Microchip sensors enable rapid, molecular-level profiling of donated lungs for transplant assessment.
Keywords: Electrochemical sensors, Molecular analysis, mRNA profiling, Organ transplantation
Abstract
Biomarker profiling is being rapidly incorporated in many areas of modern medical practice to improve the precision of clinical decision-making. This potential improvement, however, has not been transferred to the practice of organ assessment and transplantation because previously developed gene-profiling techniques require an extended period of time to perform, making them unsuitable in the time-sensitive organ assessment process. We sought to develop a novel class of chip-based sensors that would enable rapid analysis of tissue levels of preimplantation mRNA markers that correlate with the development of primary graft dysfunction (PGD) in recipients after transplant. Using fractal circuit sensors (FraCS), three-dimensional metal structures with large surface areas, we were able to rapidly (<20 min) and reproducibly quantify small differences in the expression of interleukin-6 (IL-6), IL-10, and ATP11B mRNA in donor lung biopsies. A proof-of-concept study using 52 human donor lungs was performed to develop a model that was used to predict, with excellent sensitivity (74%) and specificity (91%), the incidence of PGD for a donor lung. Thus, the FraCS-based approach delivers a key predictive value test that could be applied to enhance transplant patient outcomes. This work provides an important step toward bringing rapid diagnostic mRNA profiling to clinical application in lung transplantation.
INTRODUCTION
Lung transplantation (LTx) is a well-established lifesaving therapeutic option for patients with end-stage lung disease. The procedure has developed rapidly since its inception in the 1980s (1, 2), and clinical outcomes are continually improving (3). Unfortunately, using current clinical selection and preservation practices, life-threatening primary graft dysfunction (PGD) can occur and accounts for about one-third of all of the 30-day mortality cases after LTx (4–8). PGD occurs within the first 72 hours after transplant and is characterized by nonspecific diffuse alveolar damage, lung edema, and severe hypoxia, and is associated with an eight times higher mortality rate compared to non-PGD LTx cases (5, 6, 8–12). Upon diagnosis, PGD is graded from low (0/I) to high (III), with PGDIII being associated with significantly poorer patient outcomes (13).
Currently, lungs donated for transplant are assessed by analysis of lung function, bronchoscopic findings, and chest radiographs. These reveal abnormalities such as hypoxia, atelectasis, hemorrhagic contusions, edema, and pulmonary infiltration (13). Unfortunately, these metrics do not accurately, specifically, or reliably reflect the injury status of the lung and therefore cannot predict the risk of PGD after transplant. The demands on the rapidity of an improved classification assay are extremely high. For every additional hour that a lung is stored, graft survival is further compromised. Consequently, the survival of the recipient depends on rapid decision-making and transfer of healthy lungs from the donor to the recipient (14). As a result, most surgeons work to stay within a time window for donor lung assessment of less than 6 hours (14). Ideally, a useful classification assay must provide robust and actionable information in much less than this 6-hour time frame.
With the goal of increasing the availability of precise information relevant to clinical decision-making, an intensive search is under way for molecular biomarkers predictive of LTx outcome. A number of studies have identified mRNAs in lung samples that have been associated with episodes of acute rejection (15–20). Several studies have identified specific mRNAs in donor lung biopsies that preoperatively predict the development of severe PGD or ischemia reperfusion injury, including interleukin-6 (IL-6), IL-10, and ATP11B [ATPase (adenosine triphosphatase), class VI, type 11B] (9, 21). Because IL-6 is a proinflammatory cytokine and IL-10 is an anti-inflammatory cytokine, the IL-6/IL-10 ratio specifically reports an elevated inflammatory status of a donor lung. Donor lung tissues with a high IL-6–to–IL-10 ratio before LTx were shown to develop severe PGD and had a 20-fold relative risk of recipient death (21).
Microarray and polymerase chain reaction (PCR) technologies are powerful tools for the discovery (22) and validation (9, 17–21) of nucleic acid biomarkers for lung transplant outcomes. Unfortunately, the numerous sample purification steps and lengthy turnaround times for microarray and PCR assays [6 to 12 hours in a typical hospital workflow (23)] are unsuitable for translation to the clinic. In addition, these existing technologies require highly trained personnel and contamination-free laboratory facilities. Delays encountered with methods requiring laboratory facilities become even more significant when one accounts for the fact that many lung transplants occur outside normal laboratory operating hours.
Although the development of integrated PCR-like testing platforms has shown promise for infectious disease diagnosis in lung samples [for example, detection of tuberculosis with good sensitivity and specificity in pleural fluid and tissue biopsies (24, 25)], these current platforms are designed to amplify and detect analytes with binary outcomes (presence or absence of infection). Hence, these devices are able to actualize clinical utility at the expense of quantification. Although these platforms can make rapid diagnoses in clinical samples, it would not be possible to distinguish small differences in gene expression with a high degree of accuracy—a crucial requirement of a transplant-specific mRNA profiling test.
There has been significant progress in the area of new molecular analysis strategies. Many different electrochemical platforms are currently being studied to detect nucleic acids, including biobarcodes (26), noncovalent (27) and structural (28) reporters, DNA nanostructures (29), biodiscs (30), and sandwich assays (31, 32). Our laboratories have developed a class of electrochemical biosensors designed to rapidly capture and analyze mRNA transcripts with clinically relevant sensitivity and specificity using non-amplified samples that have been subjected to minimal cleanup (33–39).
To bring molecular diagnostics to the bedside in LTx, we posed the critical challenge of developing an electrochemical assay that would deliver a reliable and reproducible result to the clinician in less than 30 min from sample receipt. In this limited time window, tissue must be taken from its crude state and processed into a form that can be analyzed for gene expression. Herein, we report a new class of nanoscale chip-based sensors, termed fractal circuit sensors (FraCS), which enable quantitative analysis of multiple biomarkers present at relevant concentrations in human lung tissue. The lung tissue assessment chip enabled by FraCS rapidly assesses predictive mRNA biomarkers in donor lungs from a lung tissue biopsy performed by the surgeons, relying on a simplified sample preparation step to release biomarkers into solution, thus essentially requiring no sample purification (Fig. 1A). To our knowledge, there exist no previous reports of sample-to-answer processing and analysis of lung tissue biomarkers for LTx suitability using techniques requiring fewer than 12 hours.
Fig. 1. Lung transplant assessment assay.
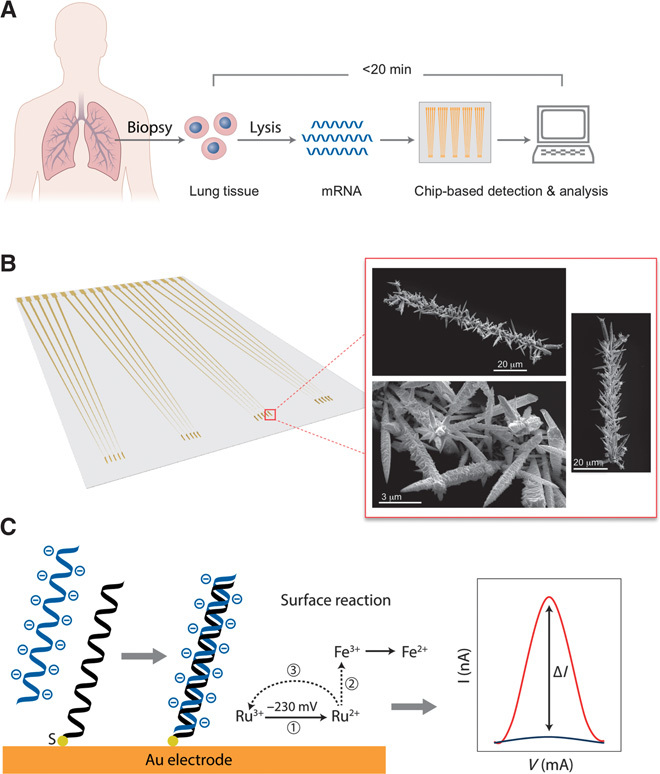
(A) The lung assessment assay workflow. A biopsy is taken from a donated lung, and the cells are homogenized and lysed. The mRNA released from the cells is analyzed using a chip-based method that delivers a gene expression profile predictive of the outcome of a transplant within 20 min of the biopsy. (B) FraCS sensor chip (left) and SEM images of sensors after electrochemical deposition (right). (C) Assay readout [PNA probe (black) and resulting differential pulse voltammetry (DPV) signal (right panel, shown in dark blue), target mRNA (light blue) hybridization, and resulting DPV signal (right panel, shown in red)].
RESULTS
Rapid nucleic acid quantification using FraCS
Here, we endeavored to create a new type of sensor with maximal surface area to reduce signal saturation. Sensors made from electrodeposited gold were templated in apertures created in an SU-8 layer patterned over gold contacts on a glass chip. Electrodeposition was performed using conditions facilitating rapid growth and the generation of spiky, fractal structures extending into solution. The linear apertures were 5 × 100 μm, a size that was selected to yield sensors with large amounts of surface area (Fig. 1B).
The electrochemical currents collected at these sensors and used for nucleic acid quantitation are generated by a label-free, redox-active, [Ru(NH3)6]3+/[Fe(CN)6]3− reporter system (Fig. 1C). The [Ru(NH3)6]3+ electron acceptor complex is positively charged and binds to the sensors at levels that correspond to the amount of negatively charged nucleic acid that binds to the sensor. The [Fe(CN)6]3− electron acceptor complex is negatively charged, and so it does not bind to the sensor, but remains in solution to accept the electrons from [Ru(NH3)6]3+ to allow multiple turnovers for this ion (Fig. 1C).
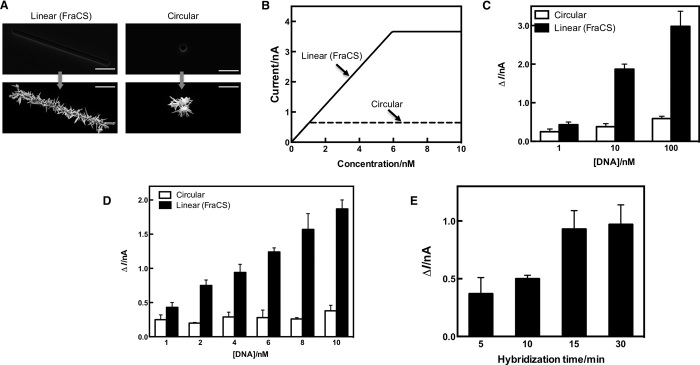
In an effort to maximize the amount of [Ru(NH3)6]3+ that could access the surface of a sensor, we moved to FraCS from the sensors described in our previous work using smaller round templates (Fig. 2A). This would theoretically allow larger electrochemical currents to be generated, and the signals would then exhibit maximal concentration dependence. A mathematical model that compared the total amount of [Ru(NH3)6]3+ (current) on sensors made with round templates to FraCS was constructed by taking into account the electrode size, hybridization efficiency, and diffusional properties of [Ru(NH3)6]3+ (the model is described in “Mathematical modeling” in Materials and Methods) (Fig. 2B). By modeling the amount of ruthenium associated with analyte on the surface of FraCS, we observed that FraCS had a twofold advantage over traditional electrodes. First, the dynamic range for currents produced by FraCS could be greatly enhanced (Fig. 2B), and second, FraCS could reach a saturation current that was significantly higher than traditional electrodes (3.67 versus 0.67 nA; Fig. 2B). We then experimentally tested whether FraCS were superior for nucleic acid quantification compared to our previously developed electrochemical sensors. Both sets of electrodes achieved signal-to-noise ratios greater than 2.0 (Fig. 2, C and D); however, as quantitative sensors, FraCS had an improved ability to achieve larger signal gains per change in analyte concentration (593% versus 136%, as shown in Fig. 2C). This was consistent with our modeling predictions in Fig. 2B, and, as expected, the FraCS signals were of greater intensity, which resulted in the ability to easily and reliably differentiate large concentration profiles over multiple orders of magnitude (Fig. 2C). This observation was even more apparent when looking at a narrow range of analyte concentrations (1 to 10 nM) (Fig. 2D). For sensors challenged with solutions of 1 to 10 nM, there was a 335% increase in signal using FraCS versus 52% using traditional electrodes (Fig. 2D). As a result, FraCS showed an enhanced ability to discriminate between very small differences in analyte concentrations (even within a single order of magnitude)—a result not possible with the traditional electrodes (Fig. 2D). As further proof-of-concept validation, we estimated other quantitative metrics of FraCS [that is, limit of detection (LOD)] and experimentally confirmed that an LOD could be improved by using a FraCS-based approach (Fig. 2, C and D).
Fig. 2. Rapid quantitation of LTx analytes using FraCS.
(A) Images collected with scanning electron microscopy for FraCS templated with linear apertures compared to those made with smaller round apertures. The scale bar shown on each image corresponds to 20 μm. (B) Mathematical modeling of FraCS (solid line) versus sensors made with circular templates (dashed line) for the current generated by the sensor as a function of DNA concentration. (C and D) Quantitative comparisons of sensors with circular apertures (white bars) and FraCS (black bars) between 1- to 100-nM target (C) and 1- to 10-nM target (D). (E) Hybridization time course for rapid RNA analysis using FraCS. Data represent n = 15 different sensors. Columns represent mean, and error bars correspond to SEM.
In addition to improved quantification capabilities, we discovered another key advantage of using FraCS for rapid analysis of mRNA. The increase in electrode size in the y direction allows for very fast accumulation of the biomarkers of interest on the surface of the detectors (Fig. 2E). On the basis of previous diffusion and hybridization studies by our group, we predicted that biomarker profiles could be collected using a FraCS approach in less than 30 min (40). Remarkably, in time-dependent rapid hybridization experiments, FraCS performed even better than anticipated and could detect mRNA transcripts in as little as 15 min (Fig. 2E).
Using these sensors, we then proceeded to develop probes against the mRNAs of three previously reported donor lung assessment markers (IL-6, IL-10, and ATP11B) (9, 21) and control sequences [human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a positive control and Drosophila melanogaster GAPDH as a negative control]. The probes for the LTx assay were successfully designed and validated using 100 nM complementary synthetic DNA oligonucleotides (fig. S1). To test whether FraCS could accurately measure the levels of the target lung assessment mRNAs in heterogeneous samples, we isolated total RNA from donor lung tissue. For all of the respective LTx assay probes, we observed concentration-dependent signals that were highly reproducible (Fig. 3). An analysis of the ratio of two important inflammatory cytokine markers, IL-6 and IL-10, illustrates the excellent precision of the detection strategy, because the ratio of IL-6 to IL-10 remained constant at various total RNA concentrations (Fig. 3C).
Fig. 3. Quantitation of RNA markers predictive of lung transplant outcome.
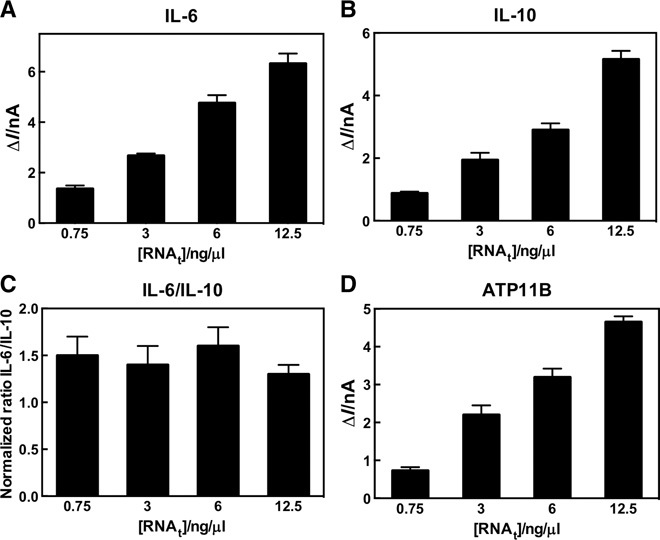
(A to D) Total RNA titration profiles for (A) IL-6, (B) IL-10, (C) IL-6/IL-10, and (D) ATP11B sensors. Data represent n = 15 different sensors. Columns represent mean, and error bars correspond to SEM.
We then validated the suitability of our GAPDH-positive control probe. The signals obtained at FraCS modified with a probe sequence complementary to human GAPDH demonstrated concentration dependence (fig. S2A) and were strongly correlated with total RNA concentrations purified from different biopsies (fig. S2B). In addition, the mRNA expression levels of GAPDH in tissue were consistent across several donor lungs irrespective of the position of sampling (fig. S2C). Hence, the GAPDH probe served as a reliable reference control for analyzing donor lung biomarker expression levels.
Donor lung assessment using multiplexed FraCS
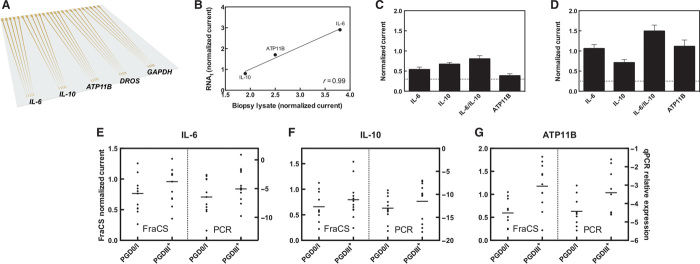
Multiplexed chips that allowed parallel analysis of the lung assessment markers were created. Each chip contained up to five sets of FraCS that could be individually functionalized with probes against the marker set of interest (Fig. 4A). Critical to minimizing turnaround time and complexity of the LTx assay is the ability to perform quantitative analysis on human lung tissue biopsies without an mRNA purification step. To achieve this, we developed a rapid (~5 min) chemical lysis method based on non-ionic detergents that were effective in lysing the lung tissue while remaining compatible with FraCS and the electrochemical reporter system. To ensure consistency for signals obtained using unpurified lysate and total RNA from the same donor lung sample, we compared the FraCS response of both types of samples (Fig. 4B). There was a strong, positive correlation of the biomarker responses for both the pure RNA and the unpurified lysate—which indicates that the assay can accommodate a notable increase in the complexity of the sample matrix.
Fig. 4. Lung assessment chip and analysis of lung tissue.
(A) A multiplexed chip that could accommodate the parallel analysis of the five markers tested as proof of principle was prepared. (B) Correlation of signals obtained from purified RNA from a lung biopsy versus unpurified lysate of the same biopsy (r indicates Pearson’s correlation coefficient). (C) Representative data obtained from a good-outcome lung. (D) Representative data obtained from a poor-outcome lung. The signals are normalized to GAPDH controls, and the nonspecific D. melanogaster signal is shown as a dashed line. Data represent n = 15 different sensors. Columns represent mean, and error bars correspond to SEM. (E to G) Comparison of the FraCS assay response (left y axis) to qPCR expression levels (right y axis) of the same biopsy [PGD0/I (n = 9 to 11), PGDIII+ (n = 9 to 12)] run on both platforms for (E) IL-6, (F) IL-10, and (G) ATP11B.
To obtain proof of principle for the use of the FraCS approach to profile assessment markers in lung biopsy samples, we assayed target gene expression levels in a biopsy from a mildly injured (Fig. 4C) and a severely injured lung (Fig. 4D). In the case of the severely injured lung (Fig. 4D), the LTx assay accurately displayed increased expression levels of the markers indicative of a poor outcome: ATP11B, IL-6, and IL-6/IL-10.
Comparison of FraCS to quantitative PCR
We then sought to compare the FraCS system with the current gold standard for quantitative gene expression profiling—quantitative PCR (qPCR). Indeed, we confirmed a significant correlation of FraCS and qPCR signals obtained from the donor lungs (n = 7, P < 0.05; fig. S3). Bland-Altman method analysis of the results obtained using FraCS and qPCR further confirmed the corroboration of both tests, because the mean difference between the two tests was zero (fig. S4 and table S1). Twenty-three donor lungs were tested on both the FraCS and qPCR platforms and compared for the expression patterns in PGD0/I and PGDIII+ donor lungs. For each of the biomarkers tested (IL-6, IL-10, and ATP11B), we observed the same expression pattern for both FraCS (left panel) and qPCR (right panel) in these donor lungs (Fig. 4, E to G). Together, these data indicate that our FraCS chip-based approach, which provides rapid assay times and does not require stringent sample purification, yields comparable data to qPCR.
Evaluation of LTx biomarkers using FraCS
We examined the expression profiles of IL-6, IL-10, IL-6/IL-10 ratio, and ATP11B in 39 distinct donor lungs that were not previously used in biomarker discovery with two objectives: (i) to confirm the predictive nature of these biomarkers and (ii) to determine whether the chip-based LTx assay could confirm the differential expression profiles of donor lungs in a set of patient samples. The population was separated on the basis of the outcome (PGD0/I and PGDIII+) (Fig. 5). We observed a significant difference between the two PGD populations for IL-6 (Fig. 5A), IL-6/IL-10 ratio (Fig. 5C), and ATP11B (Fig. 5D), but not IL-10 (Fig. 5B). To validate the ability of the LTx biomarkers to predict PGD in transplanted lungs, we performed receiver operating characteristic (ROC) curve analysis and determined a significant area under the curve (AUC) for IL-6 and ATP11B mRNA, and the IL-6/IL-10 ratio, but not IL-10 mRNA as a predictor alone (Table 1).
Fig. 5. Relative expression of LTx biomarkers.
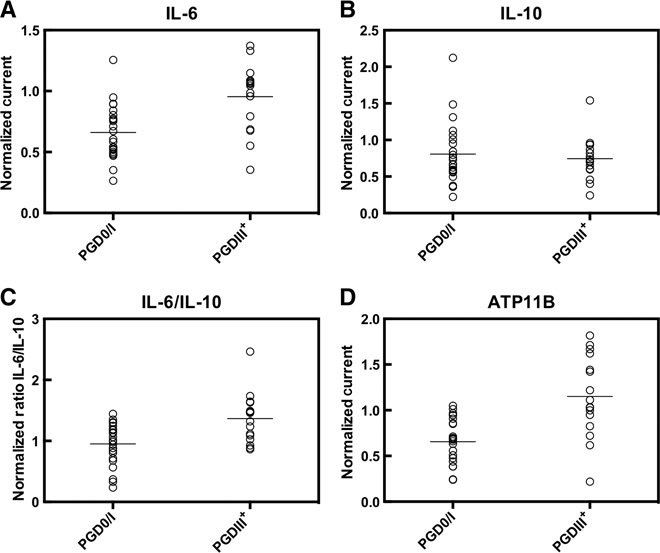
(A to D) Each circle represents the LTx biomarker signal normalized to GAPDH for an individual donor lung, and horizontal lines show the population means of PGD0/I (n = 23) and PGDIII+ (n = 16) donor lungs for (A) IL-6, (B) IL-10, (C) IL-6/IL-10, and (D) ATP11B. Data were analyzed by a two-tailed Mann-Whitney test. The P values of each comparison are as follows: IL-6, 0.0007; IL-10, 0.9833; IL-6/IL-10, 0.0027; and ATP11B, 0.0002.
Table 1. PGD predictive value of LTx biomarkers.
| Biomarker | Area under ROC curve | P |
| IL-6 | 0.74 | 0.0381 |
| IL-10 | 0.53 | 0.8177 |
| IL-6/IL-10 | 0.78 | 0.0160 |
| ATP11B | 0.84 | 0.0003 |
Development of the FraCS prediction model (FPM)
Knowing that the LTx-FraCS sensor chip could provide important information on donor lung assessment biomarkers in a transplant-relevant time frame and that the chosen biomarkers were predictive of PGD, we then sought to develop a prototype predictive model that incorporated all of the relevant biomarkers and could help to guide transplant teams in the donor lung selection process. Using the profiles from 32 donor lungs (9 PGDIII) in a development group, we performed logistic regression analysis on all marker combinations and determined that a model, referred to as the FraCS prediction model (FPM), consisting of the FraCS output for the IL-6/IL-10 ratio, IL-6, and ATP11B was the most predictive with an AUC of 0.96 (P < 0.0001) (Table 2). Using 100 rounds of ninefold cross-validation with stratification, we estimated the model to perform with an AUC of 0.88 on future samples.
Table 2. The FraCS prediction model.
The FPM was developed by logistic regression analysis and is expressed by the following equation: loge(PGDIII:PGD0/I) = −5.5669 + 0.0484*IL-6 + 2.4370*ATP11B + 2.0469*IL-6/IL-10.
| Group |
Area under ROC curve |
P |
| Development group (n = 32, PGD0/I: n = 23, PGDIII: n = 9) |
0.96 | <0.0001 |
| Cross-validation of development group | 0.88 | <0.05 |
| Validation group (n = 20, PGD0/I: n = 10, PGDIII: n = 10) |
0.87 | 0.0052 |
| All cases (n = 52, PGD0/I: n = 33, PGDIII: n = 19) |
0.88 | <0.0001 |
| Cross-validation of all cases | 0.82 | <0.05 |
We then validated this model on 20 new, previously untested, samples (10 PGDIII) and obtained an AUC of 0.87 (P < 0.0052) (Table 2). The FPM was refit to all 52 samples to generate a final set of coefficients. This final FPM had an AUC of 0.88 (P < 0.0001), representing testing on all 52 samples. Cross-validation of the final FPM was carried out using 100 rounds of stratified 10-fold cross-validation, which confirmed the significant predictive value of the FPM (AUC, 0.82) (Table 2). Furthermore, using a cutoff of −0.3575 for the FPM, we observed a diagnostic sensitivity and specificity of predicting PGD of 74 and 91%, respectively (Table 3). Thus, the FPM resulted in positive and negative predictive values (PPV and NPV) of 82 and 86%, respectively, for the probability of identifying PGDIII (poor outcome after transplant) in a donor lung (Table 3).
Table 3. Diagnostic characteristics of the FPM.
| Sensitivity | Specificity | PPV | NPV | |
| FPM | 73.7% (14/19) | 90.9% (30/33) | 82.4% | 85.7% |
DISCUSSION
The current decision-making process for donor lung selection in LTx is based on clinical evaluation of lung function that is largely qualitative. Accurate, informative, and predictive gene-level analysis of donor lungs would prove to be extremely beneficial, yet current techniques prove limiting because of the poor suitability of the required sample-to-answer time frames in the transplant setting.
We have successfully developed an LTx assay based on FraCS and an electrochemical reporter system that is capable of reporting donor lung biomarker profiles in about 20 min. Furthermore, we have assayed various gene-expression level measures of donor lung risk and have used this assessment to develop the foundation of an FPM that represents the proof of concept for a rapid diagnostic tool that successfully predicts severe graft dysfunction in donor lungs.
Two key parameters of the FraCS-based LTx assay were tested and validated: (i) a large sensor surface area that is capable of rapidly capturing slow diffusing mRNA targets and (ii) quantification of small differences in the gene expression patterns.
Sensors that are to be used for gene profiling in transplantation interrogate targets that are predominantly from inflammatory pathways and expressed on a continuum in human tissues. Thus, discerning where on the normal distribution a donor organ falls is crucial. Previous work by our group has been focused on developing metal sensors for the early detection of disease and, as such, these sensors were optimized to detect low levels of invading organisms (or altered host state), including bacteria (35, 39, 41), viruses (42), and cancers (38). They do not clearly discriminate small changes in concentration with precision because the ~100-μm2 area of existing sensors is saturated in the presence of low levels of analyte molecules. Indeed, these sensors will enter a sublinear regime in the presence of <100 complementary nucleic acid biomarkers in a sample.
Here, we have demonstrated that these novel sensors, FraCS, are superior in their ability to quantify large targets. This is due to the large surface area of functionalized FraCS that provide more binding sites for target mRNA capture and efficient electrocatalysis relative to conventional chip-based sensors while maintaining a morphology that is optimal for hybridization and electrochemical analysis (43). The advantages of FraCS were confirmed experimentally, because we observed the enhanced ability to quantitate small differences in analyte concentrations.
In rapid mRNA hybridization experiments, the FraCS-based LTx assay was able to capture and convey this genetic information in as fast as 15 min—well within our desired transplant time frame. In addition, we were able to confirm that the FraCS assay reported a consistent result regardless of whether purified RNA or unpurified lysate from a donor lung was used and that these results were consistent with the profiles obtained using qPCR as a benchmark standard. Because PCR is one of the most commonly used techniques to profile gene expression, it was imperative that our LTx assay perform equally as well as this gold standard test. Notably, the FraCS assay was able to provide a sample-to-answer from a tissue biopsy in about 20 min, whereas the qPCR procedure was completed in about 5 hours. Because FraCS do not rely on enzymatic activity, and the capture probes are composed of synthetic DNA analogs, FraCS are able to work around the lengthy sample purification steps required of traditional genetic techniques while still providing accurate and equivalent measurements to these methods. Together, these results highlight the potential clinical utility of this assay in a transplant setting.
Using the FraCS platform for gene expression analysis, we were able to confirm our biomarker discovery findings that were first reported several years ago using microarrays and qPCR. By separating the donor lungs based on the outcome for each biomarker, we observed a significant up-regulation of IL-6, IL-6/IL-10, and ATP11B mRNA in the donor lungs with poor outcomes, with the exception of IL-10. This finding was consistent with the previous work that also determined that IL-10 alone was not a predictor of poor outcome; however, as part of the IL-6/IL-10 ratio, the metric was significantly predictive of PGD (21).
Because each marker exhibited a statistically significant area under the ROC curve, we anticipated that these biomarkers could be used to develop the foundation for a PGD prediction model. It is likely that each individual biomarker itself would have limited predictive ability, because a donor lung may be consistent with a PGD0/I outcome for some markers, but not others. By combining several biomarkers, the LTx assay accounts for the individual response of each biomarker and reports a composite profile reflecting the condition of the donor lung overall, as a function of multiple biomarkers. A logistic regression model, the FPM, confirmed previous results that demonstrated that the signals from IL-6, ATP11B, and the ratio of IL-6/IL-10 were highly predictive of PGD. Both cross-validation and an independent validation group of donor lungs confirmed the predictive value of the assay and the FPM. Furthermore, this model displayed a high degree of sensitivity and specificity for detecting PGDIII in donor lungs. Ultimately, this model represents a means by which the raw gene expression output from the LTx assay biomarkers can be converted into a simple numerical score with a cutoff (for example, −0.3575) that could successfully predict donor lung outcomes—donor lungs above the cutoff are likely to develop PGDIII, and those below the cutoff are likely PGD0/I donor lungs. Having a composite numerical score with the FPM will help to facilitate ease of use for surgeons by limiting the analysis and interpretation of the data that the transplant teams would have to perform during the time-sensitive transplantation process. The NPV of the FPM (NPV 86%) represents a significant advance over current clinical practice, because all of the lungs used in the FPM were viewed as clinically suitable and transplanted; unfortunately, 19 of 52 patients developed PGDIII (NPV 63%). Thus, the use of the FPM could provide an improvement of greater than 20% in NPV. The FPM coefficients and desired cutoff will ultimately change because a larger number of donor lungs will need to be tested to improve the accuracy of the FPM; however, with the LTx assay and FraCS, we have demonstrated the possibility for this type of analysis and profiling to be performed in real time, directly in the operating rooms.
Future studies that involve large, multicenter prospective studies will be needed to further validate the FraCS approach. These trials will focus on integrating this technology within the transplant procedure, validating the prediction models, and prospectively observing patient outcomes. Additionally, standardization of the sample collection process (including biopsy size, location, etc.) will help to improve the precision of the LTx assay. With the discovery and validation of additional, informative genetic biomarkers, the scope of LTx assay biomarkers can be expanded beyond the markers studied herein, which will further increase the utility and accuracy of the FPM.
The LTx assay run using the FraCS chip could potentially aid surgeons in the decision-making process when selecting donor lungs and represents a major advance toward a personalized transplant medicine approach. The information provided by FraCS could be acted upon to treat the donor lung in a directed manner to repair a specific diagnosed injury, and studies have shown that various treatment regimes can be effective in improving donor lung function (44). Furthermore, the LTx assay could be used to guide therapy and monitor response to treatment of donor lungs that may have been previously discarded, thereby expanding the number of available organs. Studies have shown that information available at the genetic level could be used to select suitable donor lungs that would otherwise be rejected using current clinical practice standards (45). It is estimated that more than 40% of the discarded lungs may in fact be suitable for clinical transplantation (46); thus, the rapid sample-to-answer turnaround provided by the FraCS assay could have broad implications for the organ selection process. With this technology, organs can now be diagnosed, treated, repaired, and confirmed to be improved in a transplant-relevant timeline. By identifying donor lung outcomes in addition to monitoring the repair of damaged donor lungs, the LTx assay will help improve the utilization of donor lungs and the outcomes of LTx.
MATERIALS and METHODS
Experimental design
Lung tissue collection and processing
A biopsy sample (about 2 cm × 1 cm) was obtained from a peripheral part of the donor lung with a mechanical stapler at the end of the cold ischemic period (just before the implantation of the lung). The samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until use. All the patients provided informed consent for the study, and the study was approved by the institutional Human Tissue Committee and Research Ethics Board at the University Health Network, University of Toronto. A portion (~5 mm3) of each frozen tissue section was ground in liquid nitrogen and then placed in lysis buffer (2% Triton X-100, 2% NP-40; Sigma-Aldrich) with RNase (ribonuclease) inhibitor (SUPERase-In, Sigma-Aldrich) for 5 to 10 min. The lysate was subsequently spun at maximum speed for 3 min, and the supernatant was placed on the electrodes (in 1× SSC). For RNA analysis, the total RNA was purified from the lung tissues using an RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. For FPM development, the total sample size required was estimated to be n = 25 to 35 for PGD0/I donor lungs and n = 15 to 20 for PGDIII donor lungs based on previous studies by our group.
Microchip preparation
Glass microchips with exposed gold apertures were fabricated as previously described (47). FraCS were electrodeposited at room temperature using a Bioanalytical Systems Epsilon potentiostat with a three-electrode system featuring an Ag/AgCl reference electrode and a platinum wire auxiliary electrode. Gold apertures on the fabricated electrodes were used as the working electrode, and the structures were deposited using 50 mM HAuCl4 (Sigma-Aldrich) using DC potential amperometry at 0 mV for 30 s.
LTx assay protocol
PNA probe solution [0.85 μM PNA probe (PNA BIO), 0.15 μM MCH (Sigma-Aldrich), 10 mM tris-HCl (pH 7.5) (Invitrogen), 1 mM TCEP (Sigma-Aldrich), 100 mM NaCl (Sigma-Aldrich), 1 mM EDTA (Sigma-Aldrich)] was placed on freshly prepared gold electrodes (20 μl of probe solution volume) in a humidity chamber, and the deposition was allowed to occur overnight at room temperature. The electrodes were thoroughly washed with dH2O and then backfilled with 1 mM MCH (Sigma-Aldrich) for at least 2 hours. The electrodes were thoroughly washed again and then hybridized with 20 μl of the various targets (DNA, RNA, tissue lysate) in 1× SSC (Sigma-Aldrich) for 15 or 30 min at 37°C. After hybridization, the electrodes were washed and prepared for electrochemical measurements. The PNA probe sequences were as follows (N to C): GAPDH Cys-O-GTT-GTC-ATA-CTT-CTC; Drosophila Cys-O-ACC-GAA-CTC-GTT-GTC-GTA-CC; ATP11B Cys-O-ATC-CAC-TAC-CAG-CCC; IL-6 Cys-O-GCC-AGT-GCC-TCT-TTG-CT; IL-10 Cys-O-CGC-CGT-AGC-CTC-AGC-C (where O represents an ethylene glycol linker, and Cys represents the amino acid cysteine). Synthetic DNA oligonucleotides representing the complementary sequence to the respective PNA probes were ordered from Integrated DNA Technologies.
Quantitative polymerase chain reaction
For synthesis of cDNA (complementary DNA) (20 μl) from 0.5 μg of total RNA, High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used according to the manufacturer’s instructions on an Eppendorf Mastercycler pro thermocycler. qPCRs were performed on a Applied Biosystems 7500 real-time PCR system using 2 μl of cDNA, 12.5 μl of SYBR Green PCR Master Mix (2×) (Applied Biosystems), and 200 nM forward and reverse primers for a total volume of 25 μl. Conditions used for qPCR included an initial step of 15 s at 95°C followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The expression levels of target genes were normalized to the level of GAPDH. Linear regression analysis was used to compare the signals obtained from lung biopsies run on FraCS and qPCR. The equation of the regression line was used to transform qPCR values to FraCS-based signals for subsequent Bland-Altman analysis.
Mathematical modeling
MATLAB software was used to mathematically model the current generated by the FraCS as a function of DNA concentration. The parameters used in the model are shown in Table 4. For a reactant confined to the surface of the electrode (ruthenium), the peak current is given by (48):
where ip is the peak current (A), n is the number of electrons in the redox process, F is Faraday’s constant, ΓT is the total amount of redox molecule at the electrode surface (mol), v is the scan rate (V s−1), R is the gas constant, and T is temperature (K). ΓT is approximated by estimating the total amount of molecules that have reached the surface by diffusion using the equations described by Sheehan and Whitman (49). The saturation current occurs when the surface of the sensor is fully saturated with target molecules. This occurs when a target molecule is bound to each available probe. The maximum number of ruthenium molecules at a sensor is thus given by:
where A is the sensor area, ε is the hybridization efficiency, β is the ruthenium molecules per bound target molecule, and Γp is the probe density. We assumed that the number of ruthenium molecules bound per target molecule, β, is 20.
Table 4.
Electrochemical measurements
All electrochemical measurements were performed on a Bioanalytical Systems Epsilon potentiostat with a three-electrode system featuring an Ag/AgCl reference electrode and a platinum wire auxiliary electrode. The PNA-modified electrodes were incubated in 100 μM [Ru(NH3)6]3+ solution (Sigma-Aldrich) for 60 s and then rinsed in 10 mM tris-HCl (pH 7.5), and the electrochemical signals were measured in a solution containing 100 μM [Fe(CN)6]3− (Sigma-Aldrich) and 10 mM tris-HCl (pH 7.5). Differential pulse voltammetry was used to measure the current generated from the reduction of adsorbed [Ru(NH3)6]3+ at the surface of the electrode.
Statistical analysis
General
For all statistical calculations, a P value of less than 0.05 was considered statistically significant.
Biomarker validation
The signals for the lysate samples were GAPDH-normalized and FraCS-current results were plotted using GraphPad Prism 6, and the donor lungs were divided into PGD0/I (n = 23) or PGDIII+ [clinically unsuitable for transplant (n = 7) or PGDIII after transplant (n = 9)] outcomes. The population means were calculated, and a Mann-Whitney U test was used to determine statistical differences in the various groups.
FraCS prediction model
For the PGD prediction model, only the PGD0/I (proven to be non-injured by transplant) and PGDIII donor lungs (proven to have severe injury by transplant) were used in the calculations (n = 23 and 9, respectively). Logistic regression analysis on the currents generated for each individual biomarker was carried out in all combinations of measured markers and the IL-6/IL-10 ratio to develop the FPM. For validation of the FPM, an additional unique set of PGD0/I (n = 10) and PGDIII (n = 10) donor lungs were tested using FraCS and the FPM fit. Cross-validation of the FPM was carried out using 100 rounds of 10-fold (for the complete data set; 9-fold for the development set only) cross-validation with stratification. Diagnostic sensitivity, specificity, NPV, and PPV for the FPM were calculated using the PGDIII lungs as true positives and PGD0/I lungs as true negatives with a cutoff value of −0.3575.
Supplementary Material
Acknowledgments
We wish to thank all of the patients who donated the samples that were analyzed in this study. We acknowledge Y. Liang for the assistance with the illustrations used in this work. Funding: We also thank the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada for their support of this work. Author contributions: A.T.S. designed, performed, and analyzed the experiments and wrote the manuscript. J.D.B. performed the mathematical simulations, M.P. fabricated the glass microchips, L.M. contributed to the qPCR experiments, and R.Z. carried out the prediction model development and statistical analysis. M.H. and X.B. collected and managed the clinical samples used in this study. M.C., E.H.S., M.L., S.K., and S.O.K. co-supervised the project. All authors discussed the results and contributed to editing the manuscript. Competing interests: The authors declare that they have no competing interests. Data availability: Please contact the corresponding author at shana.kelley@utoronto.ca.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/7/e1500417/DC1
Fig. S1. Validation of LTx probes and FraCS approach with DNA targets.
Fig. S2. GAPDH validation with RNA.
Fig. S3. qPCR validation.
Fig. S4. Bland-Altman analysis of FraCS versus qPCR.
Table S1. Bland-Altman results of FraCS versus qPCR.
REFERENCES AND NOTES
- 1.Toronto Lung Transplant Group , Unilateral lung transplantation for pulmonary fibrosis. N. Engl. J. Med. 314, 1140–1145 (1986). [DOI] [PubMed] [Google Scholar]
- 2.Studer S. M., Levy R. D., McNeil K., Orens J. B., Lung transplant outcomes: A review of survival, graft function, physiology, health-related quality of life and cost-effectiveness. Eur. Respir. J. 24, 674–685 (2004). [DOI] [PubMed] [Google Scholar]
- 3.De Meester J., Smits J. M., Persijn G. G., Haverich A., Listing for lung transplantation: Life expectancy and transplant effect, stratified by type of end-stage lung disease, the Eurotransplant experience. J. Heart Lung Transplant. 20, 518–524 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Christie J. D., Edwards L. B., Aurora P., Dobbels F., Kirk R., Rahmel A. O., Taylor D. O., Kucheryavaya A. Y., Hertz M. I., Registry of the International Society for Heart and Lung Transplantation: Twenty-fifth official adult lung and heart/lung transplantation report—2008. J. Heart Lung Transplant. 27, 957–969 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Christie J. D., Sager J. S., Kimmel S. E., Ahya V. N., Gaughan C., Blumenthal N. P., Kotloff R. M., Impact of primary graft failure on outcomes following lung transplantation. Chest 127, 161–165 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Lee J. C., Christie J. D., Primary graft dysfunction. Proc. Am. Thorac. Soc. 6, 39–46 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Meyers B. F., de la Morena M., Sweet S. C., Trulock E. P., Guthrie T. J., Mendeloff E. N., Huddleston C., Cooper J. D., Patterson G. A., Primary graft dysfunction and other selected complications of lung transplantation: A single-center experience of 983 patients. J. Thorac. Cardiovasc. Surg. 129, 1421–1429 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Trulock E. P., Christie J. D., Edwards L. B., Boucek M. M., Aurora P., Taylor D. O., Dobbels F., Rahmel A. O., Keck B. M., Hertz M. I., Registry of the International Society for Heart and Lung Transplantation: Twenty-fourth official adult lung and heart/lung transplantation report—2007. J. Heart Lung Transplant. 26, 782–795 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Anraku M., Cameron M. J., Waddell T. K., Liu M., Arenovich T., Sato M., Cypel M., Pierre A. F., de Perrot M., Kelvin D. J., Keshavjee S., Impact of human donor lung gene expression profiles on survival after lung transplantation: A case-control study. Am. J. Transplant. 8, 2140–2148 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Suzuki Y., Cantu E., Christie J. D., Primary graft dysfunction. Semin. Respir. Crit. Care Med. 34, 305–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Perrot M., Liu M., Waddell T. K., Keshavjee S., Ischemia–reperfusion–induced lung injury. Am. J. Respir. Crit. Care Med. 167, 490–511 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Lee J. C., Christie J. D., Keshavjee S., Primary graft dysfunction: Definition, risk factors, short- and long-term outcomes. Semin. Respir. Crit. Care Med. 31, 161–171 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Christie J. D., Carby M., Bag R., Corris P., Hertz M., Weill D.; ISHLT Working Group on Primary Lung Graft Dysfunction , Report of the ISHLT Working Group on Primary Lung Graft Dysfunction part II: Definition. A consensus statement of the International Society for Heart and Lung Transplantation. J. Heart Lung Transplant. 24, 1454–1459 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Thabut G., Mal H., Cerrina J., Dartevelle P., Dromer C., Velly J. F., Stern M., Loirat P., Leseche G., Bertocchi M., Mornex J. F., Haloun A., Despins P., Pison C., Blin D., Reynaud-Gaubert M., Graft ischemic time and outcome of lung transplantation: A multicenter analysis. Am. J. Respir. Crit. Care Med. 171, 786–791 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Moudgil A., Bagga A., Toyoda M., Nicolaidou E., Jordan S. C., Ross D., Expression of Γ-IFN mRNA in bronchoalveolar lavage fluid correlates with early acute allograft rejection in lung transplant recipients. Clin. Transplant. 13, 201–207 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Ross D. J., Moudgil A., Bagga A., Toyoda M., Marchevsky A. M., Kass R. M., Jordan S. C., Lung allograft dysfunction correlates with Γ-interferon gene expression in bronchoalveolar lavage. J. Heart Lung Transplant. 18, 627–636 (1999). [DOI] [PubMed] [Google Scholar]
- 17.Shi R., Yang J., Jaramillo A., Steward N. S., Aloush A., Trulock E. P., Alexander Patterson G., Suthanthiran M., Mohanakumar T., Correlation between interleukin-15 and granzyme B expression and acute lung allograft rejection. Transpl. Immunol. 12, 103–108 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Vanaudenaerde B. M., Dupont L. J., Wuyts W. A., Verbeken E. K., Meyts I., Bullens D. M., Dilissen E., Luyts L., Van Raemdonck D. E., Verleden G. M., The role of interleukin-17 during acute rejection after lung transplantation. Eur. Respir. J. 27, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Krustrup D., Madsen C. B., Iversen M., Engelholm L., Ryder L. P., Andersen C. B., The number of regulatory T cells in transbronchial lung allograft biopsies is related to FoxP3 mRNA levels in bronchoalveolar lavage fluid and to the degree of acute cellular rejection. Transpl. Immunol. 29, 71–75 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Madsen C. B., Norgaard A., Iversen M., Ryder L. P., Elevated mRNA levels of CTLA-4, FoxP3, and granzyme B in BAL, but not in blood, during acute rejection of lung allografts. Transpl. Immunol. 24, 26–32 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Kaneda H., Waddell T. K., de Perrot M., Bai X. H., Gutierrez C., Arenovich T., Chaparro C., Liu M., Keshavjee S., Pre-implantation multiple cytokine mRNA expression analysis of donor lung grafts predicts survival after lung transplantation in humans. Am. J. Transplant. 6, 544–551 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Lande J. D., Patil J., Li N., Berryman T. R., King R. A., Hertz M. I., Novel insights into lung transplant rejection by microarray analysis. Proc. Am. Thorac. Soc. 4, 44–51 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley S. O., Mirkin C. A., Walt D. R., Ismagilov R. F., Toner M., Sargent E. H., Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nat. Nanotechnol. 9, 969–980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du J., Huang Z., Luo Q., Xiong G., Xu X., Li W., Liu X., Li J., Rapid diagnosis of pleural tuberculosis by Xpert MTB/RIF assay using pleural biopsy and pleural fluid specimens. J. Res. Med. Sci. 20, 26–31 (2015). [PMC free article] [PubMed] [Google Scholar]
- 25.Hillemann D., Rüsch-Gerdes S., Boehme C., Richter E., Rapid molecular detection of extrapulmonary tuberculosis by the automated GeneXpert MTB/RIF system. J. Clin. Microbiol. 49, 1202–1205 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding C., Zhang Q., Lin J. M., Zhang S. S., Electrochemical detection of DNA hybridization based on bio-bar code method. Biosens. Bioelectron. 24, 3140–3143 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Gasparac R., Taft B. J., Lapierre-Devlin M. A., Lazareck A. D., Xu J. M., Kelley S. O., Ultrasensitive electrocatalytic DNA detection at two- and three-dimensional nanoelectrodes. J. Am. Chem. Soc. 126, 12270–12271 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Fan C., Plaxco K. W., Heeger A. J., Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. U.S.A. 100, 9134–9137 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin M., Wen Y., Li L., Pei H., Liu G., Song H., Zuo X., Fan C., Huang Q., Target-responsive, DNA nanostructure-based E-DNA sensor for microRNA analysis. Anal. Chem. 86, 2285–2288 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Yu H. Z., Li Y., Ou L. M., Reading disc-based bioassays with standard computer drives. Acc. Chem. Res. 46, 258–268 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Song S., Zhang L., Wang L., Wu H., Pan D., Fan C., Sequence-specific detection of femtomolar DNA via a chronocoulometric DNA sensor (CDS): Effects of nanoparticle-mediated amplification and nanoscale control of DNA assembly at electrodes. J. Am. Chem. Soc. 128, 8575–8580 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Wang J., Liu G., Merkoci A., Electrochemical coding technology for simultaneous detection of multiple DNA targets. J. Am. Chem. Soc. 125, 3214–3215 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Das J., Cederquist K. B., Zaragoza A. A., Lee P. E., Sargent E. H., Kelley S. O., An ultrasensitive universal detector based on neutralizer displacement. Nat. Chem. 4, 642–648 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Fang Z., Soleymani L., Pampalakis G., Yoshimoto M., Squire J. A., Sargent E. H., Kelley S. O., Direct profiling of cancer biomarkers in tumor tissue using a multiplexed nanostructured microelectrode integrated circuit. ACS Nano 3, 3207–3213 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Lam B., Fang Z., Sargent E. H., Kelley S. O., Polymerase chain reaction-free, sample-to-answer bacterial detection in 30 minutes with integrated cell lysis. Anal. Chem. 84, 21–25 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Vasilyeva E., Lam B., Fang Z., Minden M. D., Sargent E. H., Kelley S. O., Direct genetic analysis of ten cancer cells: Tuning sensor structure and molecular probe design for efficient mRNA capture. Angew. Chem. Int. Ed. Engl. 50, 4137–4141 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Yang H., Hui A., Pampalakis G., Soleymani L., Liu F. F., Sargent E. H., Kelley S. O., Direct, electronic microRNA detection for the rapid determination of differential expression profiles. Angew. Chem. Int. Ed. Engl. 48, 8461–8464 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Ivanov I., Stojcic J., Stanimirovic A., Sargent E., Nam R. K., Kelley S. O., Chip-based nanostructured sensors enable accurate identification and classification of circulating tumor cells in prostate cancer patient blood samples. Anal. Chem. 85, 398–403 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Lam B., Das J., Holmes R. D., Live L., Sage A., Sargent E. H., Kelley S. O., Solution-based circuits enable rapid and multiplexed pathogen detection. Nat. Commun. 4, 2001 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Soleymani L., Fang Z., Lam B., Bin X., Vasilyeva E., Ross A. J., Sargent E. H., Kelley S. O., Hierarchical nanotextured microelectrodes overcome the molecular transport barrier to achieve rapid, direct bacterial detection. ACS Nano 5, 3360–3366 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Besant J. D., Das J., Sargent E. H., Kelley S. O., Proximal bacterial lysis and detection in nanoliter wells using electrochemistry. ACS Nano 7, 8183–8189 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Bhimji A., Zaragoza A. A., Live L. S., Kelley S. O., Electrochemical enzyme-linked immunosorbent assay featuring proximal reagent generation: Detection of human immunodeficiency virus antibodies in clinical samples. Anal. Chem. 85, 6813–6819 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y. G., Wan Y., Sage A. T., Poudineh M., Kelley S. O., Effect of microelectrode structure on electrocatalysis at nucleic acid-modified sensors. Langmuir. 30, 14322–14328 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Liu M., Slutsky A. S., Anti-inflammatory therapies: Application of molecular biology techniques in intensive care medicine. Intensive Care Med. 23, 718–731 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cypel M., Kaneda H., Yeung J. C., Anraku M., Yasufuku K., de Perrot M., Pierre A., Waddell T. K., Liu M., Keshavjee S., Increased levels of interleukin-1β and tumor necrosis factor-α in donor lungs rejected for transplantation. J. Heart Lung Transplant. 30, 452–459 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Ware L. B., Wang Y., Fang X., Warnock M., Sakuma T., Hall T. S., Matthay M., Assessment of lungs rejected for transplantation and implications for donor selection. Lancet 360, 619–620 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Lam B., Holmes R. D., Das J., Poudineh M., Sage A., Sargent E. H., Kelley S. O., Optimized templates for bottom-up growth of high-performance integrated biomolecular detectors. Lab Chip 13, 2569–2575 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Brown A. P., Anson F.C., Cyclic and differential pulse voltammetric behavior of reactants confined to the electrode surface. Anal. Chem. 49, 1589–1595 (1977). [Google Scholar]
- 49.Sheehan P. E., Whitman L. J., Detection limits for nanoscale biosensors. Nano Lett. 5, 803–807 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Bin X., Sargent E. H., Kelley S. O., Nanostructuring of sensors determines the efficiency of biomolecular capture. Anal. Chem. 82, 5928–5931 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Lapierre-Devlin M. A., Asher C. L., Taft B. J., Gasparac R., Roberts M. A., Kelley S. O., Amplified electrocatalysis at DNA-modified nanowires. Nano Lett. 5, 1051–1055 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/7/e1500417/DC1
Fig. S1. Validation of LTx probes and FraCS approach with DNA targets.
Fig. S2. GAPDH validation with RNA.
Fig. S3. qPCR validation.
Fig. S4. Bland-Altman analysis of FraCS versus qPCR.
Table S1. Bland-Altman results of FraCS versus qPCR.