SUMMARY
Aging has been associated with a progressive decline of proteostasis, but how this process manifests itself at the level of proteome composition remains largely unexplored. Here we profiled more than 5,000 proteins along the lifespan of the nematode C. elegans. We find that one third of proteins change in abundance at least 2-fold during aging, resulting in a severe proteome imbalance. These changes are reduced in the long-lived daf-2 mutant, but enhanced in the short-lived daf-16 mutant. While ribosomal proteins decline and lose normal stoichiometry, proteasome complexes increase. Proteome imbalance is accompanied by widespread protein aggregation, with abundant proteins that exceed solubility contributing most to aggregate load. Notably, the properties by which proteins are selected for aggregation differ in the daf-2 mutant, and an increased formation of aggregates associated with small heat shock proteins is observed. We suggest that sequestering proteins into chaperone-enriched aggregates is a protective strategy to slow proteostasis decline during nematode aging.
INTRODUCTION
Protein homeostasis (proteostasis), the state in which the proteome of a living organism is in functional balance, must be tightly controlled within individual cells, tissues and organs. Maintaining proteome balance requires a complex network of cellular factors, including the machineries of protein synthesis, folding and degradation (Balch et al., 2008; Hartl et al., 2011), as well as neuronal signaling pathways that regulate proteostasis at the organismal level (Prahlad and Morimoto, 2009; Taylor and Dillin, 2013; van Oosten-Hawle and Morimoto, 2014). An important function of these systems is to prevent the accumulation of potentially toxic misfolded and aggregated protein species (Knowles et al., 2014). However, as organisms age, quality control and the cellular response to unfolded protein stress become compromised (Ben-Zvi et al., 2009; Douglas and Dillin, 2010), and the defense against reactive oxygen species declines (Finkel and Holbrook, 2000). Indeed, aging is considered the principal risk factor for the onset of a number of neurodegenerative disorders associated with aggregate deposition, such as Alzheimer’s, Huntington’s and Parkinson’s diseases (Knowles et al., 2014). The accumulation of aberrant protein species in these pathologic states in turn places a burden on the proteostasis machinery and thus may accelerate aging by interfering with protein folding and clearance, and other key cellular processes (Balch et al., 2008; Gidalevitz et al., 2006; Hipp et al., 2014; Olzscha et al., 2011). Understanding these relationships requires systematic analyses of the changes that occur in proteome composition and balance during aging.
The nematode C. elegans is one of the most extensively studied model organisms in aging research, owing to its relatively short lifespan and the availability of genetic tools to identify pathways that regulate longevity. Inhibition of the insulin/insulin-like growth factor 1 signaling (IIS) pathway in strains carrying mutations in the DAF-2 receptor (or the downstream PI(3) kinase AGE-1) activates the DAF-16/FOXO transcription factor and leads to a dramatic lifespan extension (Kenyon et al., 1993; Murphy et al., 2003). Several lines of evidence suggest that the lifespan-prolonging effect of IIS reduction involves an improvement in cellular stress resistance and proteostasis capacity through upregulation of the machineries mediating protein folding and preventing the formation of toxic aggregate species (Morley et al., 2002; Cohen et al., 2009; Demontis and Perrimon, 2010). In addition to DAF-16 activation, the longevity phenotype in daf-2 mutants requires the function of HSF-1, the transcription factor regulating the expression of multiple heat-shock proteins and molecular chaperones (Hsu et al., 2003; Morley and Morimoto, 2004). These pathways of proteostasis maintenance appear to be conserved in evolution from worms to mammals (Cohen et al., 2009; Demontis and Perrimon, 2010).
Aging and the effect of the IIS pathway have been studied in C. elegans by transcriptome analysis (Budovskaya et al., 2008; Golden and Melov, 2004), but only limited information exists about changes at the proteome level (Dong et al., 2007). Here we exploit the recent progress in mass spectrometry-based proteomics, which now enables the identification and quantification of thousands of proteins in complex mixtures (Bensimon et al., 2012; Cox and Mann, 2011). We applied stable isotope labeling with amino acids in cell culture (SILAC) (Ong et al., 2002) to profile the abundance levels of more than 5,000 different proteins at multiple time points during the lifespan of C. elegans. We then extended our study to short-lived and long-lived strains carrying mutations in the IIS pathway and performed a detailed analysis of age-related protein aggregation. Our data show that during aging, the proteome of the animal undergoes extensive remodeling, escaping proteostasis and ultimately reaching a state of marked proteome imbalance. These changes are accompanied by widespread protein aggregation, with abundant proteins that exceed their solubility limit making the major contribution to aggregate load. Interestingly, the intrinsic aggregation propensity of proteins is modulated in long-lived daf-2 mutant worms, resulting in the enhanced formation of chaperone-containing aggregates. Thus, protein aggregation may occur not just as a consequence of proteostasis decline, but may also be induced to improve proteostasis by sequestering surplus, potentially harmful protein species.
RESULTS
Extensive Proteome Remodeling During Aging
To study proteome changes in aging nematodes in depth and with high accuracy, we established a quantitative proteomics approach using SILAC (Ong et al., 2002). Near-complete incorporation of 13C6-14N2-lysine into the proteome was achieved by feeding worms with SILAC labeled (“heavy”) E. coli cells (Larance et al., 2011). We used a pool of lysates prepared from labeled worms of different ages as internal standards for quantifying protein expression. These standards were added to lysates of synchronized worm populations, followed by digestion and peptide analysis by mass spectrometry (MS) (Figure 1A). Replicate analyses indicated a high degree of reproducibility between individual experiments (Figure S1A and Table S1A). We analyzed the proteomes of adult wild-type (WT) worms from one day up to 22 days of age, when less than 30% of the animals remain alive (L4 larval stage defined as day 0). More than 5,000 different proteins were identified and quantified at a false discovery rate of 1% (Table S1B).
Figure 1. Proteomic Analysis of Aging in C. elegans.
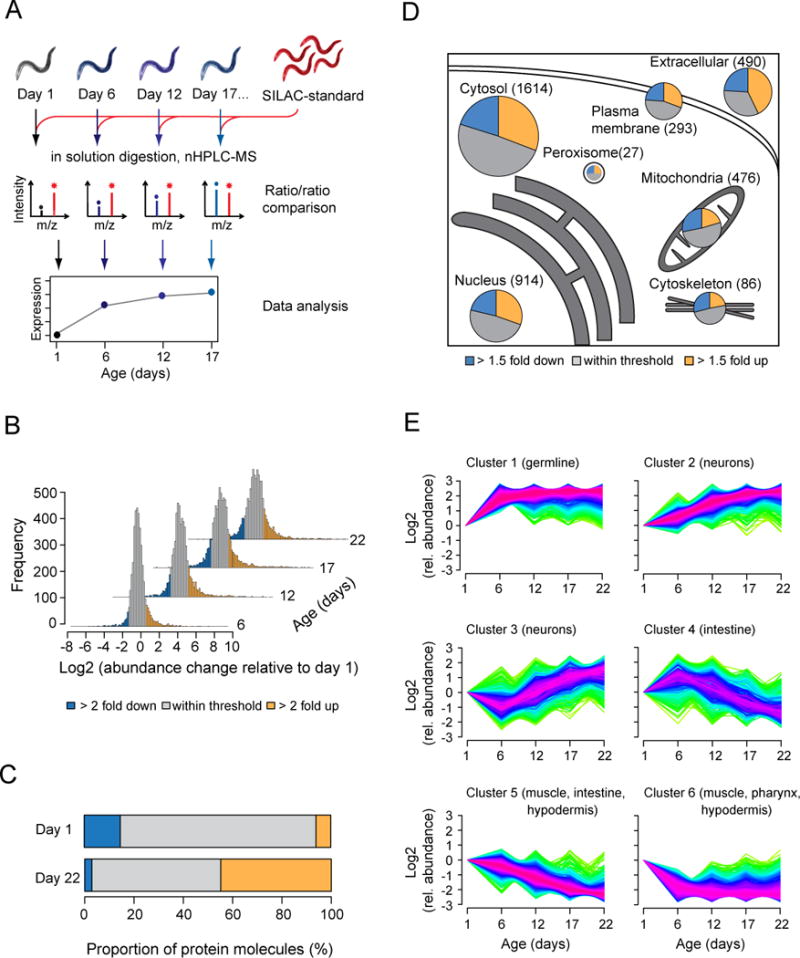
(A) Experimental design of total proteome analysis. Synchronized worm populations at different time points were lysed and mixed with a metabolically (SILAC) labeled internal protein standard. After digestion, peptides were either analyzed directly or after fractionation by isoelectric focusing, followed by nano-HPLC coupled mass spectrometry (MS).
(B) Proteome changes in WT animals 6, 12, 17 and 22 days of age relative to day 1 animals (Table S1B). The proportions of proteins that are at least 2-fold increased or decreased in abundance are marked in yellow or blue, respectively.
(C) Contribution to the total proteome of the proteins that change at least 2-fold in abundance between young (day 1) and aged (day 22) animals, as displayed in (A) and estimated by label free quantification (absolute LFQ) (Table S1B).
(D) Proteome changes in subcellular compartments. The fractions of the total proteome that increased (yellow) or decreased (blue) at least 1.5-fold in abundance in old (day 22) versus young (day 1) animals are shown. Grey, proteins that remained within the indicated abundance thresholds. Numbers of identified proteins are indicated. Protein subcellular localization was predicted using WoLF PSORT.
(E) Clustering of time course expression patterns in WT animals using the fuzzy c-means algorithm (Kumar and Futschik, 2007). Significantly enriched tissues as determined by Wilcoxon rank sum test at 2% false discovery rate against predicted expression scores (Chikina et al., 2009) are indicated for each cluster. Warm (red) and cold (blue) colors indicate low and high deviation from the consensus profile, respectively.
Our analysis reveals a broad remodeling of the C. elegans proteome during aging. About one third of the quantified proteins increased or decreased in abundance by at least 2-fold, when equal amounts of total protein were analyzed (Figure 1B and Table S1B). The proteins that increased by at least 2-fold amounted to approximately 50% of total protein in aged animals, as determined by label free absolute quantification (absolute LFQ values) (Schwanhausser et al., 2011) (Figure 1C). Protein abundance changes were progressive until day 22 (Figure 1B, S1B and Table S2A) and were observed in most cellular compartments (Figure 1D). Thus, proteome composition and the relative stoichiometries of proteins change dramatically during aging, presumably impeding overall proteostasis. A similar mechanism of proteostasis impairment has been suggested to occur as a result of aneuploidy (Oromendia et al., 2012; Stingele et al., 2012).
Changes in transcript levels previously observed during aging (Budovskaya et al., 2008; Golden and Melov, 2004) contribute to the changes in protein abundance observed here, but the overall correlation is only moderate (R = 0.3) (Figure S1C). Thus, the age-dependent accumulation of a substantial fraction of the proteome is likely to be largely due to post-transcriptional processes. Taking into consideration that miRNA-mediated translational repression of mRNAs is relieved during aging and stress (Ibanez-Ventoso et al., 2006), we compared our proteome data with a published transcriptome analysis of Dicer mutant worms with defective miRNA biogenesis (Welker et al., 2007). We find that ~30% of proteins that increased more than 2-fold between day 6 and 22 (99 of 357 proteins), i.e. after the worms have reproduced, have significantly elevated transcript levels in dicer mutants, and this proportion increases to nearly 40% for the subset of proteins with a more than 4-fold abundance change (50 out of 133 proteins) (Figure S1D). Thus, miRNA-mediated translational derepression is likely to contribute to the observed increase in protein abundance.
We analyzed the proteomic changes in C. elegans aging in terms of various criteria, including subcellular compartments, pathways and cell types. Among the proteins that increased more than 2-fold in aged worms (22 days) were 183 extracellular proteins (out of 490 extracellular proteins quantified) (Figure S1E and Table S2B). These included multiple transthyretin (TTR)-like factors, which increased up to 100-fold (Figure S1F), as well as all six of the vitellogenin egg storage proteins, despite egg formation having been completed before day 6. Likewise, proteins involved in DNA replication and repair processes were up-regulated (Figure S1E), even though all somatic cells of adult C. elegans are postmitotic. These examples suggest that many changes in protein abundance during aging do not correlate with biologically relevant activities and instead reflect proteome dysregulation. Among the proteins that declined during aging are nucleolar ribosome biogenesis factors, various peroxisomal enzymes and proteins involved in lipid glycosylation (Figure S1E and Table S2C). The levels of many mitochondrial proteins also decreased (Figure 1D). For example, subunits of respiratory chain complex I declined gradually by up to 50% during lifespan (Figure S1G), which may result in the production of reactive oxygen species.
To discern cell-type specific patterns of change, we grouped proteins into clusters using the fuzzy c-means method (Kumar and Futschik, 2007) and analyzed these by tissue-specific expression scores (Chikina et al., 2009) (Figure 1E). We find that age-dependent changes in proteome composition affect a range of tissues. For example, proteins that are predominantly expressed in the germline strongly increase during the first 6 days of adulthood (cluster 1), when the animals reproduce, but surprisingly retain constant levels later in life. Proteins enriched in neuronal cells either increase in abundance throughout lifespan or after day 6 (clusters 2 and 3). In contrast, the levels of many proteins enriched in intestine, muscle and hypodermis decline (clusters 4–6), consistent with an age-related deterioration of these tissues.
Age-related Changes in Proteostasis Network Components
Approximately 440 proteostasis network components involved in protein synthesis, folding and degradation were quantified throughout the nematode lifespan (Figure S2A and Table S3). A ~25% reduction in the median level of cytosolic ribosomal proteins occurred between day 1 and day 12 (Figure 2A, left panel). This reduction correlates with a decrease in the transcript level of ribosome proteins (Golden and Melov, 2004) and an overall age-associated reduction in polysomes (Kirstein-Miles et al., 2013). A similar decrease was observed for mitochondrial ribosomes between day 1 and day 6 (Figure 2A, right panel). Interestingly, aged animals displayed a pronounced imbalance in the relative subunit stoichiometry of cytosolic, but not mitochondrial ribosomes, with several subunits decreasing more than 60% below median subunit levels (Figure 2A, left).
Figure 2. Abundance Changes in Specific Components of the Proteostasis Network.
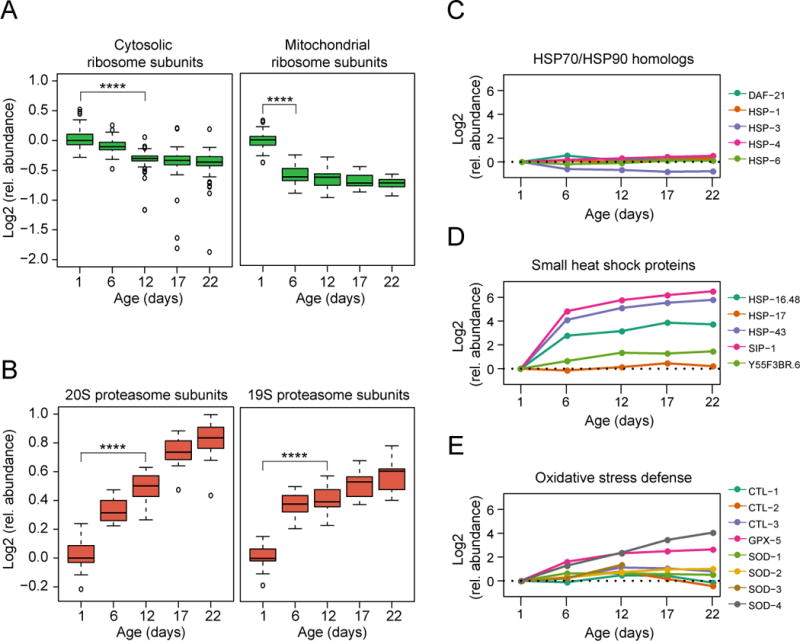
(A) Abundance changes of ribosomal proteins during the lifespan of C. elegans. 70 different cytosolic (left) and 34 mitochondrial ribosomal proteins (right) were quantified (see Table S3). Log2 values of fold-changes are shown in boxplot representation. Solid horizontal lines indicate the median values, whisker caps indicate 10th and 90th percentiles, and circles indicate outliers. ****, p-value <4.35 × 10−13 for cytosolic ribosomal proteins and 1.17 × 10−10 for mitochondrial ribosomal proteins from Wilcoxon signed rank test. Only proteins quantified at both time points tested were considered.
(B) Abundance changes of proteasome subunits during lifespan. All 14 subunits of the 20S and 17 subunits of the 19S proteasome were quantified. Only subunits quantified in at least two time points are displayed. ****, p-value <1.23 × 10−4 and ****, p-value <1.53 × 10−5 from Wilcoxon signed rank test.
(C–E) Abundance profiles of proteostasis network (PN) components along the lifespan of WT animals. Log2 relative changes in abundance are shown for HSP70 and HSP90 homologs (C), small HSPs (D), and proteins involved in oxidative stress defense (E). Only components quantified at day 1 and at least three consecutive time points are displayed.
Next, we employed SILAC to estimate protein synthesis in aging C. elegans. Pulse labeling of worms with “heavy” bacteria as the food source showed a sharp reduction in the incorporation of labeled amino acids into protein between day 1 and day 4 of adulthood (Figure S2B and Table S1C). This effect was not caused by reduced food uptake, as eat-2 mutant animals, deficient in pharyngeal pumping, showed protein labeling equivalent to WT controls, despite their reduced food uptake (data not shown). The reduction in protein synthesis between day 1 and 4 is greater than the decrease in ribosomal levels (Figure 2A and S2B) and probably reflects the reduction in growth of the animals.
In contrast to the effect on ribosomes, we observed an age-dependent increase in 20S and 19S proteasomal subunits (~2-fold at day 22 for 20S subunits) (Figure 2B), correlating with an increase in proteasome activity measured in worm lysates in vitro (Figure S2C). Many E3 ubiquitin ligases and other components of the ubiquitin proteasome system (UPS) also increased moderately (Table S3B), while there was no systematic change in the components of autophagy (Figure S2D).
Age-dependent changes in the levels of abundant cytosolic chaperones of the HSP70 and HSP90 (DAF-21) families (Figures 2C) as well as their DnaJ (DNJ/HSP40) and tetratricopeptide repeat (TPR) co-factors were limited (Figure S2E and S2F). Similarly, the subunits of the TRiC/CCT chaperonin remained unchanged (Figure S2G). In contrast, multiple small HSPs, chaperones that function by buffering aggregation, increased dramatically (~13–90-fold), mainly between day 1 and day 6 (Figure 2D). Several of these proteins are under regulation by DAF-16 and HSF-1 (Hsu et al., 2003).
Several components mediating the defense against oxidative stress, including the glutathione peroxidase isoform GPX-5 and several forms of superoxide dismutase (SOD) increased during aging (up to 12-fold) (Figure 2E and Table S3B). While changes in mitochondrial proteostasis components were generally moderate (Figure S2H and Table S3B), we observed diverse alterations in the proteostasis network of the endoplasmic reticulum (ER) during the nematode lifespan (Figure S2I). For example, protein disulfide isomerases (PDI-2, C14B9.2), the chaperone calreticulin (CRT-1) as well as the HSP70 homolog HSP-3 decreased ~2-fold, and the pro-collagen modifying enzymes lysyl hydroxylase (LET-268) and prolyl-4-hydroxylase α (DPY-18, PHY-2) decreased ~3–10-fold. These findings suggest an age-dependent decline in ER quality control and collagen synthesis capacity.
In summary, the levels and activities of two main branches of proteostasis control, protein synthesis and degradation, change in opposite directions during aging. The decrease in ribosomal subunit proteins is accompanied by a dysregulation of cytosolic ribosome assembly, while the increase in proteasome subunits is likely to reflect an attempt at removing surplus or damaged proteins. Other notable changes in the proteostasis system include an increase in the abundance of small HSP chaperones and of components involved in the defense against oxidative stress, as well as a decline in ER protein quality control machinery.
Proteome Changes in Long-lived and Short-lived Mutant Strains
To understand in more detail the relationship between the observed proteome changes during lifespan and the aging process, we next analyzed the proteomes of long-lived daf-2 (e1370), and short-lived daf-16 (mu86) and hsf-1 (sy441) mutant worms. The increase in levels of specific proteins observed during aging of WT animals was considerably less pronounced in daf-2 mutant animals and enhanced in daf-16 mutant animals (Figure S3A, left panel), indicating that the long-lived daf-2 mutant strain is more effective in controlling the accumulation of surplus proteins. The extent to which proteins decreased in abundance during aging was also greater in daf-2 mutant worms (Figure S3A, right panel).
The changes in components of the proteostasis network observed in the mutant strains occurred again predominantly in the protein synthesis and degradation pathways, but at different rates compared to WT. The upregulation of proteasomal subunits commenced earlier during the lifespan of the daf-2 mutant and was more pronounced than in the WT worms (Figure 3A and B); such upregulation was instead less prominent in the short-lived daf-16 and hsf-1 mutant strains (Figures 3C and D). These results are consistent with the DAF-16 dependent regulation of some proteasome subunits, including RPN6 which is required for 26S proteasome assembly (Vilchez et al., 2012). The decrease in ribosomal proteins occurred at a similar rate in daf-2 mutant worms as in WT (Figure 3A), but was strongly enhanced in daf-16 mutant worms (Figure 3C), suggesting that DAF-16 is involved in ribosome maintenance.
Figure 3. Remodeling of the Proteostasis Network During Aging.
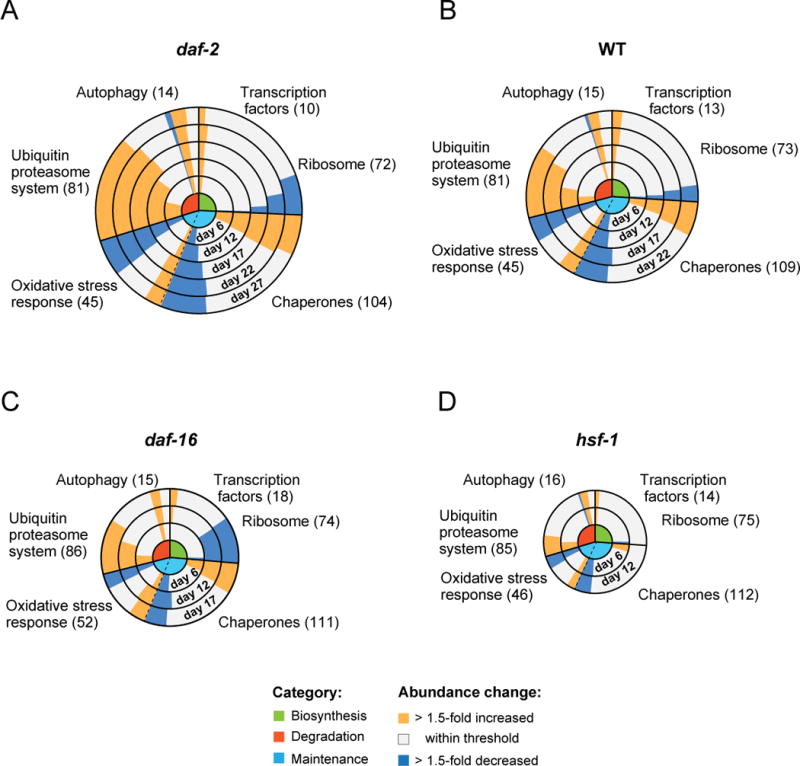
(A–D) Abundance changes in components of the PN (see Figure S2A) during aging in daf-2 (A), WT (B), daf-16 (C) and hsf-1 (D) mutant worms. Concentric circles represent increasing age in days from center to periphery. Circle size corresponds with lifespan. Functional categories of components are indicated in the center: green, biosynthesis; red, degradation; light blue, conformational maintenance (see Figure S2A). Abundance changes of components within these categories relative to day 1 of each strain (yellow, >1.5-fold up; blue, >1.5-fold down) are indicated as bars, with the length of the bar representing the number of proteins undergoing change. The total number of proteins quantified in the respective categories are indicated.
Components involved in the oxidative stress response showed marked differences in levels between WT and daf-2 mutant animals. For example, cytosolic and peroxisomal catalases (CTL-1, CTL-3 and CTL-2, respectively) were 4- to 8-fold higher in the daf-2 mutant than in WT worms throughout their lifespans (Figure S3B). SOD-1 (cytoplasmic) and SOD-2 (mitochondrial) were elevated 2-fold compared to the WT and short-lived mutant animals (Figure S3C), consistent with their DAF-16-dependent transcriptional regulation (McElwee et al., 2003; Murphy et al., 2003). Among the small HSPs, SIP-1 was already more abundant in young daf-2 mutant worms (day 1) and HSP-16.48 was markedly elevated in hsf-1 mutant animals (Figure S3D).
The earlier and more pronounced increase in proteasome abundance in daf-2 mutant animals may improve the capacity of the organism for the clearance of surplus proteins that accumulate during aging. The elevated levels of catalases and superoxide dismutase may provide improved defense against oxidative damage.
Age-dependent Protein Aggregation and its Relation to Protein Abundance
Declining proteostasis capacity is thought to result in the accumulation of protein aggregates, consistent with recent reports of age-dependent aggregate formation in C. elegans (David et al., 2010; Reis-Rodrigues et al., 2012). To analyze this process systematically, we developed a sensitive method for the quantification of aggregated proteins (see Experimental Procedures) and validated it in animals expressing muscle specific FlucDM-GFP, a conformationally unstable mutant of firefly luciferase fused to GFP (Gupta et al., 2011) (Figure S4A and B). We isolated insoluble proteins from total lysates of WT animals by centrifugation and performed MS analysis using lysate from labeled worms for quantification (Figure S4C). About 90% of the proteins that were quantified in 3 out 4 experiments (975 of 1083 proteins) accumulated significantly in the insoluble fraction of day 12 animals relative to day 1 (Table S1D). Age-dependent aggregation was most pronounced between day 6 and day 12 (Figure 4A), i.e. after the hermaphrodite animals ceased to lay eggs. Proteins with predicted transmembrane segments were not enriched in the insoluble fraction (Figure S5A), indicating that lysis was efficient.
Figure 4. Proteome-wide Analysis of Protein Aggregation during Aging.
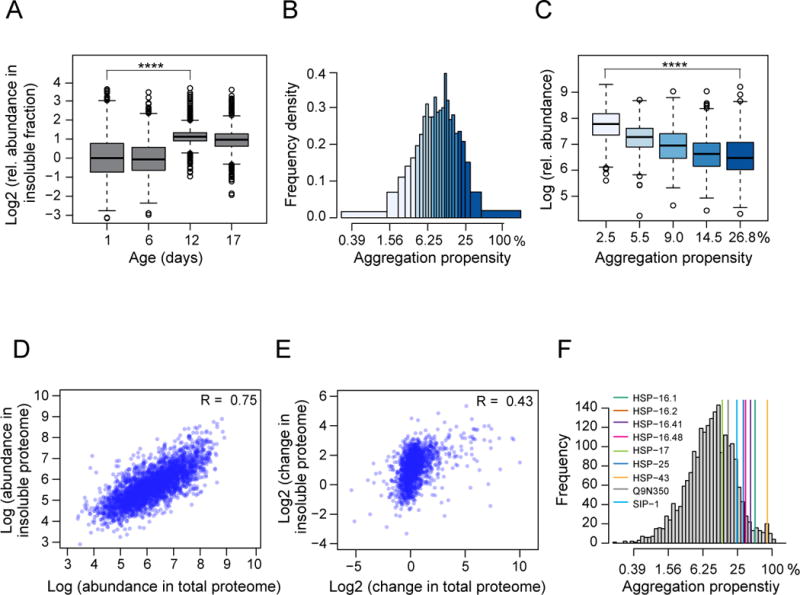
(A) Relative abundance of proteins in the insoluble fraction of WT animals during aging determined by SILAC quantification (see Figure S4C and Table S1D). At least 1355 proteins were quantified at the different time points (~3228 different proteins in total). ****, p-value <2.2 × 10−16 from Wilcoxon signed rank test.
(B) Distribution of aggregation propensities of proteins (insoluble protein as fraction of total protein) in WT animals at day 12 (median from 3 independent experiments; Table S1E). Whole worm lysates and insoluble fractions were quantified against the same SILAC standard and ratios were calculated for each protein in % of total (see Figure S4D).
(C) Relationship between aggregation propensity and total protein abundance. Proteins were divided into quantiles based on their measured aggregation propensities (median values are indicated in %). Label free absolute quantification was used to estimate total protein abundance (displayed as relative abundance values). ****, p-value <2.2 × 10−16 from Wilcoxon rank sum test.
(D) Protein abundance in the insoluble fraction is positively correlated with abundance in the total proteome (absolute LFQ values). Data for WT animals at day 12 are shown. The Pearson correlation coefficient R is indicated.
(E) Positive correlation between age-related protein abundance changes in the insoluble fraction and abundance changes for the same proteins in the total proteome. Abundance differences measured by SILAC between aged (day 12) and young (day 1) WT animals are plotted. The Pearson correlation coefficient R is indicated.
(F) Aggregation propensities of small HSP family members relative to the aggregation propensities of all quantified proteins in the proteome of day 12 WT animals.
To measure the aggregation propensities of proteins during aging, we quantified the insoluble amount of each protein as a fraction of its total amount in aged WT worms (day 12) (Figure S4D and Table S1E). The aggregation propensities of >2100 analyzed proteins varied by more than two orders of magnitude (Figure 4B), with the median insoluble fraction per individual protein amounting to ~ 9%.
Previous studies reported a negative correlation between computationally predicted aggregation propensities and protein abundance (Tartaglia et al., 2009). To investigate this dependency at the proteome scale, we grouped proteins according to their aggregation propensities measured at day 12 and estimated the total abundance of each protein in the whole cell lysate by absolute LFQ (Figure 4C). The most abundant proteins were 10-times more soluble than the least abundant proteins. An analysis of the physicochemical properties of the abundant proteins based on their amino acid sequences revealed that they were more hydrophobic (Figure S5B) and more structured (data not shown) than the less abundant ones. These results suggest that abundant proteins increase their solubility, at least in part, by stabilizing their native states through formation of a more extensive hydrophobic core. Indeed, a calculation of the aggregation propensities (Z-scores) (Tartaglia et al., 2008; Sormanni et al.,in press) (see Extended Experimental Procedures) predicts that the more abundant proteins, if correctly folded, are also more soluble (Figure S5C). This conclusion is consistent with the idea that the solubility of proteins follows their abundance (Tartaglia et al., 2009).
We found, however, that despite of their lower intrinsic aggregation propensities, the most abundant proteins contribute most to the total aggregate load. A strong correlation (R = 0.75) was observed between the abundance of specific proteins in the aggregate fraction and their level in the corresponding whole cell lysate (Figure 4D). Apparently, the high solubility of abundant proteins is insufficient to protect them from age-dependent aggregation, as eventually these proteins exceed their critical concentrations, a phenomenon referred to as ‘supersaturation’ (Ciryam et al., 2013). Notably, we also observed a medium correlation (R = 0.43) between the age-dependent change in the total abundance of proteins and their increase in the aggregate fraction (Figure 4E), and this correlation became stronger as aging progressed (data not shown). Thus, proteome remodeling during aging likely drives the aggregation of numerous proteins.
We further investigated whether aggregation also correlates with function. Gene ontology analysis showed that proteins with a relatively high aggregation propensity in aged animals are enriched in the nucleus, whereas abundant glycolytic enzymes and mitochondrial proteins tend to be highly soluble (Figure S5D and Table S4A). Interestingly, all identified small HSPs, but not other chaperones, were highly insoluble at day 12 (Figure 4F), with a high rate of accumulation in the aggregate fraction during aging (Figure S5E). The recruitment of these chaperones into the insoluble fraction may reflect an attempt of the organism to sequester protein aggregates.
Protein Aggregation in Long-lived and Short-lived Mutant Strains
Is the age-dependent formation of insoluble aggregates merely a reflection of declining proteostasis capacity, or is it a means to improve proteostasis by sequestering surplus proteins? Consistent with the former possibility are findings that aggregation-prone model proteins increasingly aggregate in proteostasis-compromised hsf-1 mutant strains (Ben-Zvi et al., 2009). Indeed, compared to WT animals, the short-lived hsf-1 mutant worms accumulated more insoluble proteins and aggregation occurred earlier during aging (between day 1 and day 6) (Figures 5A and S6A). However, in support of a beneficial role for aggregation, we found that the long-lived daf-2 mutant worms also accumulated aggregates and that their aggregate load was significantly greater than that of age-matched WT animals (Figures 5A, S6A and B). This effect was not observed in daf-16 mutant animals (Figures 5A and S6A), suggesting that age-dependent aggregation is (at least in part) an active process under regulation by DAF-16. The increased aggregation in daf-2 mutant animals comprised preferentially cytosolic proteins (Figure S6C and Table S4B) and initiated between day 6 and day 12 as in WT (Figure S6A), i.e. when the long-lived mutant worms are still youthful.
Figure 5. Protein Aggregation in Lifespan Mutant Worms During Aging.
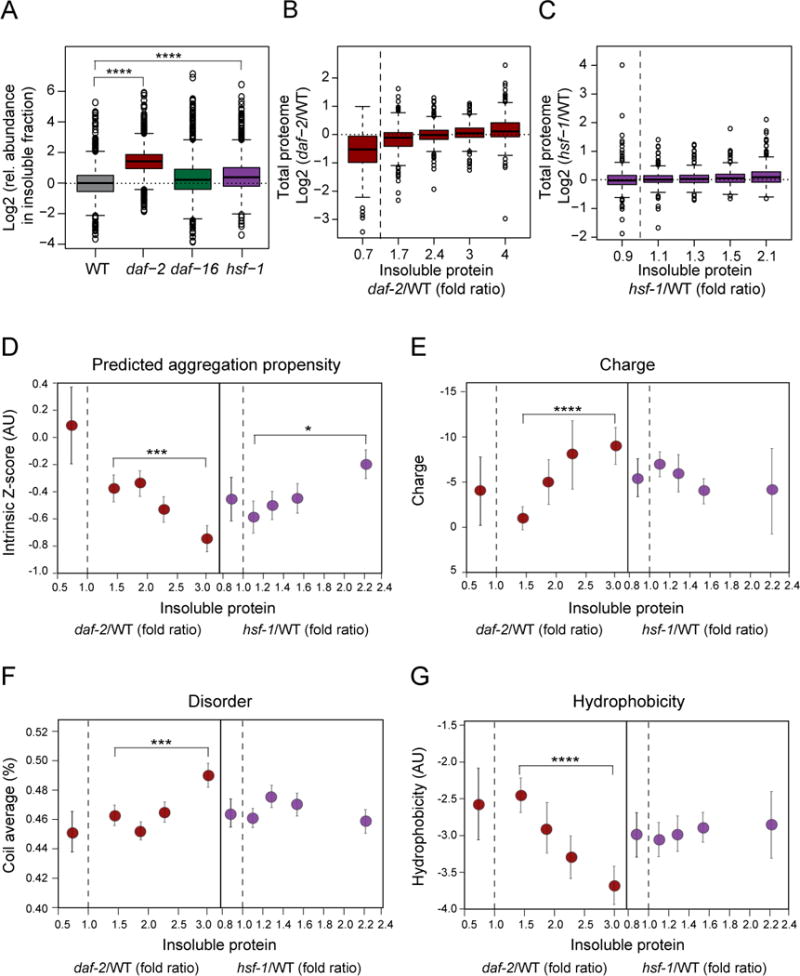
(A) Increased aggregate load in daf-2 mutant animals compared to WT, daf-16 and hsf-1 mutant animals at day 12. Relative abundance values of proteins in the insoluble fraction were determined by SILAC quantification. 1367, 1988, 1449 and 1485 proteins were quantified in WT, daf-2, daf-16 and hsf-1 mutant animals, respectively (one representative out of 4 independent experiments is displayed; Table S1F). ****, p-value <2.2 × 10−16 from Wicoxon signed rank test.
(B and C) Quantiled abundance of proteins in the insoluble fraction of daf-2 (352–354 proteins per quantile) (B) and hsf-1 mutant (292 proteins per quantile) (C) relative to WT animals at day 12 plotted against differences in total protein abundance values. Quantile median values are indicated on the X-axis. Proteins that aggregated less in the mutant strains than in the WT have been grouped separately (91 proteins in daf-2 and 259 in hsf-1 mutant).
(D–F) Physico-chemical properties of proteins enriched in the insoluble fractions of daf-2 and hsf-1 mutant relative to WT animals at day 12. (D), Aggregation propensity scores (Z-scores, see Extended Experimental Procedures). ***, p-value <1.4 · 10−4; *, p-value < 0.016, Wilcoxon rank sum test. (E), Net charge. ****, p-value <4.9 · 10−11; (F), Coil content. ***, p-value <1.1 · 10−4. (G), Overall hydrophobicity ****, p-value <2.9 · 10−7. Quantile median values are indicated on both axes and standard errors are reported on the y-axis.
While there was a large overlap between the proteins identified in the insoluble fractions, the extent to which specific proteins aggregated varied greatly in a strain specific manner. Interestingly, the proteins that showed increased aggregation in the daf-2 mutant over WT are not generally more abundant at the total proteome level (Figure 5B), indicating that abundance in this case is not the main driver of aggregation. Similar findings were made in the hsf-1 mutant (Figure 5C). On the other hand, proteins that aggregated less in the daf-2 strain than in WT are also generally less abundant (Figure 5B), which would allow these proteins to maintain solubility.
Next, we compared the physico-chemical properties of the insoluble proteins. Strikingly, the proteins that aggregate most in the daf-2 mutant animals are predicted to have significantly lower aggregation-propensity Z-scores, are more charged, display more structural disorder (coil average) and are less hydrophobic compared to the proteins aggregating in WT (Figure 5D–G). These findings strongly support the hypothesis of an extrinsic rescuing mechanism of aggregation that is activated in the daf-2 mutant, modulating the intrinsic properties of proteins that typically govern aggregation. As a result, aggregation is enhanced for a set of proteins that have certain properties resembling disease-associated proteins with structural disorder (Knowles et al., 2014). By contrast, aggregation in the hsf-1 mutant correlates with intrinsic aggregation scores (Figure 5D), consistent with a degeneration mechanism arising from the premature decline of proteostasis.
Among the proteins that were strongly increased in the insoluble fraction of daf-2 mutant animals were several small HSPs (Figure 6A). These proteins contribute ~7% to total aggregate load, suggesting that they may be involved in a ‘protective aggregation response’. Small HSPs were also enriched in the insoluble fraction of hsf-1 mutant animals, although to a lesser extent, but not in the aggregates of the daf-16 mutant (Figure 6A). Besides small HSPs, 26S proteasome complexes were enriched in the insoluble fractions (Figure 6B), most strongly in the daf-2 mutant strain, but contributed only ~1% to total aggregate load.
Figure 6. Aggregation of small HSPs and Proteasome in Lifespan Mutant Worms.
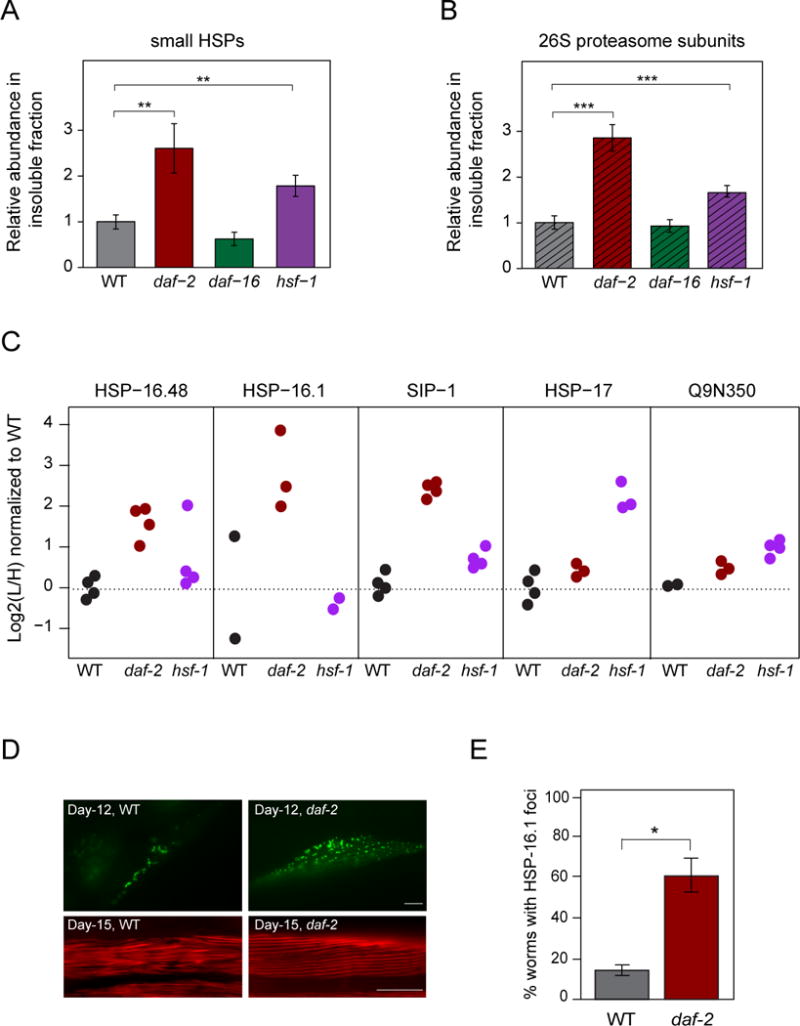
(A) Abundance of small HSPs in the insoluble fraction of daf-2, daf-16 and hsf-1 mutant relative to WT animals as determined by summed absolute LFQ values. 6–11 different small HSPs were quantified. **, p-value <0.0075 (WT vs. daf-2) and <0.0022 (WT vs. hsf-1) from Welch’s t-tests.
(B) Abundance of 26S proteasome subunits in the insoluble fraction of daf-2, daf-16 and hsf-1 mutant relative to WT animals. 19–27 subunits were quantified. ***, p-value <2.1 · 10−4 (WT vs. daf-2) and <4.6 · 10−4 (WT vs. hsf-1) from Welch’s t-tests.
(C) Enrichment of the small HSPs HSP-16.1, HSP-16.48, SIP-1, HSP-17 and Q9N350 in the insoluble fractions of day 12 WT (black circles), daf-2 mutant worms (red circles) and hsf-1 mutant worms (purple circles). Data from 2 to 4 independent experiments are shown.
(D and E) Formation of HSP-16.1 inclusions in muscle cells. (D) Representative fluorescence images of muscle cells of WT and daf-2 mutant animals expressing HSP-16.1::GFP (top). Actin was stained with rhodamine-phalloidin (bottom). Scale bar, 10μm. (E) Animals with HSP-16.1::GFP inclusions in muscle cells were quantified (20 animals per group). Averages ± SD are given in % of total. *, p-value < 0.01 from Welch’s t-test.
Interestingly, the proportion of specific small HSPs in the aggregates differed between strains. SIP-1 and HSP-16.1 made the major contribution by mass to the aggregates in the daf-2 mutant, while HSP-17 was most enriched in the aggregates of hsf-1 mutant animals (Figure 6C and Table S1F). To monitor the behavior of HSP-16.1 during aging, we used strains expressing GFP-tagged HSP-16.1 (hsp-16.1::gfp) under its endogenous promoter. HSP-16.1-GFP formed inclusions in muscle cells. This phenomenon was strongly enhanced in daf-2 mutant worms, with ~60% of animals at day 12 containing inclusions compared to ~20% in WT (Figure 6D and E). While the actin architecture of muscle cells was well preserved in daf-2 mutant animals, the muscles of day 15 WT animals showed a reduced structural integrity (Figure 6D). Indeed, the daf-2 mutant animals displayed a higher proteostasis capacity, as reflected in their ability to maintain the metastable FlucDM-GFP (Gupta et al., 2011) expressed in muscle in a functionally active state. While similar levels of total and soluble FlucDM-GFP protein were present in day 12 WT and daf-2 mutant worms, the latter contained ~4-fold more luciferase activity (Figure S6D). In contrast, a muscle specific polyQ protein construct (Q35-GFP) aggregated more extensively in daf-2 mutant worms already early in adulthood (day 2), and semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) of worm lysates revealed that the protein accumulated predominantly in higher molecular weight, SDS-resistant oligomers (Figure S6E).
Taken together these results suggest that daf-2 mutant animals drive aberrant, potentially toxic proteins into aggregates, thereby sequestering them and improving overall proteostasis. Small HSPs are likely to play a role in this process.
DISCUSSION
Age-dependent Deterioration of Proteome Balance
Organisms allocate considerable resources towards maintaining proteome composition, including the relative balance of subunits of multi-protein complexes (Li et al., 2014). Using quantitative mass spectrometric methods, we have shown here that aging in C. elegans is associated with the progressive failure to maintain protein homeostasis, resulting in extensive proteome remodeling and protein imbalances. These imbalances are largely due to changes at the level of protein translation and turnover, and give rise to the accumulation of potentially harmful, aggregation-prone species (Figure 7A and B). Our analysis of the long-lived daf-2 mutant strain revealed that the sequestration of such proteins in insoluble aggregates is a protective strategy that contributes to maintaining proteome integrity during aging.
Figure 7. Proteome Maintenance During Aging in C. elegans.
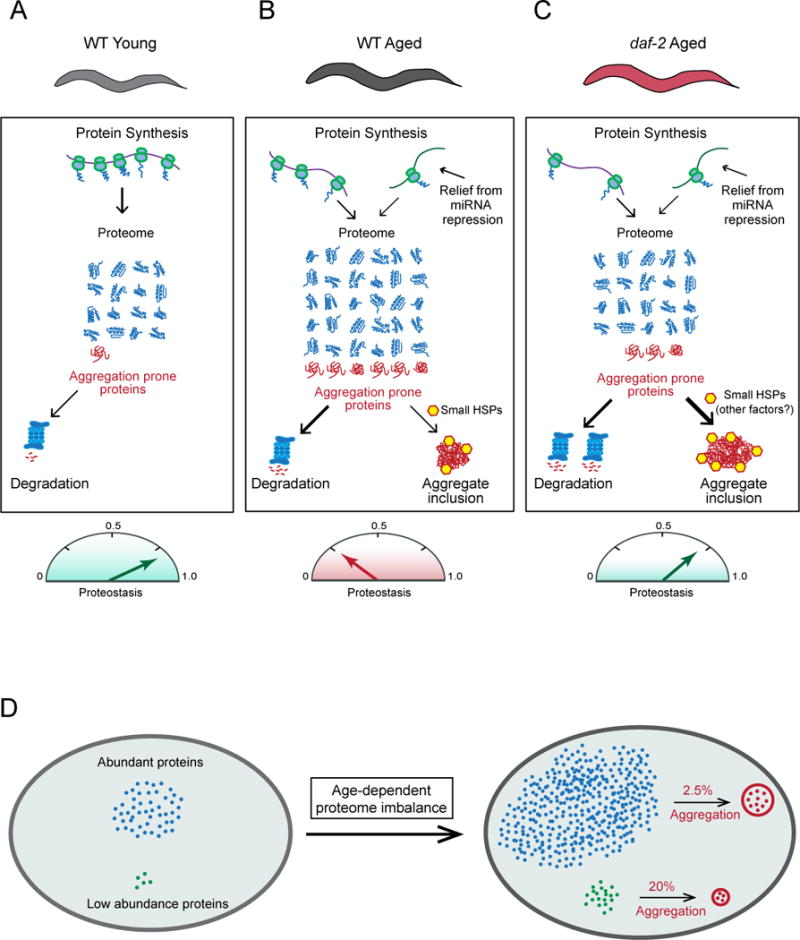
(A) The proteome of young adult WT worms is maintained in balance by the proteostasis system. Aberrant protein species, including metastable conformers and soluble aggregates (red) are efficiently cleared.
(B) In aged WT animals, numerous proteins increase in abundance and normal protein stoichiometries are lost, due in part to a relief of miRNA-mediated translational repression. The amount of aggregation-prone species exceeds clearance capacity and insoluble aggregates associated with small HSPs accumulate. Mechanisms of protective aggregate formation are partially activated. Proteostasis is strongly reduced.
(C) Proteostasis collapse is delayed in aged daf-2 mutant worms. Proteome imbalance and the soluble aggregate pool is reduced relative to age-matched WT animals, as clearance by protein degradation may be more effective and protective aggregate formation is fully activated.
(D) Protein aggregate loads increase proportionally to protein abundance. Although abundant proteins have lower aggregation propensities, they contribute more to aggregate load (see Figure 4). The age-dependent increase in expression level affects the subproteome of supersaturated proteins, which fail to maintain solubility as their levels increase and proteostasis capacity declines.
The extensive proteome remodeling during aging in C. elegans is contrary to observations in tissues of aged mice, where comparatively minor proteomic changes were detected with a similar experimental approach (Walther and Mann, 2011). Evidently mammals devote greater resources to maintaining proteome balance, resulting in a more protracted proteostasis decline. These differences correlate with different reproductive strategies in worms and mice, in which the former display a more extensive, time-restrained burst of reproduction followed by a rather rapid and massive decline of somatic functions. Future studies on a range of metazoans will be necessary to establish whether deterioration of proteome integrity during aging or proteome stability is more typical.
Changes in the Proteostasis System During Lifespan
We showed that aging in C. elegans affects multiple components of the proteostasis system, most prominently protein biosynthesis and protein degradation. A decrease in the levels of cytosolic and mitochondrial ribosomal proteins was accompanied by an overall reduction in protein synthesis. In contrast, we observed an increase in the abundance of proteasome subunits and a corresponding increase of in vitro proteasome activity. These changes may initiate as a response to the altered physiological requirements of the aging organism (Shore and Ruvkun, 2013), but ultimately may prove insufficient or even detrimental (Figure 7B). The reduction of the levels of cytosolic ribosomes was associated with a pronounced imbalance in the stoichiometry of ribosomal proteins. Thus, attenuating the translational machinery as an adaptive measure imparts the danger of dysregulation of the essential machines that ensure balance of the proteostasis network, which in turn may promote aging. In contrast, the increase in proteasomal subunits is likely to represent an attempt of the organism to remove aberrant protein species. Whether proteasome upregulation is effective in vivo is unclear, however, as a decline of proteasome function is generally thought to occur as a result of aging and protein aggregation (Hipp et al., 2014).
Protein Aggregation and Lifespan Extension
Previous studies demonstrated the formation of insoluble protein aggregates in aged worms (David et al., 2010; Reis-Rodrigues et al., 2012). Here, we performed an in-depth quantitative analysis of aggregation along the lifespan of C. elegans. We found that aggregation is a proteome-wide process that initiates mainly after day 6 of adulthood. Highly abundant proteins are generally more soluble and display lower intrinsic aggregation-propensities than less abundant ones, as previously predicted (Tartaglia et al., 2009). However, this higher solubility is still not sufficient in the end, as abundant proteins make by far the major contribution by mass to the age-dependent aggregate load. Importantly, proteome remodeling acts as a driver of aggregation by raising the level of a subset of proteins beyond a critical solubility limit (‘supersaturation’) (Ciryam et al., 2013) (Figure 7D).
While protein aggregation may be merely a consequence of declining proteostasis capacity, our results provide evidence that a protective aggregation response is also an important mechanism of the aging organism to improve proteostasis and mitigate the effects of proteome imbalance. We observed that long-lived daf-2 mutant animals accumulate increasing amounts of insoluble protein during aging, and that such accumulation correlates with their ability to maintain proteome composition more effectively (Figure 7C). Whereas the proteins that aggregate most in the short-lived hsf-1 mutant are predicted to be more aggregation-prone, the enhanced aggregation in the long-lived daf-2 mutant is much less dependent on intrinsic properties: the proteins that are most enriched in the insoluble fraction have lower aggregation scores, are less hydrophobic, more charged and contain more structural disorder, arguing for the existence of an active, proteome-wide mechanism in promoting aggregation. This conclusion is consistent with the view that soluble oligomers are the major proteotoxic species in neurodegenerative diseases and that their sequestration into insoluble aggregates reduces proteotoxicity (Arrasate et al., 2004; Cohen et al., 2006; Cohen et al., 2009). Interestingly, several highly toxic disease-associated proteins are rich in disordered structure and have low overall hydrophobicity (Knowles et al., 2014; Vendruscolo et al., 2011), properties resembling those of the proteins with enhanced aggregation in the daf-2 mutant. Indeed, a mechanism of aggregate deposition under regulation of the insulin signaling pathway has been proposed for disease-related protein species, such as toxic Aβ peptide (Cohen et al., 2006). However, that an overall ‘protective aggregate response’ operates at the proteome-scale during aging was not anticipated.
We assume that this protective aggregation response is only partially activated during normal C. elegans aging. As a result, WT worms are expected to accumulate a larger soluble pool of aberrant, potentially toxic proteins than daf-2 mutant animals, eventually exhausting the available chaperone capacity needed for protein folding and conformational maintenance, and the clearance of misfolded polypeptides (Figure 7B and C). Formation of insoluble aggregates may also be an, albeit insufficient, rescue attempt in the short-lived hsf-1 mutant strain.
Among the proteostasis components with strongly enhanced, age-dependent insolubility were multiple small HSPs, a specific class of chaperones known to associate with aggregation-prone proteins (Haslbeck et al., 2005). The small HSPs were most enriched in the insoluble fraction of daf-2 mutant worms, consistent with the view that they may play a role as ‘extrinsic’ promoters of aggregation. In support of this possibility, individual RNAi knock down of several small HSPs, including SIP-1, caused a 25 % shortening of lifespan in WT and daf-2 mutant worms (Hsu et al., 2003) and overexpression resulted in lifespan extension (Walker and Lithgow 2003). Having multi-valent binding ability for aberrant proteins, the small HSPs may act to seed and concentrate aggregate material, consistent with findings in vivo (Escusa-Toret et al., 2013; Kaganovich et al., 2008; Specht et al., 2011) and in vitro (Haslbeck et al., 2005; Jiao et al., 2005). The co-existence of multiple small HSPs suggests that different forms may vary in their structural specificity for endogenous proteins. Notably, small HSPs are also transcriptionally induced in the aging brain, while most other major chaperone components are down-regulated (Brehme et al., 2014). The association of the 26S proteasome with the aggregates may also be functionally relevant. Although aggregates can inhibit the proteolytic activity of the proteasome (Andersson et al., 2013; Hipp et al., 2014), evidence has been presented that the proteasomal ATPase chaperones may promote aggregation (Rousseau et al., 2009).
Collectively, our data suggest that aging in C. elegans is associated with a progressive loss of proteome balance, which drives the accumulation of surplus and aberrant protein species that overburden the proteostasis system. As the maintenance of protein solubility imposes stringent constraints on proteome composition, effective aggregate management appears to be critical in determining lifespan.
EXPERIMENTAL PROCEDURES
C. elegans Strains and Growth Conditions
A list of strains used in this study is provided in Extended Experimental Procedures. The Bristol strain N2 was used as wild-type. The L4 larval stage was considered as day 0. Bacterial cultures (ET505) for SILAC labeling were grown in 13C6-14N2-lysine (heavy lysine) containing M63 minimal media (Krijgsveld et al., 2003) (see Extended Experimental Procedures for details).
Sample Preparation for Total Proteome Measurements
Briefly, worms were suspended in lysis buffer (4% SDS, 0.1 M Tris/HCl pH 8.0, 1 mM EDTA), incubated at 95°C for 5 min and further treated by ultra-sonication. Typically, an aliquot of lysate containing 40 μg of protein was mixed with an equal amount of a heavy lysine labeled lysate pool (Figure S1A). Proteins were reduced, alkylated and digested with endoproteinase LysC using the FASP method (Wisniewski et al., 2009). Peptide mixtures were either analyzed without fractionation or after fractionation by isoelectric focusing, as described in Extended Experimental Procedures.
Isolation of Protein Aggregates
Worms were resuspended in lysis buffer (50 mM Tris/HCl pH 8.0, 0.5 M NaCl, 4 mM EDTA, 1% (v/v) Igepal CA630, Complete proteinase inhibitor cocktail; Roche Diagnostics), disrupted by sonication and clarified by low-speed centrifugation (1 min, 1,000 rcf). Insoluble proteins were sedimented by ultracentrifugation (500,000 rcf at 4°C, 10 min), washed twice with lysis buffer containing 0.15 M NaCl and 0.5% sodium deoxycholate, and solubilized in SDS sample buffer for 10 min at 95°C. For quantitative proteome measurements of aggregated proteins, an aliquot of pooled total lysate from heavy lysine labeled animals was added prior to ultracentrifugation. For experiments measuring aggregation propensities, unlabeled worm lysates were first fractionated and subsequently supplemented with SILAC-labeled whole cell lysate.
Mass Spectrometry and Data Analysis
Peptides were separated by reversed phase nano-HPLC and sprayed online into LTQ-Orbitrap XL, LTQ-Orbitrap Velos or Orbitrap-Elite mass spectrometers (Thermo Fisher). In each scan cycle, fragmentation spectra of the 10 most intense peptide precursors in the survey scan were acquired in the higher-energy collisional dissociation (HCD) mode. Raw data was processed using the MaxQuant software environment (Cox and Mann, 2008) and peak lists were searched with Andromeda (Cox et al., 2011) against a database containing the translation of all predicted proteins listed in Uniprot (release January 15, 2012) as well as a list of commonly observed contaminants and the NCBI protein database of E. coli strain K12. The minimal required peptide length was set to seven amino acids and both protein and peptide identifications were accepted at a false discovery rate of 1%. To identify aggregation-prone proteins that were significantly affected by aging, those proteins that were quantified in at least 3 out of 4 biological replicate experiments at day 1 and day 12 were subjected to a Welch’s t-test and filtered based on a 5% permutation-based false discovery rate threshold.
Miscellaneous
Proteasome activity assays, detection of polyQ aggregates by SDD-AGE, light microscopy and methods used for bioinformatic analyses are described in Extended Experimental Procedures.
Supplementary Material
Highlights.
Proteome profiling reveals loss of proteome balance during C. elegans aging.
Proteome imbalances alter protein stoichiometries and cause proteostasis stress.
Changes in protein abundance drive extensive protein aggregation during aging.
Sequestration of surplus proteins in chaperone-associated aggregates is protective.
Acknowledgments
We thank the Caenorhabditis Genetics Center for providing most of the strains used in this study. hsp-16.1::gfp transgenic worms were provided by Dr. Junho Lee, Seoul National University. This work was supported by the European Commission under FP7 GA n°ERC-2012-SyG_318987 – ToPAG, the Wellcome Trust (094425/Z/10/Z), the Center for Integrated Protein Science Munich (CIPSM) and the Munich Cluster for Systems Neurology (SyNergy). We would further like to thank Igor Paron and Korbinian Mayr for excellent technical assistance with MS instrumentation as well as Daniel Hornburg, Manajit Hayer-Hartl and members of the Hartl lab for critically reading this manuscript.
Footnotes
ACCESSION NUMBERS
Proteomics raw data and selected MaxQuant output files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD001364 and can be accessed using the following login data: Username: reviewer14957@ebi.ac.uk; Password: 7OsXFIP1
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and four tables.
AUTHOR CONTRIBUTIONS
F.U.H., M.M., D.M.W. and P.K. conceived the approach. D.M.W. and P.K. designed and performed the experiments with contributions from M.Z. D.M.W. and S.P. performed bioinformatics analyses with contributions from P.K. G.V. and P.C. analyzed the protein aggregation data and interpreted the results together with M.V., C.M.D. and R.I.M. F.U.H.. D.M.W. and P.K. wrote the manuscript with contributions from M.M., M.V., C.M.D. and R.I.M.
References
- Andersson V, Hanzen S, Liu B, Molin M, Nystrom T. Enhancing protein disaggregation restores proteasome activity in aged cells. Aging. 2013;5:802–812. doi: 10.18632/aging.100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci USA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Heck AJ, Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. doi: 10.1146/annurev-biochem-072909-100424. [DOI] [PubMed] [Google Scholar]
- Brehme M, Voisine C, Rolland T, Wachi S, Soper JH, Zhu Y, Orton K, Villella A, Garza D, Vidal M, Ge H, Morimoto RI. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014;9:1135–1150. doi: 10.1016/j.celrep.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C.elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina MD, Huttenhower C, Murphy CT, Troyanskaya OG. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLoS Comp Biol. 2009;5:e1000417. doi: 10.1371/journal.pcbi.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–790. doi: 10.1016/j.celrep.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, Masliah E, Dillin A. Reduced IGF-1 Signaling Delays Age-Associated Proteotoxicity in Mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. Quantitative, high-resolution proteomics for data-driven systems biology. Annu Rev Biochem. 2011;80:273–299. doi: 10.1146/annurev-biochem-061308-093216. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Prot Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread Protein Aggregation as an Inherent Part of Aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- Donnelly N, Passerini V, Durrbaum M, Stingele S, Storchova Z. HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 2014;33:2374–2387. doi: 10.15252/embj.201488648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Dillin A. Protein homeostasis and aging in neurodegeneration. J Cell Biol. 2010;190:719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Golden TR, Melov S. Microarray analysis of gene expression with age in individual nematodes. Aging Cell. 2004;3:111–124. doi: 10.1111/j.1474-9728.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- Gupta R, Kasturi P, Bracher A, Loew C, Zheng M, Villella A, Garza D, Hartl FU, Raychaudhuri S. Firefly luciferase mutants as sensors of proteome stress. Nat Methods. 2011;8:879–884. doi: 10.1038/nmeth.1697. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: the structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Hsu A-L, Murphy CT, Kenyon C. Regulation of Aging and Age-Related Disease by DAF-16 and Heat-Shock Factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging cell. 2006;5:235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Jiao W, Li P, Zhang J, Zhang H, Chang Z. Small heat-shock proteins function in the insoluble protein complex. Biochem Biophys Res Commun. 2005;335:227–231. doi: 10.1016/j.bbrc.2005.07.065. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C.elegans mutant that lives twice as long as wild-type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kirstein-Miles J, Scior A, Deuerling E, Morimoto RI. The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J. 2013;32:1451–1468. doi: 10.1038/emboj.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Krijgsveld J, Ketting RF, Mahmoudi T, Johansen J, Artal-Sanz M, Verrijzer CP, Plasterk RHA, Heck AJR. Metabolic labeling of C-elegans and D-melanogaster for quantitative proteomics. Nat Biotechnol. 2003;21:927–931. doi: 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- Kumar L, Futschik ME. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larance M, Bailly AP, Pourkarimi E, Hay RT, Buchanan G, Coulthurst S, Xirodimas DP, Gartner A, Lamond AI. Stable-isotope labeling with amino acids in nematodes. Nat Methods. 2011;8:849–851. doi: 10.1038/nmeth.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–284. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad V, Morimoto RI. Integrating the stress response: lessons for neurodegenerative diseases from C. elegans. Trends Cell Biol. 2009;19:52–61. doi: 10.1016/j.tcb.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Rodrigues P, Czerwieniec G, Peters TW, Evani US, Alavez S, Gaman EA, Vantipalli M, Mooney SD, Gibson BW, Lithgow GJ, Hughes RE. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell. 2012;11:120–127. doi: 10.1111/j.1474-9726.2011.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau E, Kojima R, Hoffner G, Djian P, Bertolotti A. Misfolding of proteins with a polyglutamine expansion is facilitated by proteasomal chaperones. J Biol Chem. 2009;284:1917–1929. doi: 10.1074/jbc.M806256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Shore DE, Ruvkun G. A cytoprotective perspective on longevity regulation. Trends Cell Biol. 2013;23:409–420. doi: 10.1016/j.tcb.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sormanni P, Aprile FA, Vendruscolo M. The CamSol Method of Rational Design of Protein Mutants with Enhanced Solubility. J Mol Biol. doi: 10.1016/j.jmb.2014.09.026. in press. [DOI] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M. Prediction of aggregation-prone regions in structured proteins. J Mol Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Tartaglia GG, Pechmann S, Dobson CM, Vendruscolo M. A Relationship between mRNA Expression Levels and Protein Solubility in E. coli. J Mol Biol. 2009;388:381–389. doi: 10.1016/j.jmb.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosten-Hawle P, Morimoto RI. Organismal proteostasis: role of cell-nonautonomous regulation and transcellular chaperone signaling. Genes Dev. 2014;28:1533–1543. doi: 10.1101/gad.241125.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M, Knowles TP, Dobson CM. Protein solubility and protein homeostasis: a generic view of protein misfolding disorders. Cold Spring Harb Perspect Biol. 2011;3:a010454. doi: 10.1101/cshperspect.a010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Walther DM, Mann M. Accurate quantification of more than 4000 mouse tissue proteins reveals minimal proteome changes during aging. Mol Cell Proteomics. 2011;10:M110.004523. doi: 10.1074/mcp.M110.004523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker NC, Habig JW, Bass BL. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 2007;13:1090–1102. doi: 10.1261/rna.542107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


