Abstract
Nrf2 is the major transcription factor that regulates many of the cytoprotective enzymes involved in the adaptive stress response. Modulation of Nrf2 could be therapeutically useful in a number of disease states. Activation can occur through either an electrophilic or non-electrophilic mechanism. To date, most of the research has focused on electrophilic Nrf2 activators, but there is increasing interest in non-electrophilic modulators of Nrf2. This Digest examines the current selection of small molecules that modulate Nrf2 through non-electrophilic mechanisms, and it highlights new opportunities for this important therapeutic target.
Graphical Abstract
Introduction
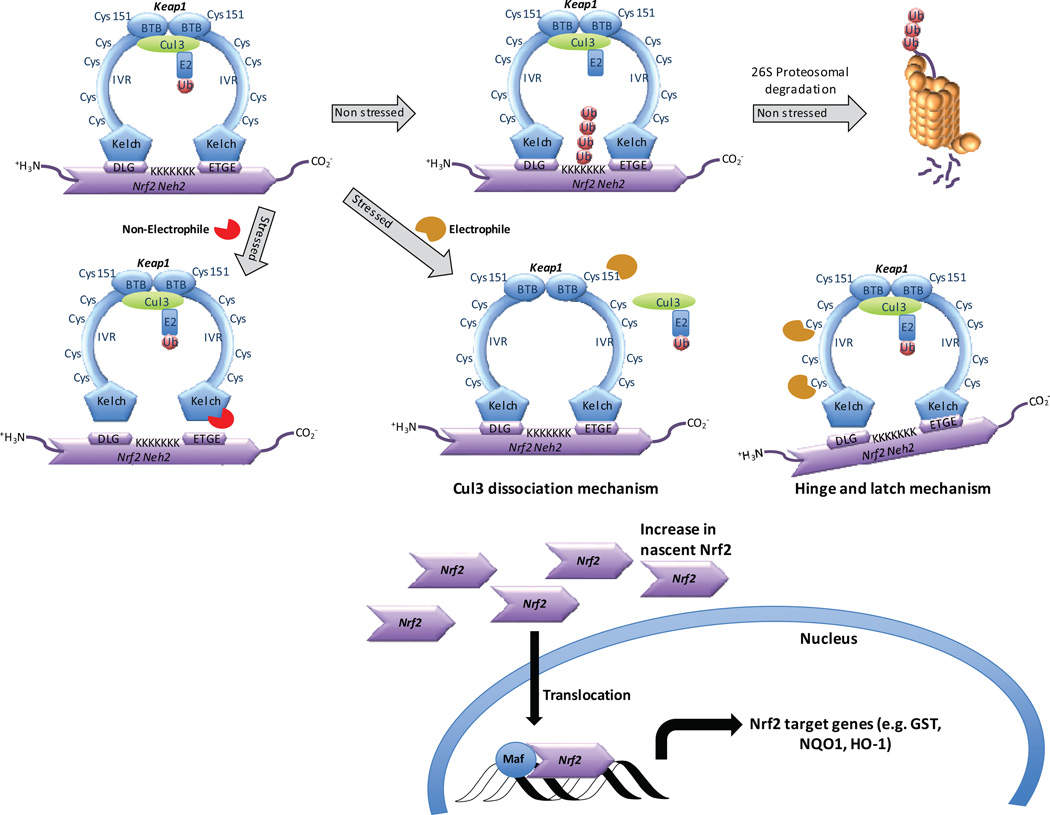
Cellular factors that sense the presence of oxidative and electrophilic stressors regulate transcription of genes encoding for cytoprotective enzymes and proteins that are crucial in maintaining cellular homeostasis.1 One such factor is Nrf2 (Nuclear factor-erythroid 2 (NF-E2)-related factor 2). Comprising seven domains (Neh1-Neh7), Nrf2 is a member of the cap ‘n’ collar family of basic leucine zipper transcription factors.2–5 Upon activation, Nrf2 is translocated to the nucleus, forms a heterodimer with small Maf proteins, and binds to an antioxidant response element (ARE) consensus sequence in the promoter region of Nrf2 target genes (See Figure 1).6 This heterodimeric complex stimulates transcription of genes such as glutathione S-transferase (GST),7 heme-oxygenase-1 (HO-1), NAD(P)H:quinone oxidoreductase (NQO), UDP-glucuronosyltransferase (UGT), aldo-keto reductase (AKR), glutamate cysteine ligase (GCL) and many other cytoprotective proteins that confront oxidative insults encountered by cells.8,9 Under normal stress-free conditions, cytosolic free Nrf2 concentration is regulated by Keap1 (Kelch-like erythroid cell-derived protein with CNC homology (ECH)-associated protein 1).10 Keap1 is a 69 kDa protein that consists of five distinct domains—namely, an N-terminal domain (amino acids 1–60), the BTB domain (amino acids 61–178), an intervening region (IVR; amino acids 179–321), the Kelch repeat domain (amino acids 322–608), and a C-terminal domain (amino acids 609–625). Keap1 ensures that free Nrf2 in the cell is at appropriately low levels11 and acts as a cytoplasmic “gatekeeper” for free Nrf2. Keap1 allows Cul3 (Cullin-dependent E3 ubiquitin ligase) to carry out ubiquitination of the Neh2 domain of Keap1-bound Nrf2, which leads to 26S proteasome-mediated degradation.12–15 The BTB region of Keap1 serves as a dimerization domain and thus effectively assists Keap1 function.13,16,17
Figure 1.
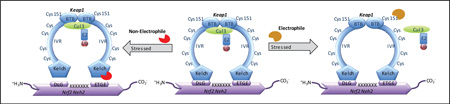
Schematic representation of modulation of the transcription factor Nrf2 by substrate adaptor protein Keap1. Under non-stressed conditions, Nrf2 is bound to Keap1 and ubiquitinated by Cul3, eventually leading to degradation through the 26S proteasome pathway. Activation of Nrf2 may occur through either a non-electrophilic mechanism that inhibits the Nrf2/Keap1 pathway, or by an electrophilic mechanism whereby cysteines of Keap1 react with electrophiles. Two models that may explain electrophilic activation of Nrf2 are shown (Cul3 dissociation or hinge-and-latch). In both non-electrophilic and electrophilic mechanisms, nascent Nrf2 production leads to translocation of Nrf2 to the nucleus, where it binds with small Maf proteins and leads to transcription of Nrf2 target genes.
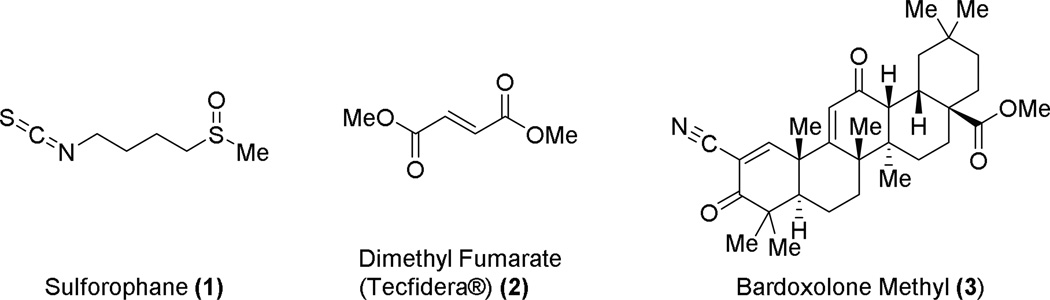
Some diseases, such as some cancers, have a misregulated Nrf2 pathway, so that repressors of Nrf2 activity may be useful; such molecules are discussed in the final portion of this Digest. The vast majority of research into Nrf2’s role in disease, however, has been in Nrf2 activation. Given the effects caused by oxidative stress to various tissues, Nrf2 activation has been suggested as a promising therapeutic strategy for a number of inflammatory and injury repair diseases,18 including diabetes, neurodegenerative diseases, atherosclerosis, and fibrotic diseases.19,20 In fact, there are a number of electrophilic small molecules that are known to activate Nrf2, and these have been reviewed in detail elsewhere.21,22 Some of the more prominent members of this class include the isothiocyanate sulforaphane (1), derived from cruciferous vegetables like broccoli; and dimethyl fumarate (2), a recently approved therapeutic for multiple sclerosis (see Chart 1). Another member of this class is the cyanoenone bardoxolone methyl (3), an agent currently in clinical trials for pulmonary arterial hypertension in the United States and for diabetic nephropathy in Japan. Bardoxolone methyl (3) is a very potent Nrf2 inducer, and its therapeutic index is greater than that of other electrophiles like sulforaphane (1) and dimethyl fumarate (2).23 These electrophilic agents clearly hold promise for the treatment of several diseases, but there is a selectivity issue with these electrophiles; they may react with a number of nucleophiles within the cell, and this pleiotropic nature may be responsible for the biological activities seen with these molecules. As an example, a bardoxolone analog was shown to react with over 500 molecular targets.24 In fact, many of the activators of Nrf2 share structural similarities with some of the pan-assay interfering compounds (PAINS) that Baell and coworkers have recently reported on.25,26 Researchers have used electrophilic Nrf2 activators to study Nrf2 and other pathways in isolation, but the therapeutic effects of these molecules are likely due to activities at a multitude of molecular targets, some of which may be interdependent. Because of the inherent lack of selectivity seen with the electrophilic activators, it is difficult to understand the consequences of pharmacological activation of Nrf2. One approach to this issue has been to develop non-electrophilic activators of Nrf227 that directly inhibit the interaction between Nrf2 and Keap1. It is important to note that the non-electrophilic activators described here bind at a completely different site (Kelch domain) than the electrophilic activators (BTB and/or IVR domains; see below and Figure 1 for depiction of approach). These compounds may serve as more selective pharmacological probes that will allow a better understanding of this important transcription factor and that may lead to new therapeutics.
Chart 1.
Representative electrophilic Nrf2 activators.
Nrf2/Keap1 interaction
Before examining the non-electrophilic mechanism of Nrf2 activation, it is worth pointing out the two models proposed for electrophilic Nrf2 activation: 1) the hinge and latch mechanism,28,29 and 2) the Cul3 dissociation mechanism31,32 (See Figure 1). Both models ultimately achieve attenuation of both ubiquitination and proteasomal degradation of Nrf2.
According to the hinge and latch theory, oxidative stress causes electrophilic modification of the cysteine residues in the IVR region of the Keap1 dimer, bringing about a slight conformational modification that disrupts binding of the DLG motif of Nrf2’s Neh2 domain while keeping the ETGE motif intact with Keap130–32 (See Figure 1). The DLG motif disruption relocates Nrf2 from its ideal position for ubiquitination, making Cul3 inaccessible to cause polyubiquitination of seven lysine residues at the C-terminal region and thus preventing degradation of Nrf2. The half-life of Nrf2 in the cytoplasm has been reported to be as short as 15 minutes,15 and Nrf2 is constantly being produced. Preventing degradation of the Nrf2 pool causes increased accumulation and nuclear translocation of Nrf2, although it is not clear whether there is a proportional increase in target gene expression with free Nrf2 concentration.
In the Cul3 dissociation mechanism, Cys151, in particular, in the BTB domain of Keap1 undergoes modification by electrophiles or oxidants33 (See Figure 1). This leads to the disruption of the Keap1-Cul3 complex which, in turn, halts ubiquitination of Nrf2; thus, Nrf2 is not degraded and remains bound to Keap1. The newly formed Nrf2 is thus available for nuclear translocation and to mediate transcription of target genes. Recent studies seem to support the Cul3 dissociation mechanism,34 although the hinge and latch mechanism has not been ruled out as a possible mechanism for Nrf2 activation.
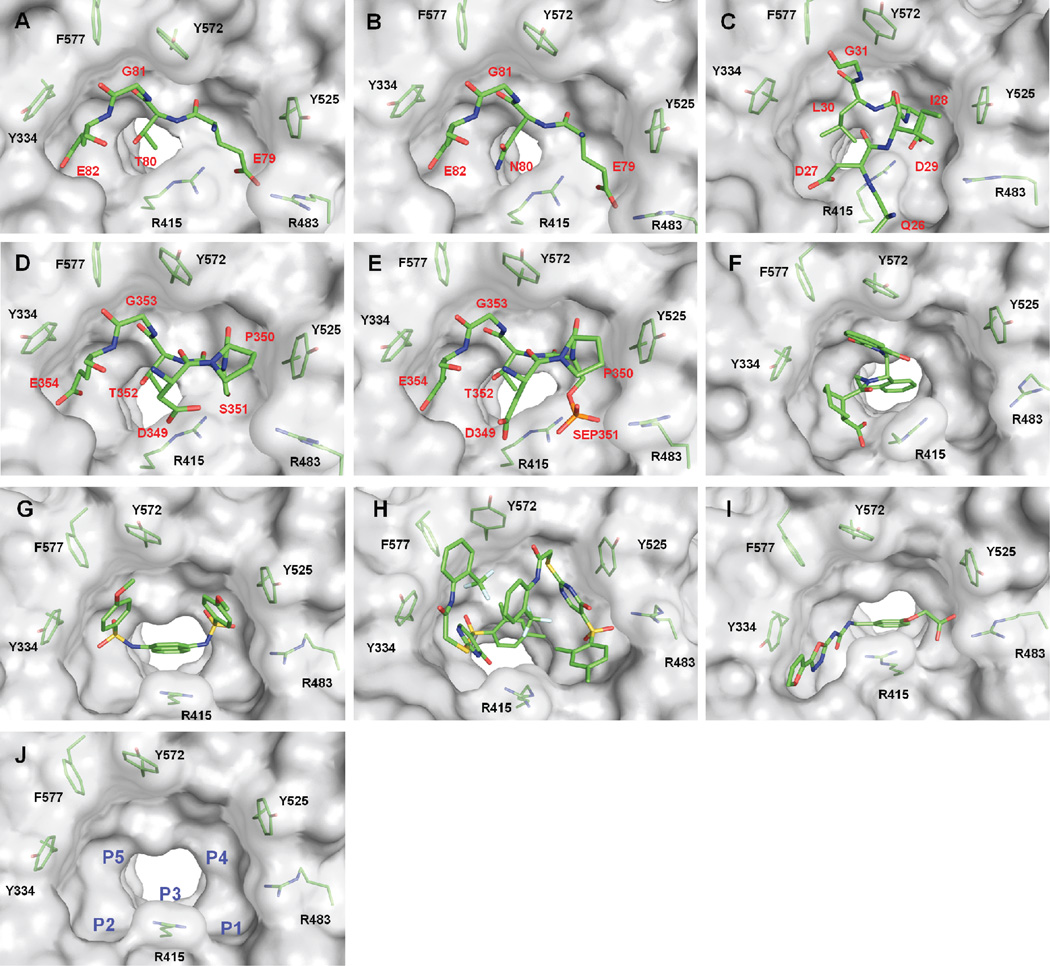
Although the reactive cysteines of Keap1 are located in the BTB and IVR domains, Keap1 binds Nrf2 and other proteins (see below) through its Kelch domain.35,36 The Kelch domain contains a pocket that recognizes and binds Nrf2 through two motifs in the Neh2 domain: the ETGE and DLG motifs.21 Jiang et al.37 have described five subpockets within the Kelch domain where the ETGE and DLG motifs bind (See Figure 2A and 2C, respectively). These subpockets are referred to by Jiang et al. as P1, P2, P3, P4, and P537 (See Figure 2J). In this Digest we use the same nomenclature. P1, formed by residues Ser 508, Phe 478, Ile 461, Arg 483, Arg 415, and Gly 462, contains positively charged Arg residues which form electrostatic interactions with substrates of Keap1. P2 comprises Ser 363, Asn 382, Asn 414, and positively charged Arg 380. As such, P1 and P2, through their electrostatic interactions with substrates, account for a significant portion of the binding energy of these peptide ligands. P3 is the central subpocket and is composed of residues with relatively small side chains: Gly 509, Ala 556, Ser 555, Ser 602, Gly 603, and Gly 573. P4 and P5 are more hydrophobic than the other three subpockets and are rich in aromatic side chains; P4 comprises Tyr 525, Gln 530 and Tyr 572, while P5 is made up of Tyr 334, Phe 577, and Tyr 572. This collection of subpockets P1-P5 binds both the ETGE and DLG motifs of Nrf2.38 The ETGE motif has a higher affinity (KD = 5 nM) than the DLG motif (KD = 1 µM).39,40 This difference in affinities stems from the motifs’ interactions with the pocket, detailed below.
Figure 2.
Crystal structures of A) the E79TGE82 motif of Nrf2 complexed with Keap1 (PDB ID 2FLU)a; B) the E79NGE82 motif of ProTα (PDB ID 2Z32)a; C) the DLG motif of Nrf2 (PDB ID 3WN7)a; D) the KIR region of p62 (3ADE)a; E) the phosphorylated KIR region of p62 (PDB ID 3WDZ)a; F) tetrahydroisoquinoline 4 (PDB ID 4L7B); G) naphthalene 7 (PDB ID 4IQK); H) thiopyrimidine 6 (PDB ID 4IN4); I) urea 10 (PDB ID 3VNH); J) binding pocket of the Kelch domain of Keap1 with the naphthalene ligand 7 removed, demonstrating the five subpockets P1-P5 of the Kelch domain (PDB ID 4IQK).
aThe remainder of the residues have been removed for clarity.
The high affinity E79TGE82 motif of Nrf2 that binds to the Keap1 Kelch domain is located on a type 1 β-turn22 (See Figure 2A). Glu 79 forms the i + 1 residue of the β-turn, and it occupies the positively charged P1 subpocket, making multiple hydrogen bonds and salt bridges with the subpocket’s polar residues. Glu 82, the i + 4 residue of the turn, occupies the relatively positively charged P2 subpocket. Similarly to Glu 79, it forms multiple hydrogen bonds and salt bridges with the polar residues. These two glutamate residues contribute significantly to the high affinity seen with the ETGE motif. The rather constrained P3 subpocket is occupied by the peptide backbone. To demonstrate the size limitations of P3, mutating Gly 81 to Ala results in a near total loss of binding.38 The hydrophobic subpockets P4 and P5 are both occupied by the sidechains of Leu 76, Glu 78, and Phe 83.37 The combination of these interactions, and Glu 79 and Glu 82 in particular, lead to the high affinity of the ETGE motif. This high affinity is crucial in the recognition and binding of Nrf2 to Keap1.
Approximately fifty amino acid residues to the N-terminal side of the ETGE motif is found the sequence, LxxQDxDLG, known as the DLG motif.41 A major difference between the two motifs is the lack of Glu in the DLG motif, which begins to account for the three orders of magnitude difference seen in binding affinities between the two motifs. Instead, the DLG motif utilizes Gln 26 to occupy subpocket P1. Gln 26 lacks the carboxylate group of Glu 79, leading to an inability to form key hydrogen bonds and salt bridges that the ETGE motif forms. P2 is occupied by Asp 27, allowing for interactions similar to the ETGE motif, as both substrates contain similar carboxylate groups. P3 still contributes more to steric control/selectivity than binding affinity, and in the DLG motif, P4 and P5 are left nearly unoccupied.38 In comparison to the ETGE motif, this motif is obviously less fit for the binding pocket; it does not contribute as many hydrogen bonds or salt bridges, and it fails to occupy two of the five subpockets. This results in a binding affinity much lower than the one observed for ETGE.
The differing binding affinities play a major role in the mechanism of activation of Nrf2. According to the hinge and latch mechanism,29,42 the DLG motif, with its lower binding affinity, is released upon introduction of electrophilic stressors (see above). When DLG is bound, it locks Nrf2 into a favorable conformation for polyubiquitination and subsequent degradation; however, when DLG is released, this “latch” is undone. ETGE, on the other hand, retains its occupancy of the binding pocket, due to its higher binding affinity. This leads the ETGE motif to act as the “hinge” that swings Nrf2 out of the optimal conformation for polyubiquitination.
Molecules that disrupt the Keap1/Nrf2 protein-protein interaction show great promise as Nrf2 activators. Protein-protein interactions are often referred to as “intractable” or “undruggable” drug targets.43 While there are now several examples of clinically used agents that target protein-protein interactions, there are certainly reasons to refer to some protein-protein interactions as “undruggable”: they often occur over large areas on the surfaces of proteins, and they can be rather featureless.44 The Keap1/Nrf2 interaction, however, is a rather unique proteinprotein interaction in that it relies on a well-defined binding pocket on the Kelch domain that utilizes both polar and hydrophobic interactions for anchoring small molecules and peptides. The size of the binding site41 (420 Å2) is more typical of a small molecule binding site on a receptor or enzyme (300–1,000 Å2),45 than of a protein-protein interaction (1,500–3,000 Å2).46 Given these characteristics, it is perhaps unsurprising that substantial progress has been made on inhibiting the Keap1/Nrf2 interaction with small molecules. In this digest, we will review small molecule Nrf2/Keap1 inhibitors, followed by peptide/protein inhibitors. Lastly, we will review molecules that seem to repress Nrf2’s activity.
Non-electrophilic Nrf2 activators
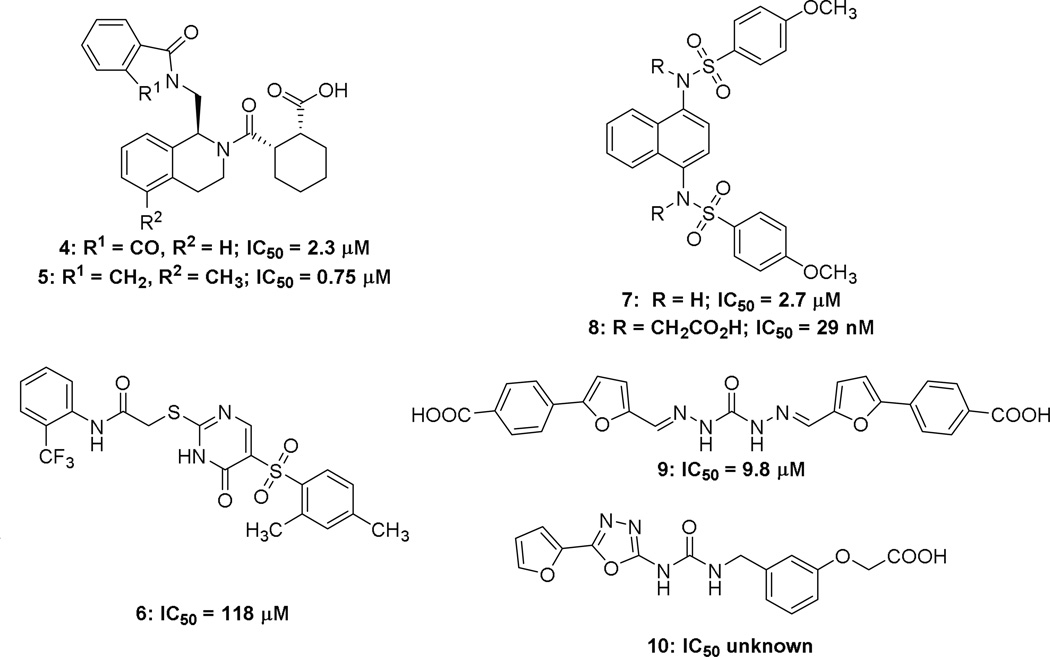
Several high throughput screens have yielded a number of hits as direct inhibitors of Nrf2. These hits include, tetrahydroisoquinoline (THIQ) 4 (IC50 = 2.3 µM),47,48 thiopyrimidine 6 (IC50 = 118 µM),49 naphthalene 7 (IC50 = 2.7 µM),49 carbazone 9 (IC50 = 9.8 µM),50 and urea 10 (affinity unknown).51 Some of these molecules have been further elaborated, yielding structure activity relationships and crystal structures from which more information can be gathered about non-electrophilic Nrf2 activators.
The tetrahydroisoquinoline 4, discovered by Hu et al.47 in an HTS of the MLPCN library was cocrystallized48 with Keap1, and this crystal structure drove an SAR study of the hit scaffold (See Figure 2F).41,47,52 The crystal structure showed that the aromatic ring of THIQ settles at the top of the binding pocket with potential space to incorporate new interactions with the pocket: the cyclohexanecarboxylic acid moiety makes interactions with Tyr 572, Ser 602, Arg 415, and Asn 414, and the phthalimide forms hydrogen bonds and a π-stacking complex with Ser 602 and Tyr 572, respectively. The parent compound possesses moderate affinity for Keap1, IC50 = 2.3 µM with similar potencies seen in reporter gene assays, but linked to the THIQ core is a phthalimide moiety, which was removed by Jnoff et al.,48 presumably because of its poor physicochemical and pharmaceutical properties. To this end, it was found that isoxindole could be used to replace the phthalimide while retaining high levels of potency, even though it sacrificed a hydrogen bond acceptor from the phthalimide. Further exploration around the molecule found that the cyclohexane ring can be replaced with a cyclopentane, while maintaining similar potency. Additionally, the carboxylic acid cannot be replaced by a carboxamide or cyano group, but it can be replaced by a tetrazole with a small decrease in potency. Lastly, the possibility of extending the molecule into the free space near the THIQ phenyl ring was explored. The endeavor yielded promising results with methylation of the 5-position increasing the potency to yield the most potent molecule in this series 5, at 0.75 µM.48
A separate HTS by Marcotte et al.49 using the Evotec Lead Discovery library yielded two hits: thiopyrimidine 6 and naphthalene 7 with EC50 values of 118 µM and 2.7 µM, respectively.49 Though there are crystal structures of both hits bound to Keap1 (See Figure 2H and 2G) no further research has been done on 6. Interestingly, both crystallographic and biophysical studies suggested that two molecules of 6 bind to the Kelch domain. It would be interesting to know whether linking these two molecules might serve to increase the affinity of the conjoined molecule; however, the large size of such a molecule might preclude its usefulness. More generally, it raises the possibility of a fragment-based approach53 to discovering Keap1 binders, something that has thus far not been undertaken.
In contrast to 6, several studies have been performed on 7. In one study, structure-based virtual screening was utilized to find compounds similar to 7.27 From these similar compounds, analogues were found, purchased, and screened using an established fluorescence polarization assay. An issue with the study is that the compounds and structural classes reported may be pan assay interference compounds (PAINS),25,26 and their activity may not be confined to Nrf2 activation. Of the scaffolds analyzed, none were able to produce a noticeably more potent compound than the parent compound 7, IC50 = 2.7 µM. A different study by Jiang et al.37 used molecular modeling to analyze Keap1’s key interactions with its natural ligand, the ETGE motif, to design a small molecule that utilized as much of the binding pocket as possible.37 Analysis of the crystal structure of 7 showed that it effectively occupied subpockets P3, P4, and P5 only (See Figure 2G). The naphthalene core stabilizes the binding conformation by occupying P3, and the para-methoxy benzene rings are capable of occupying the hydrophobic P4 and P5. The molecule made slight interactions with P1 and P2 through hydrogen bonding with the sulfonamide hydrogens, but the interactions do not penetrate deeply into the subpockets. Thus, Jiang et al. designed a variant that formed more interactions with the P1 and P2 pockets in hopes of increasing the potency. The study culminated in the placement of two acetate functionalities on the scaffold, yielding 8 and decreasing the compound’s IC50 ca. 100-fold to 29 nM. Although there are currently no reported crystal structures of this compound with Keap1, the increased potency of bis-acid 8 is believed to stem from its acetate moieties forming ionic interactions with the arginines of P1 and P2 in a similar manner to the i + 1 and i + 4 glutamates of the ETGE motif. An issue with this compound is that the multiple carboxylic acids may hinder the cellular permeability of this molecule, although Western blot studies showed that the compound was able to activate Nrf2 target genes after 24 hours. It is not clear, however, how the gene expression profile of this compound compares to other known Nrf2 activators.
The carbazone 9 was found via structure-based screening of the Specs database. To create a focused library out of the initial 251,774 compounds found in the database, certain compound traits were selected based on the crystal structures of the Nrf2 ETGE and DLG motifs. Both motifs display acidic residues that form salt bridges with Keap1. Thus, the selection criteria were set to include only molecules calculated to have a formal charge ≤ −1; furthermore, a pharmacophore was generated from two separate crystal structures of the ETGE motif. The pharmacophore consisted of one hydrogen bond donor, two hydrogen bond acceptors, and three negative ionizable centers. Screening the pharmacophore through the focused library, while allowing omission of two features to account for simplicity of small molecules, produced carbazone 9 with a moderately high potency, as determined by a fluorescence polarization assay (IC50 = 9.8 µM). The presence of multiple carboxylic acids and a carbazone moiety hinders the compound’s cellular activity, most likely due to poor cell permeability.
The urea 10 has a reported crystal structure with no biological data associated with it (See Figure 2I).51 It does seem to bind to Keap1 in the Nrf2 binding pocket and is therefore very likely an inhibitor of the Keap1/Nrf2 complex.
All of the hits that have been discovered thus far have been found through screening approaches. There are a number of β-turn mimetics in the literature that have been developed as generalizable inhibitors of other targets,54 but, to date, no designed inhibitors have been reported in the literature for the Keap1/Nrf2 interaction. Part of the reason for this may be because many of these peptidomimetics have been designed to mimic the i + 1 through i + 3 residues of a β-turn, and the ETGE motif comprises the i + 1 through i + 4 residues,41 so that it is unclear how to include a mimic of the important Glu 82 residue.
Peptide- and protein-based Nrf2 activators
Prothymosin α (ProTα) is a small acidic nuclear oncoprotein initially identified as a precursor of the thymus hormone thymosin α1. It is implicated in cell proliferation and protection against apoptosis and has been studied as such extensively; however, recently it was found to also bind to Keap1 competitively with Nrf2 in vivo. 55 Karapetian et al.55 proposed that ProTα associates with Keap1 through its β-propeller Kelch domain. This proposition implied that ProTα and Nrf2 may share overlapping binding sites. As these interactions were not clear, X-ray crystallographic analysis studies were performed on ProTα to ascertain precisely how ProTα complexes with Keap1.56
The results of the structural analysis on a crystal structure of ProTα complexed with Keap1 (See Figure 2B) confirmed that not only does ProTα interact with Keap1, but it utilizes the same binding pocket as Nrf2.56 Within the pocket, an ENGE motif from ProTα binds in a manner similar to that of the Nrf2 ETGE fragment. A superposition of the complexes of Keap1/ProTα and Keap1/Nrf2 gave an root mean squared deviation value of 0.19 Å, suggesting that the ligands interact with the Kelch domain similarly. Comparison of the complexes shows that the ETGE in Nrf2 is roughly equivalent to the ENGE in ProT_; the substitution of Asn for Thr does not hinder interactions with Keap1 as the residues in both peptides act to stabilize the backbone conformations of the proteins instead of contributing to binding. The structural analysis coupled with the biological findings of Nrf2 activation demonstrates that ProTα competes either with the ETGE motif and/or the DLG motif of Nrf2.
The protein p62 is a selective substrate for macroautophagy and regulates the formation of protein aggregates in autophagy-deficient mice and flies. Previous genetic studies on an autophagy-essential protein (Atg7) in mice showed that loss of autophagy caused accumulation of p62 along with induction of antioxidant proteins such as NQO1 and GST, both regulated by Nrf2.57 Mouse livers deficient in the autophagy protein show an accumulation of Nrf2 in the nucleus coupled with a loss of p62.57 These findings led the Komatsu group to postulate that Nrf2 could be activated in a p62-dependent manner.57 Subsequent investigation led to the identification of Keap1 as a p62-interacting protein; furthermore, p62 binds in the Kelch domain, as is the case for Nrf2.56 The findings imply that p62 activation of Nrf2 does not follow an electrophilic pathway, as the key cysteine residues modified in the electrophilic pathway are found in the IVR region of Keap1.
The crystal structure of Keap1 in complex with p62 (See Figure 2D) not only confirms the implication that p62 activates Nrf2 through a non-electrophilic pathway, but also that the mechanism is likely one of competitive inhibition, as p62 shares the same binding pocket as Nrf2. P62 contains a conserved 13-mer sequence, referred to by the Komatsu group as the Keap1-interacting region (KIR).57 In this region there is a sequence, DPSTGE, that forms the key interactions with the binding pocket. Hydrogen bond analysis of the crystal structure shows overlap in interactions between the Keap1/ETGE complex and the Keap1/p62 complex. The KIR of p62 forms hydrogen bonds with Tyr 334, Ser 363, Arg 380, Asn 382, Arg 415, Gln 530, Ser 555, and Ser 602, all of which are involved in Keap1 recognition of the ETGE motif.38,57 p62- KIR interacts with the Keap1 pocket in a manner similar to the previously mentioned peptides. The STGE forms almost identical interactions as the ETGE motif. The i + 4 Glu also forms a salt bridge with the Arg of P2, the i + 3 Gly interacts with P3, and the i − 1 Asp interacts with the Arg of P1, mirroring the interactions of the ETGE motif very closely. The importance of these interactions was verified by point mutations which showed loss of Keap1 binding when any of these residues were lost. Further evidence that the KIR interacts similarly as the previous peptides can be found upon phosphorylation of the i + 2 Ser. The phosphorylated Ser forms stronger interactions with the Keap1 pocket due to its interacting with Arg 483 and Ser 508 in P1, though with fewer hydrogen bonds than the ETGE motif (See Figure 2E).58
To confirm that p62 can competitively inhibit the Keap1/Nrf2 interaction, its affinity for Keap1 must be comparable to either the ETGE and/or the DLG motif, KD= 5 nM and 1 µM, respectively. The binding energy, determined by isothermal titration calorimetry (ITC), showed that the affinity of p62 for Keap1 was similar to that of the DLG motif, KD = 2 µM.57 Upon phosphorylation, the increased interactions result in an increase in affinity, KD = 118 nM.58 This result indicates that overproduction of p62 will counteract interaction between Keap1 and the DLG motif, but not between Keap1 and the ETGE motif. The authors posit that this, however, is all that is needed to activate Nrf2, as removal of the “latch” DLG motif will cause Nrf2 to swing out of the optimal conformation for polyubiquitination. The biological data combined with the crystal structure analysis confirm that p62 competitively inhibits the Keap1/Nrf2 interaction.
There has been a presumption within the literature that non-electrophilic Nrf2 activators should be more selective than electrophilic activators, but it is worth reiterating that other proteins (like p62 and prothymosin-α) bind to the Kelch domain, as well. Blocking the Kelch domain may activate Nrf2, but it may also disrupt the interaction between Keap1 and these other proteins. Thus far, the consequences of blocking these other interactions have not been thoroughly examined, and it is unclear whether they might be beneficial or detrimental within a given disease state. Keeping these other Keap1 binders in mind will become increasingly important as we learn more about the other partners that interact with Keap1.
Several ETGE-containing Nrf2 peptides have been reported to inhibit the Keap1/Nrf2 complex. These small peptides are truncated versions of the Nrf2 Neh2 domain, often used for competition fluorescence polarization assays, and contain the ETGE binding motif along with several amino acid residues on either side of the motif.39 The 16mer Nrf2 peptide 69AFFAQLQLDEETGEFL84 is the most potent of the peptides (KD = 28.7 nM); however, because the size of the labeled peptide is directly proportional to the rotational correlation time,59 the larger peptide does not provide as large a dynamic range for fluorescence polarization assays as a smaller peptide would. The 14mer peptide 74LQLDEETGEFLPIQ87 experiences a drop in potency (KD = 61.9 nM).39 The 10mer peptide 76LDEETGEFLP85, was less effective than the 16mer at inhibiting the complex (KD = 30.1 nM) but still managed to displace Nrf2 from the complex.39 Upon further deletion of residues from the 16mer peptide to increase its dynamic range, it was found that the 9mer 76LDEETGEFL84 (KD = 352 nM) is the minimal Nrf2 peptide sequence required for the complex to be inhibited, as the 8mer peptide no longer bound to Keap1 with any affinity (KD = ~1000 nM).22,39 One can increase the affinity of the 9mer peptide to 25.6 nM by removing the positive charge at the N-terminus of the peptide via acetylation, making the 9mer peptide the truncated peptide of choice when performing fluorescence polarization assays with Keap1, as it is extremely potent while also possessing a high dynamic range.22,39 Hancock, et al., carried out a phage display library screening to find 7mers, some of which have IC50 values below 1 µM.60 They have developed a fluorescence polarization assay based on these peptides, 60, and they have also developed a FRET-based assay to interrogate the interaction between Keap1 and Nrf2.61 Steel et al. have utilized truncated Nrf2 peptides, combined with a cell-penetrating-peptide, as an activator of Nrf2.62 In an alternative approach, Hancock, et al. have appended benzoyl or stearoyl groups onto their previously discovered peptides to develop cell-permeable Nrf2 peptides that regulate Nrf2 target genes;63 additionally, they have seen activity with a peptide that reduced the overall negative charge by converting acidic amino acids to their amide counterparts.63 In both reports,62,63 the concentrations needed to induce Nrf2’s activity in cells is much higher than the in vitro concentrations needed to inhibit the Keap1/Nrf2 interaction, but the results suggest that it may be possible to develop potent and cell-permeable peptidic Nrf2 activators. It is noteworthy that there is now at least one commercially available fluorescence polarization kit for assaying the Nrf2/Keap1 interaction;64 there are also other commercially available kits for measuring regulation of Nrf2 target genes65 and nuclear localization.66
An alternate route of Nrf2 activation to be posited would be to target the Cul3 ubiquitin ligase responsible for polyubiquitinating Nrf2 along with Keap1.67 It has been reported that there are sites on Cul3 that one could possibly target to disrupt the Keap1/Cul3 complex.67 Thus, through a mechanism similar to the Cul3 dissociation mechanism, Nrf2 would become activated. Research into this route of activation is constrained, however, by inability to identify these druggable targets. More research into this possible route of activation is warranted to test the plausibility of the theory.
Nrf2 repressors
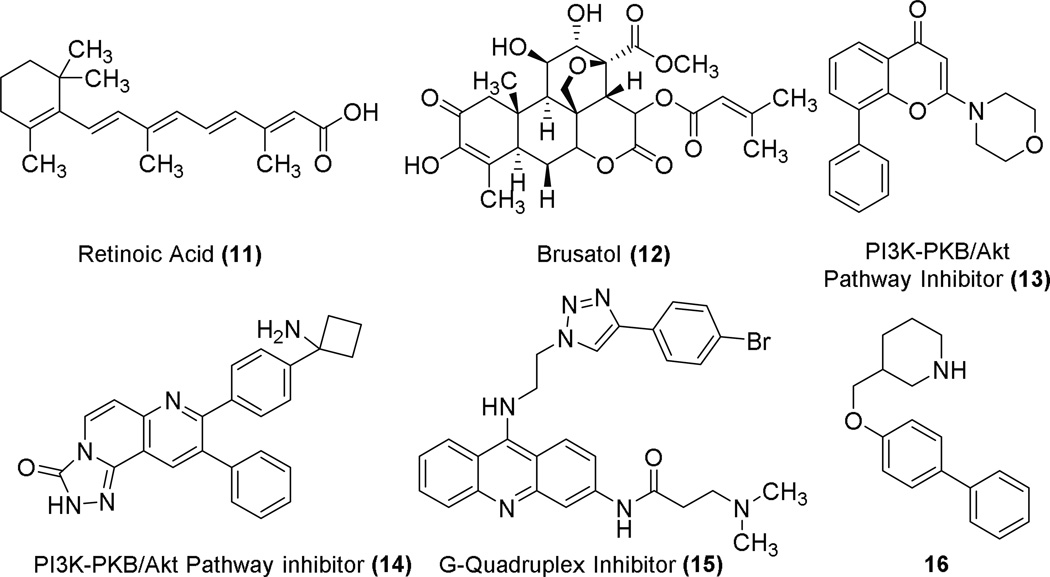
The physiological benefits of Nrf2 activation are well documented; however, in the case of existing cancers, Nrf2 overexpression leads to enhanced chemoresistance.68 Additionally, there are mutants of Keap1 that lead to constitutive activation of Nrf2.40 As such, research into repression of Nrf2 has begun and several different routes are under investigation. Because it is more straightforward to activate Nrf2 than to repress it, possible targets for Nrf2 repression have focused on the Nrf2 pathway rather than a particular interaction of Nrf2. This is unlike activators that block the Nrf2/Keap1 interaction. The Neh7 domain of Nrf2 binds to retinoic X receptor alpha, causing downregulation of Nrf2 and sensitization of chemoresistant cells to cancer therapeutics, and this effect is enhanced in the presence of all-trans retinoic acid 11.2 A small molecule, brusatol 12 (see Chart 3), has been found to sensitize chemoresistant cells to the therapeutic cisplatin through enhanced ubiquitination and degradation of Nrf2.69 In addition, PI3K inhibitors 13 and 14 also repress Nrf2’s activity in cancer cells.70,71 Along a similar route of targeting Nrf2 to achieve inhibition, Waller et al. recently reported identification of a putative G-quadruplex-forming sequence in the promoter region of Nrf2 which, if targeted, could halt production of Nrf2 and inhibit its function.72 G-quadruplex ligand 1573 may bind to this possible Nrf2 G-quadruplex, but its selectivity is unknown. A patent from Korea Institute of Radiological and Medical Sciences describes biphenyl 16 that seems to inhibit Nrf2 through an unknown mechanism.74
Chart 3.
Chemical structures of Nrf2 pathway repressors.
In conclusion, there are a number of Nrf2 modulators that have been described in the literature. Although they may eventually lead to drug candidates, non-electrophilic activators of Nrf2 may immediately allow for a better understanding of Nrf2 pharmacology, and there are a number of these compounds in the literature now that are known to potently activate Nrf2. These non-electrophilic compounds show activity in vitro and in cell culture, but they have not yet been shown to demonstrate efficacy in animal models. Most of the compounds that have been disclosed thus far are not optimized, and the structure-activity relationships carried out in this class have been limited. More expansive SARs are needed to prepare more druglike compounds that can be used in in vivo efficacy models and, eventually, in the clinic. As these molecules advance, it will become important to demonstrate that activation of Nrf2 indeed occurs through inhibition of the Keap1/Nrf2 interaction and not through metabolic activation to reactive electrophiles. Much less studied are molecules that repress Nrf2’s transcriptional activity, but these molecules may be useful, particularly with respect to cancer.
Supplementary Material
Chart 2.
Structures of small molecule inhibitors of Keap1/Nrf2 complex.
Acknowledgments
The authors thank the University of Illinois at Chicago College of Pharmacy and the University of Illinois Cancer Center for their support of this work. T.E.S. was supported by a T32 training grant funded by the Office of the Director, National Institute of Health National Center for Complementary and Integrative Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Mol. Cell. Biol. 2003;23:8786. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Liu K, Geng M, Gao P, Wu X, Hai Y, Li Y, Li Y, Luo L, Hayes JD, Wang XJ, Tang X. Cancer Res. 2013;73:3097. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 3.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. Genes Cells. 2001;6:857. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 4.Ohtsubo T, Kamada S, Mikami T, Murakami H, Tsujimoto Y. Cell Death Differentiation. 1999;6:865. doi: 10.1038/sj.cdd.4400566. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Hosoya T, Maruyama A, Nishikawa K, Maher JM, Ohta T, Motohashi H, Fukamizu A, Shibahara S, Itoh K, Yamamoto M. Biochem. J. 2007;404:459. doi: 10.1042/BJ20061611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nioi P, Nguyen T, Sherratt PJ, Pickett CB. Mol. Cell. Biol. 2005;25:10895. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes JD, Chanas SA, Henderson CJ, McMahon M, Sun C, Moffat GJ, Wolf CR, Yamamoto M. Biochem. Soc. Trans. 2000;28:33. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 8.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. Biochem. Biophys. Res. Commun. 1997;236:313. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 9.Morimitsu Y, Nakagawa Y, Hayashi K, Fujii H, Kumagai T, Nakamura Y, Osawa T, Horio F, Itoh K, Iida K, Yamamoto M, Uchida K. J. Biol. Chem. 2002;277:3456. doi: 10.1074/jbc.M110244200. [DOI] [PubMed] [Google Scholar]
- 10.McMahon M, Itoh K, Yamamoto M, Hayes JD. J. Biol. Chem. 2003;278:21592. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 11.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Genes Dev. 1999;13:76. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Mol. Cell. Biol. 2004;24:7130. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa M, Xiong Y. Mol. Cell. Biol. 2005;25:162. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canning P, Cooper CD, Krojer T, Murray JW, Pike AC, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Ayinampudi V, Marsden BD, Gileadi O, Knapp S, von Delft F, Bullock AN. J. Biol. Chem. 2013;288:7803. doi: 10.1074/jbc.M112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. J. Biol. Chem. 2003;278:4536. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 16.Zipper LM, Mulcahy RT. J. Biol. Chem. 2002;277:36544. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 17.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. J. Biol. Chem. 2006;281:24756. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 18.Crunkhorn S. Nat. Rev. Drug Disc. 2012;11:96. doi: 10.1038/nrd3655. [DOI] [PubMed] [Google Scholar]
- 19.Fox RJ, Miller DH, Phillips JT, Hutchinson M, Havrdova E, Kita M, Yang M, Raghupathi K, Novas M, Sweetser MT, Viglietta V, Dawson KT. N. Eng. J. Med. 2012;367:1087. doi: 10.1517/14656566.2013.826190. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi N, Ishii Y, Morishima Y, Yageta Y, Haraguchi N, Itoh K, Yamamoto M, Hizawa N. Respir. Res. 2010;11:31. doi: 10.1186/1465-9921-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson AJ, Kerns JK, Callahan JF, Moody CJ. J. Med. Chem. 2013;56:7463. doi: 10.1021/jm400224q. [DOI] [PubMed] [Google Scholar]
- 22.Magesh S, Chen Y, Hu L. Med. Res. Rev. 2012;32:687. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Copple IM, Shelton LM, Walsh J, Kratschmar DV, Lister A, Odermatt A, Goldring CE, Dinkova-Kostova AT, Honda T, Park BK. Toxicol. Sci. 2014;140:462. doi: 10.1093/toxsci/kfu080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yore MM, Kettenbach AN, Sporn MB, Gerber SA, Liby KT. PloS one. 2011;6:e22862. doi: 10.1371/journal.pone.0022862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baell J, Walters MA. Nature. 2014;513:481. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 26.Baell JB, Holloway GA. J. Med. Chem. 2010;53:2719. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang C, Miao Z, Sheng C, Zhang W. Curr. Med. Chem. 2014 doi: 10.2174/0929867321666140217104648. [DOI] [PubMed] [Google Scholar]
- 28.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Biol. Chem. 2006;387:1311. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 29.Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M. Mol. Cell. Biol. 2007;27:7511. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11908. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukutomi T, Takagi K, Mizushima T, Ohuchi N, Yamamoto M. Mol. Cell. Biol. 2014;34:832. doi: 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baird L, Lleres D, Swift S, Dinkova-Kostova AT. Proc. Natl. Acad. Sci. U.S.A. 2013;110:15259. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. Mol. Cell. Biol. 2004;24:8477. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleasby A, Yon J, Day PJ, Richardson C, Tickle IJ, Williams PA, Callahan JF, Carr R, Concha N, Kerns JK, Qi H, Sweitzer T, Ward P, Davies TG. PLoS One. 2014;9:e98896. doi: 10.1371/journal.pone.0098896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. Nature. 2003;425:316. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 36.Zhang DD. Antiox. Redox Signal. 2010;13:1623. doi: 10.1089/ars.2010.3301. [DOI] [PubMed] [Google Scholar]
- 37.Jiang ZY, Lu MC, Xu LL, Yang TT, Xi MY, Xu XL, Guo XK, Zhang XJ, You QD, Sun HP. J. Med. Chem. 2014;57:2736. doi: 10.1021/jm5000529. [DOI] [PubMed] [Google Scholar]
- 38.Jiang ZY, Xu LL, Lu MC, Pan Y, Huang HZ, Zhang XJ, Sun HP, You QD. J.Comput. Aided Mol. Des. 2014;28:1233. doi: 10.1007/s10822-014-9799-y. [DOI] [PubMed] [Google Scholar]
- 39.Inoyama D, Chen Y, Huang X, Beamer LJ, Kong AN, Hu L. J. Biomol. Screen. 2012;17:435. doi: 10.1177/1087057111430124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Mol. Cell. 2006;21:689. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. EMBO J. 2006;25:3605. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keum YS, Choi BY. Molecules. 2014;19:10074. doi: 10.3390/molecules190710074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.London N, Raveh B, Schueler-Furman O. Curr. Opin. Chem. Biol. 2013;17:952. doi: 10.1016/j.cbpa.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Wells JA, McClendon CL. Nature. 2007;450:1001. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 45.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, Salzberg AC, Huang ES. Nat. Biotechnol. 2007;25:71. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 46.Jones S, Thornton JM. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu L, Magesh S, Chen L, Wang L, Lewis TA, Chen Y, Khodier C, Inoyama D, Beamer LJ, Emge TJ, Shen J, Kerrigan JE, Kong AN, Dandapani S, Palmer M, Schreiber SL, Munoz B. Bioorg. Med. Chem. Lett. 2013;23:3039. doi: 10.1016/j.bmcl.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jnoff E, Albrecht C, Barker JJ, Barker O, Beaumont E, Bromidge S, Brookfield F, Brooks M, Bubert C, Ceska T, Corden V, Dawson G, Duclos S, Fryatt T, Genicot C, Jigorel E, Kwong J, Maghames R, Mushi I, Pike R, Sands ZA, Smith MA, Stimson CC, Courade JP. ChemMedChem. 2014;9:699. doi: 10.1002/cmdc.201300525. [DOI] [PubMed] [Google Scholar]
- 49.Marcotte D, Zeng W, Hus JC, McKenzie A, Hession C, Jin P, Bergeron C, Lugovskoy A, Enyedy I, Cuervo H, Wang D, Atmanene C, Roecklin D, Vecchi M, Vivat V, Kraemer J, Winkler D, Hong V, Chao J, Lukashev M, Silvian L. Bioorg. Med. Chem. 2013;21:4011. doi: 10.1016/j.bmc.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 50.Sun H-P, Jiang Z-Y, Zhang M-Y, Lu M-C, Yang T-T, Pan Y, Huang H-Z, Zhang X-J, You Q-d. MedChemComm. 2014;5:93. [Google Scholar]
- 51.Sato M, Aoki T, Inoue H, Tanaka T, Kunishima N. Japan: Toray Industries, Inc.: 2013. p. 65. [Google Scholar]
- 52.Beamer LJ, Li X, Bottoms CA, Hannink M. Acta Crystallogr. Sect. D. 2005;61:1335. doi: 10.1107/S0907444905022626. [DOI] [PubMed] [Google Scholar]
- 53.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Science. 1996;274:1531. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 54.Kee KS, Jois SD. Curr. Pharm. Des. 2003;9:1209. doi: 10.2174/1381612033454900. [DOI] [PubMed] [Google Scholar]
- 55.Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, Sukhacheva EA, Melnikov SV, Schneider U, Wanker EE, Vartapetian AB. Mol. Cell. Biol. 2005;25:1089. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Padmanabhan B, Nakamura Y, Yokoyama S. Acta Crystallogr. Sect. F. 2008;64:233. doi: 10.1107/S1744309108004995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura S, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, Yamamoto M. Nat. Cell Biol. 2010;12:213. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 58.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, Hoshii T, Hirao A, Takagi K, Mizushima T, Motohashi H, Lee MS, Yoshimori T, Tanaka K, Yamamoto M, Komatsu M. Mol. Cell. 2013;51:618. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Wu G. Assay Development : Fundamentals and Practices. Hoboken, NJ: Wiley; 2010. Chapter 2. [Google Scholar]
- 60.Hancock R, Bertrand HC, Tsujita T, Naz S, El-Bakry A, Laoruchupong J, Hayes JD, Wells G. Free Radical Biol. Med. 2012;52:444. doi: 10.1016/j.freeradbiomed.2011.10.486. [DOI] [PubMed] [Google Scholar]
- 61.Schaap M, Hancock R, Wilderspin A, Wells G. Protein Sci. 2013;22:1812. doi: 10.1002/pro.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steel R, Cowan J, Payerne E, O'Connell MA, Searcey M. ACS Med. Chem. Lett. 2012;3:407. doi: 10.1021/ml300041g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hancock R, Schaap M, Pfister H, Wells G. Org. Biomol. Chem. 2013;11:3553. doi: 10.1039/c3ob40249e. [DOI] [PubMed] [Google Scholar]
- 64. [accessed on 29 March, 2015]; http://bpsbioscience.com/keap1-nrf2-inhibitor-screening-assay-kit-72020.
- 65. [accessed on 29 March, 2015]; http://indigobiosciences.com/human-nrf2-assay-system-kits/
- 66. [accessed on 29 March, 2015]; https://www.caymanchem.com/app/template/Product.vm/catalog/600590.
- 67.Canning P, Bullock AN. Biochem. Soc. Trans. 2014;42:103. doi: 10.1042/BST20130215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, Zhang DD. Carcinogenesis. 2008;29:1235. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1433. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Cancer Cell. 2012;22:66. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 71.Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Oncogene. 2013;32:3765. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waller ZA, Howell LA, Macdonald CJ, O'Connell MA, Searcey M. Biochem. Biophys. Res. Commun. 2014;447:128. doi: 10.1016/j.bbrc.2014.03.117. [DOI] [PubMed] [Google Scholar]
- 73.Murat P, Singh Y, Defrancq E. Chem. Soc. Rev. 2011;40:5293. doi: 10.1039/c1cs15117g. [DOI] [PubMed] [Google Scholar]
- 74.Song JY, Yoon YS, Ahn JY, Jung IS, Lee SRU, Park SR, Lim MJ, Kim MH. 1114544. S. Korean Patent. 2012
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.