figure 3.
Correlation between age and UGT2B15 expression in liver (r=0.416, P=0.020). x axis indicates age while y axis UGT2B15 expression (normalized to 18S and log2 transformed). Each point denotes one individual.
Correlation between SNP and CNV genotype and expression levels
The common deletion of the UGT2B17 gene was previously found to be associated with its expression levels in prostate25 and LCL.44, 45 In LCLs, a significant association was also detected between the UGT2B17 CNV and the expression levels of the UGT2B7, UGT2B10 and UGT2B11 genes.45 We tested whether the UGT2B17 CNV is also associated with expression levels in breast and liver; in addition, we tested for an association between CNV and all UGT2B gene expression in both tissues. In breast, UGT2B17 CNV is correlated only with its own expression (r2=0.403, P<0.0001; Supplementary Figure S5) but not that of other UGT2B members (all P>0.22, data not shown). In liver, besides its own expression (r2=0.337, P=0.003; Supplementary Figure S6a), the UGT2B17 CNV was also significantly associated with UGT2B4 (r2=0.234, P=0.024; Supplementary Figure S6b).
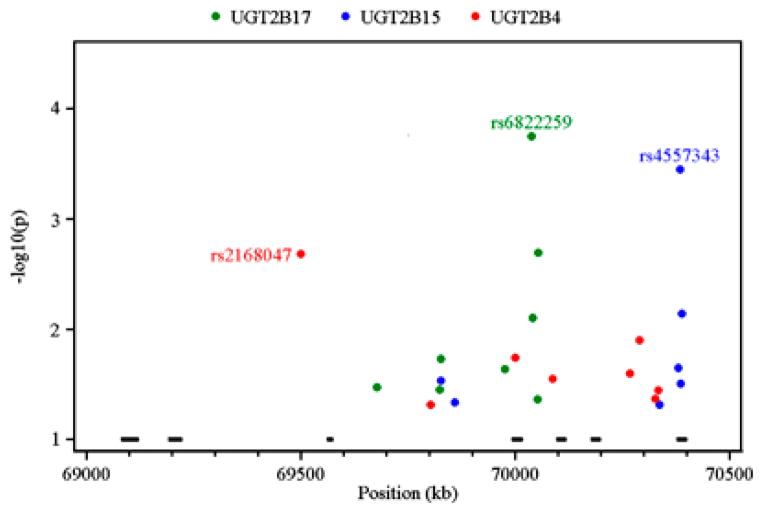
We further performed linear regression analysis between tagging SNP genotype and UGT2B expression levels in breast (Figure 4) and liver (Figure 5; see Supplementary Table S5 for detail). By this approach, 62 SNPs were identified to be significantly (P<0.05) correlated with the transcript levels of at least one UGT2B gene in at least one tissue. No SNP passed multiple test correction in liver, which may be due to the low power afforded by the relatively small sample size. Except for rs4860985 in UGT2B15 and rs17671289 in UGT2B4, none of the SNPs was significantly associated with the same gene in both tissues (see Supplementary Table S5), thus supporting the proposal that the regulation of this gene family is tissue specific.28 It was also interesting to note that multiple SNPs are associated with the expression of more than one gene. For example, rs2736483 is associated with UGT2B4 (P=0.000007429), UGT2B7 (P=0.000001492), UGT2B10 (P=0.0001018), UGT2B15 (P=0.0000715) and UGT2B17 (P=0.00352) expression in liver and rs17147073 is associated with UGT2B4 (P=0.04566), UGT2B15 (P=0.03499) and UGT2B17 (P=0.003779) in breast (see Supplementary Table S5).
Figure 4.
Association tests between tagging single-nucleotide polymorphism (SNP) genotype and UGT2B4 (red), UGT2B15 (blue) and UGT2B17 (green) expression in breast samples based on additive linear models. The x axis indicates the SNP position in chromosome 4 (build 36) while the y axis denotes P-value (–log10 transformed) for each SNP with P-value <0.05. The bars across the bottom indicate the location of the UGT2B genes in the order, from left to right: UGT2B17, UGT2B15, UGT2B10, UGT2B7, UGT2B11, UGT2B28 and UGT2B4.
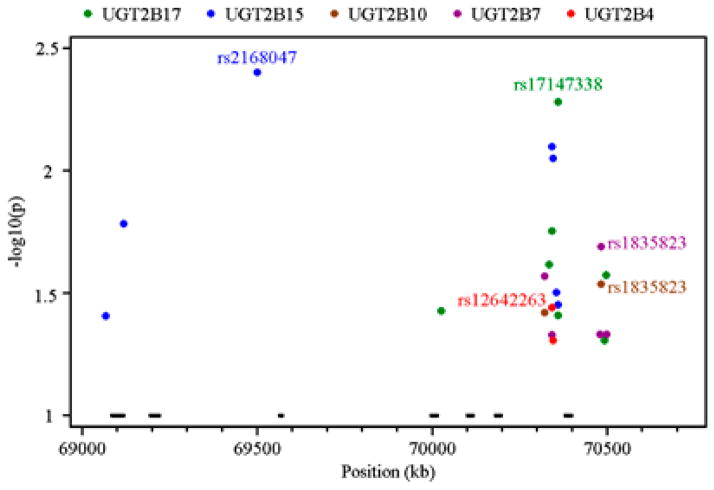
Figure 5.
Association tests between tagging single-nucleotide polymorphism (SNP) genotype and UGT2B4 (red), UGT2B7 (purple), UGT2B10 (brown) UGT2B15 (blue) and UGT2B17 (green) expression in liver samples based on additive linear models. The x axis indicates the SNP position in chromosome 4 (build 36) while the y axis denotes P-value (–log10 transformed) for each SNP with P-value <0.05. The bars across the bottom of each plot indicate the location of the UGT2B genes in the order, from left to right: UGT2B17, UGT2B15, UGT2B10, UGT2B7, UGT2B11, UGT2B28 and UGT2B4.
The most significant correlations in breast and liver are shown in Table 2. As all these SNPs are located far (>380 kb) from UGT2B genes, we hypothesized that they lie within enhancer regions and might alter the binding affinity of transcription factors regulating UGT2B gene expression. To investigate this possibility, we retrieved all the SNPs within the tag bins associated with the expression of at least one UGT2B gene and used the Match program in the TRANSFAC database (http://www.gene-regulation.com/cgi-bin/pub/programs/match/bin/match.cgi) to search for predicted transcription factor binding sites in the regions near the SNPs. By this approach, multiple canonical binding sites for transcription factors were identified (Supplementary Table S6). As shown in the table, some transcription factors, such as forkhead box D3 (FoxD3), forkhead box A2 (FoxA2 or HNF3B), HNF4A, forkhead box J2 (FoxJ2), paired box 4 (PAX4) and POU class 2 homeobox 1 (POU2F1 or OCT1), are common among those with predicted binding sites near SNPs associated with gene expression levels. This is consistent with the idea that these transcription factors are involved in the regulation of this gene family.16
Table 2.
The most significant association between SNP genotype and UGT2B transcript levels in breast and liver based on two degree of freedom linear models
| Tissue and gene | SNP positiona | rs ID | Genotype | No. | Mean log2-transformed transcript levels (s.d.) | P-value | R2 |
|---|---|---|---|---|---|---|---|
| Breast | |||||||
| UGT2B4 | 69500987 | rs2168047 | CC | 29 | −7.2 (1.81) | 0.0012 | 0.160 |
| CT | 33 | −10.16 (3.88) | |||||
| TT | 18 | −10.05 (3.81) | |||||
| UGT2B15 | 70384595 | rs4557343 | GG | 29 | −8.34 (2.16) | 0.0018 | 0.150 |
| GT | 41 | −6.82 (2.46) | |||||
| TT | 11 | −5.38 (2.86) | |||||
| UGT2B17 | 70038552 | rs6822259 | CC | 2 | −13.55 (7.11) | 0.0006 | 0.190b |
| TC | 14 | −12.48 (3.48) | |||||
| TT | 58 | −9.47 (2.41) | |||||
| CNV and rs6822259 | 0 copy | 7 | −18.44 (0.38) | 8.3E-13 | 0.510 | ||
| 1/2 copies+CC/TC | 16 | −12.61 (3.74) | |||||
| 1/2 copies+TT | 58 | −9.47 (2.41) | |||||
| Liver | |||||||
| UGT2B4 | 70342103 | rs12642263 | AA | 18 | −1.85 (1.96) | 0.013 | 0.269 |
| AG | 12 | 0.23 (1.46) | |||||
| GG | 1 | −2.31 | |||||
| UGT2B7 | 70482723 | rs1835823 | GG | 21 | 0.12 (2.01) | 0.047 | 0.196 |
| GC | 9 | 1.36 (2.11) | |||||
| CC | 1 | 4.90 | |||||
| 70320917 | rs10007427 | CC | 14 | −0.59 (2.07) | 0.019 | 0.263 | |
| CT | 14 | 1.62 (1.79) | |||||
| TT | 1 | −0.29 | |||||
| UGT2B10 | 70482723 | rs1835823 | GG | 21 | −0.04 (2.37) | 0.06 | 0.182 |
| GC | 9 | 1.19 (1.95) | |||||
| CC | 1 | 5.12 | |||||
| 70320917 | rs10007427 | CC | 14 | −0.66 (2.32) | 0.056 | 0.199 | |
| CT | 14 | 1.52 (2.23) | |||||
| TT | 1 | 0.36 | |||||
| UGT2B15 | 69500987 | rs2168047 | CC | 19 | −0.49 (1.87) | 0.0074 | 0.296 |
| CT | 10 | −2.98 (2.14) | |||||
| TT | 2 | −2.95 (2.00) | |||||
| 70342103 | rs12642263 | AA | 18 | −2.50 (2.18) | 0.0027 | 0.345 | |
| AG | 12 | 0.20 (1.35) | |||||
| GG | 1 | −2.30 | |||||
| UGT2B17 | 70360475 | rs17147338 | CC | 24 | −1.78 (3.20) | 0.039 | 0.214b |
| CT | 5 | −6.01 (3.13) | |||||
| TT | 1 | −3.39 | |||||
| CNV and rs17147338 | 0 copy or 1 copy+CT | 4 | −7.57 (1.35) | 2.5E-05 | 0.530 | ||
| 2 copies+CT/TT | 16 | −3.55 (2.46) | |||||
| 2 copies+CC | 11 | 0.25 (2.88) | |||||
Abbreviations: CNV, copy number variation; SNP, single-nucleotide polymorphism.
Only SNPs with call rate >90% were considered.
After excluding subjects with homozygous deletion of corresponding gene.
Discussion
To identify cis-regulatory variants for the human UGT2B genes, we re-sequenced candidate regulatory elements in ethnically diverse populations. These data, together with publicly available sequence variation information in the same individuals, were used to select tagging SNPs. Then, we quantified UGT2B transcript levels and genotyped the tagging SNPs in normal breast and liver samples to test for associations between genotypes and expression levels. As a result, we found 62 SNPs that are significantly correlated with the expression of at least one UGT2B gene, and 17 SNPs are significantly correlated with the expression of more than one gene. This is the first systematic investigation of sequence variation, breast and liver expression spectrum in unaffected individuals and cis-regulatory variation for the UGT2B family. Our results will undoubtedly provide a firm foundation for studies aimed at investigating the role of UGT2B variation in hormone-dependent diseases and inter-individual variation in drug response.
Our re-sequencing survey identified 162 common (MAF greater than or equal to 5%) SNPs and among them, only 22 (13.6%) were included in HapMap project; this is a substantially lower rate compared with the 25–35% coverage of common SNPs in the genome.29 The low SNP coverage in this genomic region in the HapMap is probably due to the difficulties of sequencing and genotyping regions containing highly similar duplicated genes like the UGT2B gene cluster. Therefore, our re-sequencing study significantly extends the information provided by the HapMap project for this particular region of the genome.
Recent genome-wide association studies uncovered many disease susceptibility SNPs located in non-coding regions distant from genes (for example, >100 kb). Some of the disease SNPs has been validated from the functional standpoint. For example, SNPs rs7903146, rs4939827 and rs6983267 are associated with type 2 diabetes,46 colorectal cancer47 and multiple cancers (as reviewed by reference48), respectively. Further functional in vitro and in vivo assays showed that these SNPs are located within enhancers and that they influence the activity of these enhancers thereby affecting target gene expression.48, 49, 50, 51 Many of the SNPs that we find to be associated with variation in UGT2B gene expression levels are far from the corresponding genes; as for SNPs found in genome-wide association studies, further work is necessary to elucidate the mechanism underlying their effect on mRNA levels.
The 1000 Genomes Project (http://1000genomes.org) currently underway seeks to discover essentially all genetic variation (>1% frequency) in each of the major continental populations using next-generation sequencing technologies.52 To understand whether such technologies are capable of capturing genetic variation in duplicated gene regions, we analyzed depth of coverage statistics around the UGT2B gene cluster generated during pilot 1 of the 1000 Genomes Project. Specifically, for each gene in the UGT2B gene cluster, we quantified the average depth of coverage per individual for the entire gene region, and the distribution of coverage across chromosome 4 using 1000 randomly chosen regions with matched length (Supplementary Figure S7). We found that UGT2B4, UGT2B7, UGT2B10 and UGT2B11 have coverage statistics that are consistent with coverage patterns from the rest of chromosome 4. However, UGT2B15, UGT2B17 and UGT2B28 each had much lower coverage than expected for the rest of chromosome 4 (in the 0.1, 0.5 and 2.5% tail, respectively). While the low coverage in UGT2B17 and UGT2B28 may be due to the common polymorphic deletions of these genes, no common CNVs are known for the UGT2B15 gene. Such low coverage statistics suggest that some genetic variants may be missed in the 1000 Genomes Project data for these UGT2B genes. More generally, these results suggest that some duplicated genes can indeed be re-sequenced successfully using next-generation technologies. However, other duplicated gene regions may receive little benefit from next-generation technologies, and will require more robust technologies (such as those used in this study) to discover genetic variation within them.
In addition to SNPs, CNVs may also influence gene expression, as shown in previous studies25, 44, 45 and by the findings reported here. Stranger et al.45 proposed that CNV of the UGT2B17 gene influences the expression of other UGT2B members, including UGT2B7, UGT2B10 and UGT2B11. However, we could not detect transcripts for any of these genes in LCL by qPCR (data not shown), suggesting that the previous observation was a false positive possibly due to the cross-hybridization of probes to different UGT2B genes in the array. Indeed, UGT2B17 (data not shown) and UGT2B7_v4,31 a splicing variant of UGT2B7, are highly expressed in LCLs and the sequence similarity between them is high. Probe cross-hybridization may also explain partly the failure to replicate the UGT2B cis-regulatory variants detected in a genome-wide association study in liver9 in both our study and a recent one focusing on UGT2B7.53 Furthermore, it highlights the value of using qPCR, or other high resolution approaches, to investigate the expression of duplicated genes with high sequence similarity.
There are still three shortcomings in our study. First, our liver sample size is relatively small, which resulted in low power to detect associations between SNP genotypes and expression levels. Association studies on a larger sample size or in vitro functional assays, for example, reporter gene assays, will be necessary to identify regulatory variants in liver. Second, to identify the potential regulatory element most efficiently, we used comparative genomics coupled with computational predictions of transcription factor binding sites. Although this approach has been widely used and proven to be useful,54, 55, 56, 57, 58, 59 recent studies have demonstrated that not all functional regions are conserved across species;60 therefore, we may have missed some important regulatory elements. Third, although we obtained genotype data for 189 tagging SNPs in our breast and liver tissues, these SNPs only accounted for 66.3% of the total tag bins; this implies that some regulatory variants may have been missed. Considering these caveats, it is unlikely that our survey has characterized the full repertoire of regulatory variants for this gene family. Nonetheless, our results represent the most extensive and systematic survey of sequence and expression variation in this important family of genes. As a consequence, we identified a large number of strong candidate regulatory variants for the UGT2B genes, especially in breast samples, thus significantly advancing our understanding of the genetic bases of inter-individual variation in UGT2B gene expression.
Breast cancer is the most common cancer in women and cumulative estrogen exposure over a lifetime is a likely risk factor.61, 62, 63 Indeed, despite the decrease of serum estrogen levels in postmenopausal compared with premenopausal women, the estrogen concentration in breast remains relatively constant during aging.64 Moreover, estrogen levels in breast tumor tissue are much higher than in normal breast tissue.65, 66 These observations suggest that variation in steroid hormone metabolism, especially in the target tissues like breast, may have a role in the susceptibility to breast cancer.64, 67 UGT2B15 and UGT2B17 are good candidates for breast cancer risk due to their high expression in breast (current study).18 In addition, UGT2B15 and UGT2B17 have significant activity against estrogen metabolites, especially 2- and 4-hydroxylated ones.68 Although UGT2B7 is not expressed in the breast,31 considering its high activity against estrogens,68 it might also contribute to breast cancer risk by altering the systemic estrogen levels. Previous studies aimed at detecting UGT2B variants associated with breast cancer risk mainly focused on the coding regions,20, 69 especially on amino-acid changes that alter enzyme activity or specificity, such as D85Y in UGT2B15 and H268Y in UGT2B7. Little attention has been paid to SNPs that alter UGT2B gene expression. Therefore, the SNPs identified in this study as potential cis-regulatory variants affecting the expression of the UGT2B15 and UGT2B17 genes in breast and of the UGT2B7 gene in liver represent strong new candidate variants for breast cancer risk. In this sense, the results of our study provide valuable information to investigate the role of the UGT2B genes in hormone-dependent diseases and in drug response.
Supplementary Material
Acknowledgments
We thank Ms Maria Tretiakova and Ying Sun for technical assistance. We also thank three anonymous reviewers for their helpful comments. This research was supported by the University of Chicago Breast SPORE NCI Grant CA125183 and by grant U01 GM61393.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Georges M, Coppieters W, Charlier C. Polymorphic miRNA-mediated gene regulation: contribution to phenotypic variation and disease. Curr Opin Genet Dev. 2007;17:166–176. doi: 10.1016/j.gde.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- Duan S, Huang RS, Zhang W, Bleibel WK, Roe CA, Clark TA, et al. Genetic architecture of transcript-level variation in humans. Am J Hum Genet. 2008;82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen VN, Edvardsen H, Tsalenko A, Nordgard SH, Sorlie T, Sharan R, et al. Genetic variation in putative regulatory loci controlling gene expression in breast cancer. Proc Natl Acad Sci USA. 2006;103:7735–7740. doi: 10.1073/pnas.0601893103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- Belanger A, Pelletier G, Labrie F, Barbier O, Chouinard S. Inactivation of androgens by UDP-glucuronosyltransferase enzymes in humans. Trends Endocrinol Metab. 2003;14:473–479. doi: 10.1016/j.tem.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 2003;3:136–158. doi: 10.1038/sj.tpj.6500171. [DOI] [PubMed] [Google Scholar]
- King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Curr Drug Metab. 2000;1:143–161. doi: 10.2174/1389200003339171. [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, et al. Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics. 2005;15:677–685. doi: 10.1097/01.fpc.0000173483.13689.56. [DOI] [PubMed] [Google Scholar]
- Gardner-Stephen DA, Mackenzie PI. Liver-enriched transcription factors and their role in regulating UDP glucuronosyltransferase gene expression. Curr Drug Metab. 2008;9:439–452. doi: 10.2174/138920008784746409. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T. Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines. Drug Metab Dispos. 2008;36:1461–1464. doi: 10.1124/dmd.108.021428. [DOI] [PubMed] [Google Scholar]
- Ohno S, Nakajin S. Determination of mRNA expression of human UDP-glucuronosyltransferases and application for localization in various human tissues by real-time reverse transcriptase-polymerase chain reaction. Drug Metab Dispos. 2009;37:32–40. doi: 10.1124/dmd.108.023598. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Sparks R, Ulrich CM, Bigler J, Tworoger SS, Yasui Y, Rajan KB, et al. UDP-glucuronosyltransferase and sulfotransferase polymorphisms, sex hormone concentrations, and tumor receptor status in breast cancer patients. Breast Cancer Res. 2004;6:R488–R498. doi: 10.1186/bcr818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdinjak T, Zagradisnik B. Prostate cancer and polymorphism D85Y in gene for dihydrotestosterone degrading enzyme UGT2B15: frequency of DD homozygotes increases with Gleason Score. Prostate. 2004;59:436–439. doi: 10.1002/pros.20024. [DOI] [PubMed] [Google Scholar]
- MacLeod SL, Nowell S, Plaxco J, Lang NP. An allele-specific polymerase chain reaction method for the determination of the D85Y polymorphism in the human UDP-glucuronosyltransferase 2B15 gene in a case-control study of prostate cancer. Ann Surg Oncol. 2000;7:777–782. doi: 10.1007/s10434-000-0777-3. [DOI] [PubMed] [Google Scholar]
- Park J, Chen L, Shade K, Lazarus P, Seigne J, Patterson S, et al. Asp85tyr polymorphism in the udp-glucuronosyltransferase (UGT) 2B15 gene and the risk of prostate cancer. J Urol. 2004;171:2484–2488. doi: 10.1097/01.ju.0000117748.44313.43. [DOI] [PubMed] [Google Scholar]
- Park J, Chen L, Ratnashinge L, Sellers TA, Tanner JP, Lee JH, et al. Deletion polymorphism of UDP-glucuronosyltransferase 2B17 and risk of prostate cancer in African American and Caucasian men. Cancer Epidemiol Biomarkers Prev. 2006;15:1473–1478. doi: 10.1158/1055-9965.EPI-06-0141. [DOI] [PubMed] [Google Scholar]
- Karypidis AH, Olsson M, Andersson SO, Rane A, Ekstrom L. Deletion polymorphism of the UGT2B17 gene is associated with increased risk for prostate cancer and correlated to gene expression in the prostate. Pharmacogenomics J. 2008;8:147–151. doi: 10.1038/sj.tpj.6500449. [DOI] [PubMed] [Google Scholar]
- Nagar S, Remmel RP. Uridine diphosphoglucuronosyltransferase pharmacogenetics and cancer. Oncogene. 2006;25:1659–1672. doi: 10.1038/sj.onc.1209375. [DOI] [PubMed] [Google Scholar]
- Desai AA, Innocenti F, Ratain MJ. UGT pharmacogenomics: implications for cancer risk and cancer therapeutics. Pharmacogenetics. 2003;13:517–523. doi: 10.1097/01.fpc.0000054116.14659.e5. [DOI] [PubMed] [Google Scholar]
- Sun C, Southard C, Witonsky DB, Olopade O, Di Rienzo A. Allelic imbalance (AI) identifies novel tissue specific cis-regulatory variation for human UGT2B15. Hum Mutat. 2010;31:99–107. doi: 10.1002/humu.21145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Li MC, Weng Z. Cluster-buster: finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 2003;31:3666–3668. doi: 10.1093/nar/gkg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Di Rienzo A. UGT2B7 is not expressed in normal breast. Breast Cancer Res Treat. 2009;117:225–226. doi: 10.1007/s10549-008-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izukawa T, Nakajima M, Fujiwara R, Yamanaka H, Fukami T, Takamiya M, et al. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab Dispos. 2009;37:1759–1768. doi: 10.1124/dmd.109.027227. [DOI] [PubMed] [Google Scholar]
- Ramirez J, Mirkov S, Zhang W, Chen P, Das S, Liu W, et al. Hepatocyte nuclear factor-1 alpha is associated with UGT1A1, UGT1A9 and UGT2B7 mRNA expression in human liver. Pharmacogenomics J. 2008;8:152–161. doi: 10.1038/sj.tpj.6500454. [DOI] [PubMed] [Google Scholar]
- Harrington WR, Sengupta S, Katzenellenbogen BS. Estrogen regulation of the glucuronidation enzyme UGT2B15 in estrogen receptor-positive breast cancer cells. Endocrinology. 2006;147:3843–3850. doi: 10.1210/en.2006-0358. [DOI] [PubMed] [Google Scholar]
- Bao BY, Chuang BF, Wang Q, Sartor O, Balk SP, Brown M, et al. Androgen receptor mediates the expression of UDP-glucuronosyltransferase 2 B15 and B17 genes. Prostate. 2008;68:839–848. doi: 10.1002/pros.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger A, Hum DW, Beaulieu M, Levesque E, Guillemette C, Tchernof A, et al. Characterization and regulation of UDP-glucuronosyltransferases in steroid target tissues. J Steroid Biochem Mol Biol. 1998;65:301–310. doi: 10.1016/s0960-0760(97)00183-0. [DOI] [PubMed] [Google Scholar]
- Guillemette C, Levesque E, Beaulieu M, Turgeon D, Hum DW, Belanger A. Differential regulation of two uridine diphospho-glucuronosyltransferases, UGT2B15 and UGT2B17, in human prostate LNCaP cells. Endocrinology. 1997;138:2998–3005. doi: 10.1210/endo.138.7.5226. [DOI] [PubMed] [Google Scholar]
- Hum DW, Belanger A, Levesque E, Barbier O, Beaulieu M, Albert C, et al. Characterization of UDP-glucuronosyltransferases active on steroid hormones. J Steroid Biochem Mol Biol. 1999;69:413–423. doi: 10.1016/s0960-0760(99)00061-8. [DOI] [PubMed] [Google Scholar]
- Straub RH, Miller LE, Scholmerich J, Zietz B. Cytokines and hormones as possible links between endocrinosenescence and immunosenescence. J Neuroimmunol. 2000;109:10–15. doi: 10.1016/s0165-5728(00)00296-4. [DOI] [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- Harismendy O, Frazer KA. Elucidating the role of 8q24 in colorectal cancer. Nat Genet. 2009;41:868–869. doi: 10.1038/ng0809-868. [DOI] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman AM, Naranjo S, Webb E, Broderick P, Lips EH, van Wezel T, et al. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res. 2009;19:987–993. doi: 10.1101/gr.092668.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti F, Liu W, Fackenthal D, Ramirez J, Chen P, Ye X, et al. Single nucleotide polymorphism discovery and functional assessment of variation in the UDP-glucuronosyltransferase 2B7 gene. Pharmacogenet Genomics. 2008;18:683–697. doi: 10.1097/FPC.0b013e3283037fe4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitland ML, Grimsley C, Kuttab-Boulos H, Witonsky D, Kasza KE, Yang L, et al. Comparative genomics analysis of human sequence variation in the UGT1A gene cluster. Pharmacogenomics J. 2006;6:52–62. doi: 10.1038/sj.tpj.6500351. [DOI] [PubMed] [Google Scholar]
- Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–1069. doi: 10.1086/426406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Tao H, Osoegawa K, de Jong PJ, Chen X, Doherty MF, et al. Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res. 2004;14:367–372. doi: 10.1101/gr.1961204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, et al. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key TJ. Serum oestradiol and breast cancer risk. Endocr Relat Cancer. 1999;6:175–180. doi: 10.1677/erc.0.0060175. [DOI] [PubMed] [Google Scholar]
- Thomas HV, Reeves GK, Key TJ. Endogenous estrogen and postmenopausal breast cancer: a quantitative review. Cancer Causes Control. 1997;8:922–928. doi: 10.1023/a:1018476631561. [DOI] [PubMed] [Google Scholar]
- Cauley JA, Lucas FL, Kuller LH, Stone K, Browner W, Cummings SR. Elevated serum estradiol and testosterone concentrations are associated with a high risk for breast cancer. Study of Osteoporotic Fractures Research Group. Ann Intern Med. 1999;130:270–277. doi: 10.7326/0003-4819-130-4_part_1-199902160-00004. [DOI] [PubMed] [Google Scholar]
- Guillemette C, Belanger A, Lepine J. Metabolic inactivation of estrogens in breast tissue by UDP-glucuronosyltransferase enzymes: an overview. Breast Cancer Res. 2004;6:246–254. doi: 10.1186/bcr936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijssen JH, Blankenstein MA. Endogenous oestrogens and androgens in normal and malignant endometrial and mammary tissues. Eur J Cancer Clin Oncol. 1989;25:1953–1959. doi: 10.1016/0277-5379(89)90377-5. [DOI] [PubMed] [Google Scholar]
- Mady EA, Ramadan EE, Ossman AA. Sex steroid hormones in serum and tissue of benign and malignant breast tumor patients. Dis Markers. 2000;16:151–157. doi: 10.1155/2000/305940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon D, Carrier JS, Levesque E, Hum DW, Belanger A. Relative enzymatic activity, protein stability, and tissue distribution of human steroid-metabolizing UGT2B subfamily members. Endocrinology. 2001;142:778–787. doi: 10.1210/endo.142.2.7958. [DOI] [PubMed] [Google Scholar]
- Yong M, Schwartz SM, Atkinson C, Makar KW, Thomas SS, Newton KM, et al. Associations between polymorphisms in glucuronidation and sulfation enzymes and mammographic breast density in premenopausal women in the United States. Cancer Epidemiol Biomarkers Prev. 2010;19:537–546. doi: 10.1158/1055-9965.EPI-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.