ABSTRACT
During meiosis I (MI) in oocytes, the maturation-associated decrease of histone acetylation is critical for normal meiotic progression and accurate chromosome segregation. RBBP4 is a component of several different histone deacetylase containing chromatin-remodeling complexes, but RBBP4's role in regulating MI is not known. Depleting RBBP4 in mouse oocytes resulted in multipolar spindles at metaphase (Met) I with subsequent perturbed meiotic progression and increased incidence of abnormal spindles, chromosome misalignment, and aneuploidy at Met II. We attribute these defects to improper deacetylation of histones because histones H3K4, H4K8, H4K12, and H4K16 were hyperacetylated in RBBP4-depleted oocytes. Importantly, we show that RBBP4-mediated histone deacetylation is essential for regulating bipolar spindle assembly, at least partially, through promoting Aurora kinase (AURK) C function. To our knowledge, these results are the first to identify RBBP4 as a regulator of histone deacetylation during oocyte maturation, and they provide evidence that deacetylation is required for bipolar spindle assembly through AURKC.
Keywords: Aurora kinase, chromatin, histone, histone deacetylation, meiotic maturation, meiotic spindle, oocyte, oocyte maturation, RBBP4, spindle
INTRODUCTION
Inaccurate chromosome segregation during oocyte maturation leads to aneuploidy, which is a major cause for the decline in female fertility with age [1, 2]. Emerging evidence indicates a correlation between oocyte aging and histone acetylation in which inadequate histone deacetylation contributes to the high incidence of aneuploidy in older female mice [3]. Consistent with this conclusion, acetylation of H3K14, H4K8, and H4K12 gradually increases during mouse oocyte postovulatory aging, and inhibition of histone deacetylases (HDACs) results in an accelerated progression of postovulatory aging [4]. These observations indicate that histone acetylation is a main factor that regulates the aging process in mammalian oocytes, and they provide a strong rationale to understand how HDACs are regulated.
Acetylation and deacetylation of histones are catalyzed by histone acetyltransferases (HATs) and HDACs, respectively. In mitosis, a balance of HAT and HDAC activity exists. Although HATs are active, they cannot acetylate histone H3 and H4 during meiosis I (MI) in mouse oocytes [5]. Upon resumption of meiosis, oocytes undergo a maturation-associated decrease of histone acetylation [5]. Perturbing HDACs during oocyte maturation results in histone hyperacetylation, altered meiotic progression, abnormal spindle formation, and subsequent aneuploidy [6]. Collectively, these data support the model that HDACs control the acetylation status of histones during oocyte maturation. How HDACs are regulated during oocyte maturation is not fully understood.
Recently, we identified retinoblastoma binding protein P46 (RBBP7) as a regulator of HDAC activity in mouse oocytes [7]. Perturbing RBBP7 function phenocopies oocytes treated with trichostatin A (TSA), a global inhibitor of HDACs. These phenotypes include chromosome misalignment, cytokinesis defects, and aneuploidy at metaphase (Met) II [7]. Furthermore, we found that the changes in histone acetylation caused delocalization of the chromosomal passenger complex (CPC), which is consistent with the observed phenotypes. Whether RBBP7 is the sole regulator of HDACs during oocyte maturation is not known.
Another candidate that could regulate HDACs is retinoblastoma binding protein P48 (RBBP4). RBBP4 is a ubiquitously expressed nuclear protein that belongs to the WD-40 family [8]. Although the functions of RBBP4 and RBBP7 can overlap—both are subunits of many HDAC complexes, including SIN3A [9, 10] and NuRD [11, 12]—they also have nonoverlapping functions. For example, RBBP4 functions in the chromatin assembly factor 1 (CAF-1) complex, whereas RBBP7 functions in HAT-containing complexes [8, 13, 14]. Incorporation of RBBP4 into Sin3A- and NuRD-HDAC complexes implicates RBBP4 as another regulator of histone deacetylation in mouse oocytes.
Using a knockdown (KD) approach, we show that RBBP4 is a regulator of histone deacetylation during mouse oocyte meiotic maturation. Depleting RBBP4 results in abnormal spindle formation at metaphase of MI with subsequent first polar body extrusion (PBE) defects, chromosome misalignment, and aneuploidy at Met II. These phenotypes likely are a consequence of perturbed Aurora kinase (AURK) C-CPC localization. KD of both RBBP4 and RBBP7 indicates that the functions of these proteins do not overlap, but their combined functions are required for successful completion of MI (i.e., histone deacetylation is regulated through multiple pathways in mammalian oocytes).
MATERIALS AND METHODS
In Vitro cRNA Synthesis
The DNA linearization and purification were carried out as described previously [7, 15]. In vitro transcription to generate Aurka-Gfp, Aurkc-Gfp, and H2b-mCherry was carried out using the mMessage mMachine T7 Ultra Kit (Ambion). Finally, the cRNA was purified using an RNAEasy Kit (Qiagen) and resolved in a denaturing agarose gel to confirm length.
Oocyte Collection, Microinjection, Treatment, and In Vitro Maturation
Cumulus-oocyte complexes were obtained from equine chorionic gonadotropin-primed, 6-wk-old, female CF-1 mice (Harlan Laboratories) as previously described [16]. Full-grown, denuded, germinal vesicle (GV)-intact oocytes were obtained by mechanical removal of cumulus cells. Milrinone (2.5 μM) was added to the collection and injection medium (bicarbonate- free minimal essential medium [Earle salt]) to prevent meiotic resumption [17]. All animal experiments were approved by the institutional animal use and care committees of the University of Pennsylvania and Rutgers University and were consistent with National Institutes of Health (NIH) guidelines.
Oocytes were microinjected with 10 pl of a combination of short interfering (si) RNA (25 μM) and morpholino (1 mM). The siRNAs used to target Rbbp4 (5′-UAU UGU UUG AAC GAG UGU CCC-3′) and Rbbp7 (5′-UUU CAG AUU ACG CAG GUC CCA-3′; both from Ambion, Inc.) were diluted with Milli-Q water (Millipore Corp.) to a final concentration of 100 μM and stored at −80°C. The morpholino oligonucleotide spanning the start codon of the Rbbp4 transcript (5′-CAA AGG CCG CTT CCT TGT CAG CCA T-3′) and Rbbp7 transcript (5-CTT CAA ACA TCT CTT TAC TCG CCA T-3′) were purchased from Gene Tools. Control oocytes were injected with a combination of a siRNA (Luciferase GL2 Duplex; D-001100-01-05; Thermo Fisher Scientific) and morpholino (5′-CCT CTT ACC TCA GTT ACA ATT TAT A-3′). Following microinjection, the oocytes were cultured in Chatot, Ziomek, and Bavister (CZB) medium [18] containing 2.5 μM milrinone under 5% CO2 in air at 37°C for 12–14 or 1 h, followed by in vitro maturation (IVM) in milrinone-free CZB medium for either 8 or 18 h, respectively, in 5% CO2 in air at 37°C.
The HDAC inhibitor TSA (1 μM; T8552; Sigma) was added to the IVM medium under the same culture conditions for 8 h.
Real-Time RT-PCR
Fifty oocytes or embryos at the indicated developmental stage were isolated and frozen at −80°C before processing. After thawing, 2 ng of Gfp cRNA were added to each sample as an external control. Total RNA was purified using the PicoPure RNA Isolation Kit (Arcturus) according to the manufacturer's instructions. RT was performed using random hexamers and Superscript II Reverse Transcriptase (Invitrogen) following the manufacturer's protocol. TaqMan probes specific for Rbbp4 transcript (Mm00771401_g1; Applied Biosystems) were used, and the comparative threshold cycle method was employed to determine the differences in genes expression among different stages. An ABI Prism 7000 (Applied Biosystems) was used to acquire the data. Exogenous Gfp was used to normalize the amounts of RNA in each sample. The relative abundance of Rbbp4 transcript in GV-intact oocytes was set to one.
Immunoblotting
Fifty oocytes were lysed in 1% SDS, 1% β-mercaptoethanol, 20% glycerol, and 50 mM Tris-HCl (pH 6.8) and denatured at 95°C for 5 min. Next, 10% gradient SDS-polyacrylamide precast gels were used to separate the proteins by electrophoresis depending on their molecular weight. Stained proteins of known molecular mass (range, 14–200 kDa) were run simultaneously as standards. The electrophoretically separated proteins were transferred to nitrocellulose membranes, followed by incubation in 2% blocking solution (ECL; Amersham) in Tris-buffered saline with 0.1% Tween 20 (TBS-T) for 1 h. The membranes were then incubated with primary antibodies at 4°C overnight. After washing five times (8 min each time) with TBS-T, the membranes were incubated with a secondary antibody labeled with horseradish peroxidase for 1 h, followed by five washes with TBS-T as before. The signal was detected using the ECL Advance Western Blotting Detection Reagents (Amersham) following the manufacturer's instructions.
Immunocytochemistry
The cold stable kinetochore-microtubule assay was carried out as described previously [7, 19]. In brief, oocytes were incubated for 10 min on ice in minimal essential medium containing polyvinylpyrrolidone and then fixed for 25 min on a warm plate with 3.7% formaldehyde in 100 mM PIPES (pH 6.8) containing 10 mM ethyleneglycoltetra-acetic acid, 1 mM MgCl2, and 0.2% Triton X-100. For all other experiments, oocytes or embryos were fixed in 2.5% paraformaldehyde solution in PBS for 20 min at room temperature. After fixation, the oocytes or embryos were permeabilized with 0.1% Triton X-100 in PBS for 15 min, followed by blocking (PBS + 0.3% bovine serum albumin + 0.01% Tween 20) for 15 min. Immunostaining was performed by incubating the fixed cells with the primary antibody overnight at 4°C, followed by secondary antibodies for 1 h; omission of the primary antibody served as negative control. DNA was stained and mounted with TO-PRO-3 (1:500; T3605; Life Technologies) diluted in VECTASHELD (Vector Laboratories). Fluorescence was detected on a Leica TCS SP or a Zeiss 510 meta laser-scanning confocal microscope
For RBBP4 immunostaining, all samples (i.e., oocytes, eggs, and embryos) were processed at the same time. The laser power was adjusted to a level at which the signal intensity was just below saturation for the developmental stage that displayed the highest intensity, and all images were then scanned at that laser power. The intensity of fluorescence was quantified using ImageJ software (NIH). In brief, background was subtracted from all z-projected images using identical parameters. Next, we cropped the images to the same dimensions. After cropping, the fluorescence intensity of a region of interest was measured. Three independent region measurements were taken per image and then averaged. Background fluorescence was subtracted by taking an average of three background intensity measurements per image. The final measurement was determined by subtracting the background average from the fluorescence average. To normalize the data, the fluorescence intensity of each group was averaged. The experimental group average was then divided by the average value of the control group in each replicate. The same procedure was performed for the control group, making each replicate value one.
In Situ Chromosome Counting
Immunocytochemical detection of kinetochores and chromosome counting were performed using Monastrol treatment as previously described [20]. Briefly, eggs were cultured for 1 h in CZB containing 100 μM Monastrol (Sigma) to disperse the chromosomes. Eggs were then fixed in 2.5% paraformaldehyde and stained for kinetochores (CREST antibody) and DNA (A11012; 1:5000; SYTOX Green; Invitrogen). Images were scanned at 0.4-μm intervals to span the entire region of the Met II spindle (16–20 μm in total) using a Leica DM6000 microscope with 100×, 1.4 NA oil-immersion objective. Serial confocal sections were analyzed to determine the total number of kinetochores and quantified using ImageJ software.
Live Cell Imaging
The cRNAs encoding an AURKA-GFP fusion protein and a histone H2B-mCherry fusion protein were microinjected in GV-intact oocytes and then transferred into separate drops of CZB medium covered with mineral oil in a FluoroDish (World Precision Instruments). GFP and mCherry image acquisition was started at the GV stage using a Leica DM6000 microscope with a 63×, 1.25 NA oil immersion objective and a charge-coupled device camera (Orca-AG; Hamamatsu Photonics) controlled by MetaMorph Software (Molecular Devices). A microenvironment chamber (PeCon) was used to maintain 5% CO2 and the microscope stage at 37°C using an airstream incubator (ASI 400; Nevtek). Images of each oocyte were acquired every 20 min and processed using ImageJ software.
Antibodies
The following antibodies were used in immunofluorescence (IF) and/or Western blotting (WB): anti-RBBP4 (WB, 1:2000; IF, 1:100; 2566-1; Epitomics), anti-β-actin (WB, 1:10 000; ab20272; Abcam), anti-β-tubulin (IF, 1:75; 3623, Cell Signaling Technology), anti-α-tubulin-Alexa Fluor 488 conjugate (IF, 1:100; 322588; Life Technologies), anti-γ-tubulin (IF, 1:100; T6557; Sigma), CREST autoimmune serum (IF, 1:40; Immunovision), anti-AURKC (IF, 1:30; A400-023A-BL1217; Bethyl), anti-survivin (IF, 1:500; 2808S; Cell Signaling Technology), phospho-specific S893/S894 inner centromere protein (INCENP; IF, 1:1000; gift from Michael Lampson, University of Pennsylvania [21]), and anti-acetyl-histone H3 (Lys4) (IF, 1:200; 39382; Active Motif). Anti-acetyl-histone H4 (Lys8) (06–760), anti-acetyl-histone H4 (Lys12) (06–761), and anti-acetyl-histone H4 (Lys16) (06–762) were all from Millipore and were used in IF at 1:300.
Statistical Analysis
Student t-test and one-way ANOVA were used to evaluate the differences between groups as indicated in the figure legends. Differences of P < 0.05 were considered to be significant.
RESULTS
Temporal and Spatial Pattern of RBBP4 Expression During Oocyte Maturation and Preimplantation Embryo Development
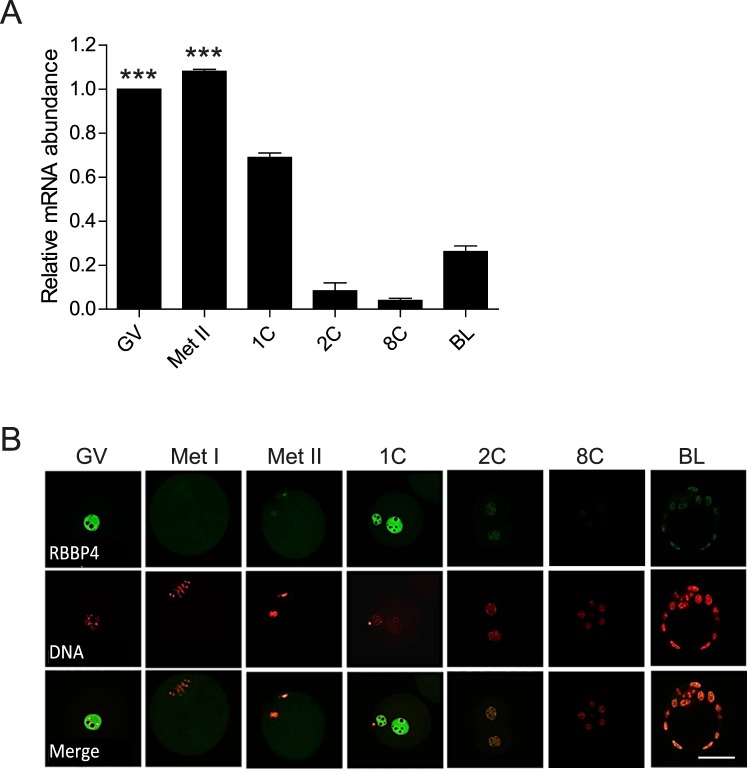
We first assessed the temporal pattern of Rbbp4 mRNA expression during oocyte maturation and preimplantation embryonic development using quantitative RT-PCR (Fig. 1A). Rbbp4 is a maternal transcript that is degraded following fertilization, with zygotic expression occurring between the 8-cell and blastocyst stages. Immunocytochemistry results showed that RBBP4 protein localized in the nucleus (GV) as anticipated; upon resumption of maturation, RBBP4 diffused into the cytoplasm (Fig. 1B). Failure to detect a signal in Met I and Met II oocytes presumably reflects the approximately 30-fold dilution following GV breakdown (GVBD) because the protein returns to the pronuclei in the 1-cell embryo. However, when the laser power was increased, we detected faint signals of chromatin-localized protein (Supplemental Fig. S1; Supplemental Data are available online at www.biolreprod.org). These results indicate that RBBP4 is expressed in mouse oocytes and suggest a role of RBBP4 during oocyte maturation.
FIG. 1.
Temporal and spatial pattern of RBBP4 expression in oocytes and preimplantation embryos. A) Relative abundance of Rbbp4 transcripts in oocytes and preimplantation embryos was determined by quantitative RT-PCR. The experiment was performed twice, and the data are expressed as the mean ± SEM and relative to that in oocytes. One-way ANOVA was performed to assess statistical significance. ***P < 0.001. B) Immunocytochemical detection of RBBP4 in oocytes and preimplantation embryos. Representative images are shown. The experiment was performed three times, and at least 20 oocytes/embryos were analyzed from each replicate. GV, germinal vesicle oocyte; Met II, metaphase II egg; 1C, 1-cell embryo; 2C, 2-cell embryo; 8C, 8-cell embryo; BL, blastocyst. Bar = 50 μm.
KD of RBBP4 Perturbs Meiotic Progression and Chromosome Segregation
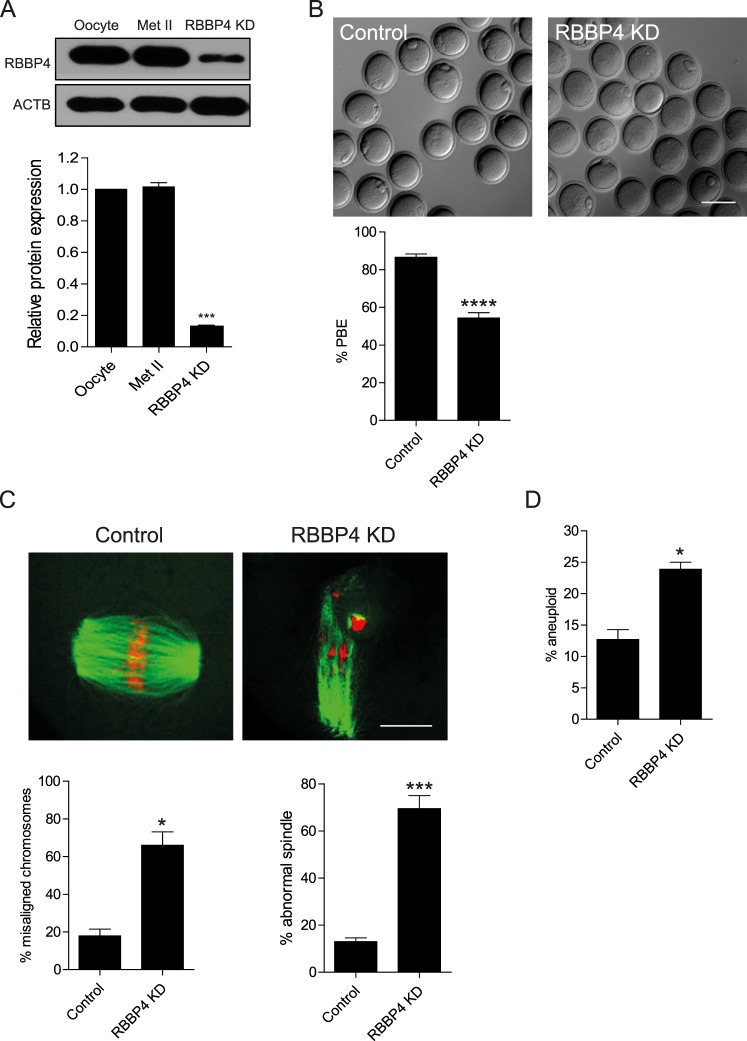
To address the function of RBBP4 during oocyte maturation, full-grown, GV-intact oocytes were injected with a 1:1 mixture of siRNA and morpholino oligonucleotides. This combined approach, which inhibits Rbbp4 mRNA translation and leads to degradation of Rbbp4 mRNA, leads to an approximately 87% decrease in RBBP4 protein (Fig. 2A).
FIG. 2.
Knockdown of RBBP4 perturbs meiotic progression and chromosome segregation. A) Full-grown, GV-intact oocytes were injected with the siRNA and morpholino cocktail, followed by maturation in vitro for 18 h. A) Fifty oocytes and Met II eggs were collected for immunoblot analysis, and β-actin (ACTB) was used as a loading control. The experiments were carried out five times. The quantification of the abundance of RBBP4 is shown. The data are expressed as the mean ± SEM, and one-way ANOVA was used to analyze the data. B) First PBE was scored to assess meiotic progression. C) Fixation and staining of spindle with an anti-β-tubulin antibody (green) and DNA with TOPRO-3 (red) were performed using confocal microscopy. Representative images are shown. The confocal images were analyzed for both the percentage of misaligned chromosomes and abnormal spindle morphology (below). D) Met II eggs were treated with Monastrol for 2 h to induce monopolar spindles, followed by kinetochore staining with CREST antiserum and DNA staining with TOPRO-3. Met II eggs with greater or less than 40 kinetochores were considered to be aneuploid. The experiments were carried out at least three times, and at least 20 oocytes were examined for each treatment group. The data are expressed as the mean ± SEM. Student t-test was used to analyze the data. *P < 0.05, ***P < 0.001, ****P < 0.0001. Bar = 100 μm (B) and 10 μm (C).
To ascertain whether RBBP4 is required for oocyte maturation, we first decreased the amount of RBBP4 protein using the siRNA/morpholino cocktail, followed by IVM for a time in which controls extruded a polar body and reached Met II. Although the percentage of oocytes that failed to undergo GVBD did not vary significantly (9% and 5% for control and RBBP4 KD oocytes, respectively), targeting RBBP4 inhibited first PBE in approximately 50% of oocytes (Fig. 2B), suggesting that RBBP4 is required to complete meiotic maturation.
To further explore the effect of RBBP4 depletion on Met II eggs, we examined spindle formation and chromosome alignment. Analysis of meiotic spindle configuration and chromosome alignment revealed a significant increase in the proportion of oocytes with misaligned chromosomes and abnormal spindles in the RBBP4 KD group when compared to injected controls (Fig. 2C). Chromosome misalignment and spindle defects at Met II are indicative of segregation errors during MI. To assess MI segregation errors, we conducted an in situ chromosome spread at Met II and found that the percentage of aneuploidy was significantly higher in RBBP4 KD eggs compared to controls (Fig. 2D). Thus, RBBP4 function is required for accurate chromosome segregation during MI in mouse oocytes.
RBBP4 Depletion Causes Defects in Spindle Formation
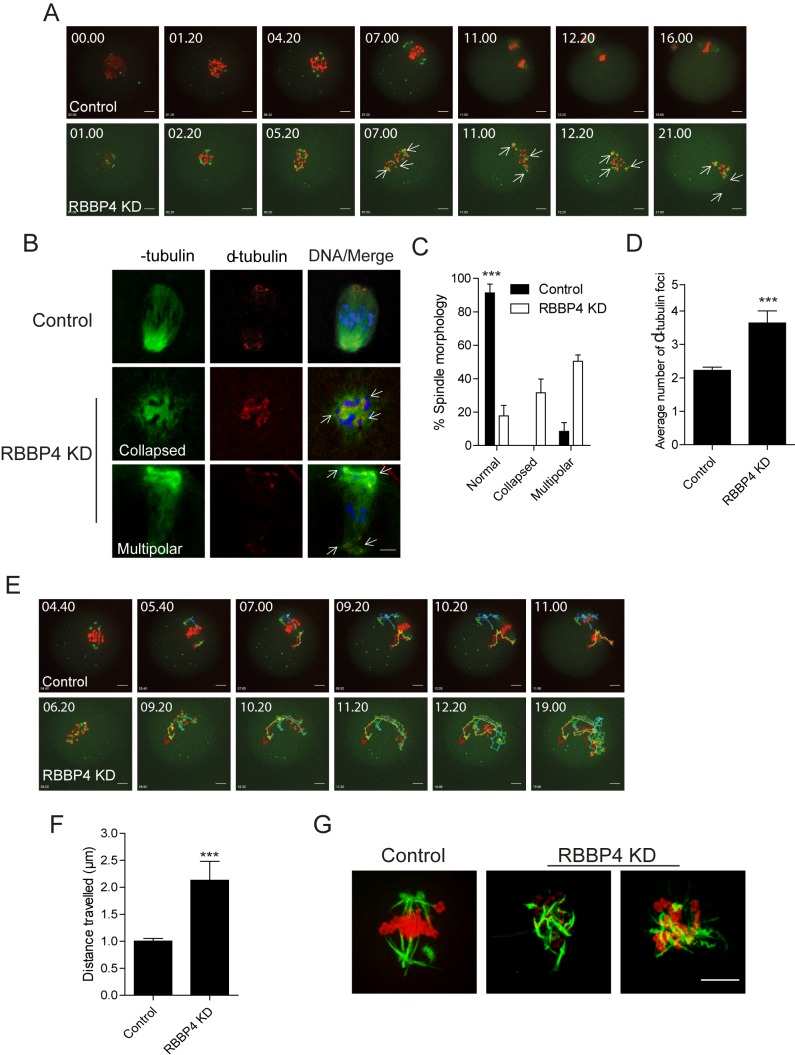
To explore how RBBP4 KD perturbs MI, we monitored control and RBBP4 KD oocytes using time-lapse confocal imaging. To mark spindles, we injected oocytes with Aurka-Gfp cRNA that localizes to microtubule-organizing centers (MTOCs; spindle poles) [15], and to monitor chromosome movement, we coinjected H2b-mCherry. In contrast to control-injected oocytes, chromosomes in RBBP4 KD oocytes failed to congress on the Met I plate. This failure may be due to the presence of multiple spindle poles because the number of AURKA-GFP foci increased (Fig. 3A and Supplemental Movies S1 and S2). These oocytes (∼46%) did not progress to anaphase I and arrested at Met I. It is unlikely that the effect of RBBP4 KD on the spindle poles was due to a cell-cycle delay because AURKA-GFP foci failed to aggregate into two dominant poles even after spending more than 14 h at Met I (Fig. 3A and Supplemental Movie S2). We next examined the Met I spindle in fixed oocytes using anti-α-tubulin and anti-γ-tubulin antibodies. Depleting RBBP4 resulted in a significant increase of abnormal spindles at Met I with two main spindle phenotypes: either collapsed or multipolar (Fig. 3, B and C). Consistent with the time-lapse imaging, the average number of spindle poles as determined by the number of γ-tubulin foci significantly increased in RBBP4 KD oocytes compared to injected controls (Fig. 3D).
FIG. 3.
RBBP4 KD perturbs bipolar spindle assembly. A and E–F) Full-grown, GV-intact oocytes were injected with siRNA and morpholino cocktail along with cRNAs encoding H2b-mCherry (red) and Aurka-Gfp (green), followed by IVM. A and E) Fluorescence images were captured every 20 min. Representative images from a time course are shown. Arrows indicate spindle poles. B) Met I oocytes were fixed and stained with anti-α-tubulin antibody (green) to detect spindle microtubules and anti-γ-tubulin antibody (red) to detect spindle poles. Arrows indicate spindle poles. Representative images are shown. C) Quantification of spindle morphology. D) Quantification of the average number of γ-tubulin foci. E) Tracking AURKA-GFP foci movement from the data set shown in A (see Supplemental Movie S2). Each color indicates the tracking of different foci. F) The distance that AURKA-GFP foci traveled in A from GVBD to Met I (4 h). G) Confocal images of cold-stable kinetochore microtubules. DNA was stained with TOPRO-3 (red) and kinetochore microtubule fibers with an anti-β-tubulin antibody (green). The experiments were carried out at least twice, and at least 20 oocytes were examined for each sample. The data are expressed as the mean ± SEM. Student t-test was used to analyze the data except in C, where one-way ANOVA was used. ***P < 0.001. Bar = 10 μm.
Another interesting observation in RBBP4-depleted oocytes was that the torsional movements of the spindle poles increased and appeared uncontrolled when compared to control oocytes (Fig. 3E and Supplemental Movies S2–S4). In addition, our analysis of MTOC trajectories showed a significant increase in the distance that they travelled over a 4-h period after GVBD in RBBP4-depleted oocytes compared to the distance traveled in control oocytes (Fig. 3F). In mitosis, abnormal torsional movement is caused by a perturbed spindle matrix; the improper assembly of a bipolar spindle and an imbalance of spindle forces resulted in curling and twisting of spindle microtubules similar to what we observed in RBBP4-depleted oocytes (∼37%) [22, 23]. Moreover, when we conducted an assay to visualize microtubules that formed stable attachments to kinetochores, we observed more microtubules that curled and interlaced with each other in RBBP4 KD oocytes as compared to controls, where the microtubules most often emanated as straight rods from the poles (Fig. 3G). Therefore, the mechanism regulating spindle assembly is perturbed in RBBP4-depleted oocytes.
RBBP4 Is a Regulator of Histone Deacetylation During Oocyte Maturation
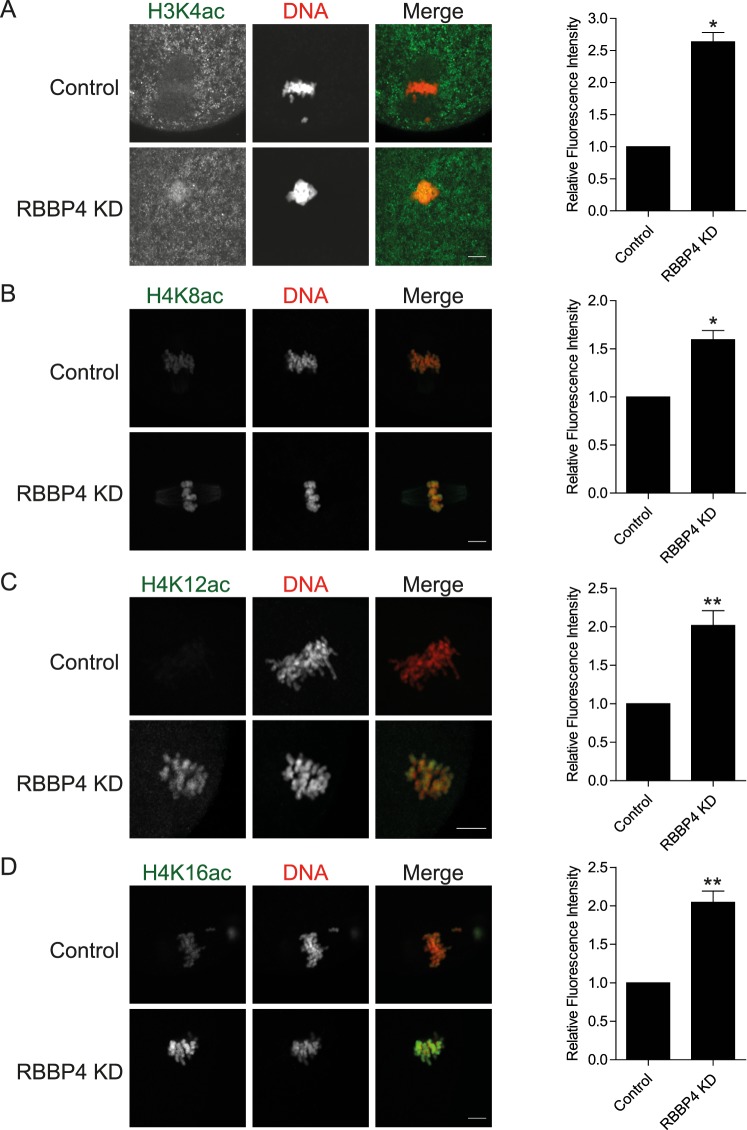
The phenotypes (Met I arrest, Met II chromosome misalignment, defective spindles, and aneuploidy) we observed in RBBP4 KD oocytes were identical to those of oocytes lacking HDAC2 or RBBP7 or when cultured in the presence of TSA, a class I and II HDAC inhibitor [6, 7, 24]. Given that RBBP4 is a regulatory component of HDAC complexes, we postulated that RBBP4 might also function in this capacity in mouse oocytes. To assess a role in regulating histone deacetylation, we assayed the presence of several histone acetylation marks during oocyte maturation using immunocytochemistry. Although no apparent effect of RBBP4 KD on acetylated H3K9, H3K14, or H4K5 has been found (unpublished observations), acetylation of H3K4, H4K8, H4K12, and H4K16 significantly increased when compared to control-injected oocytes (Fig. 4). These results implicate RBBP4 as a regulator of histone deacetylation during meiotic maturation.
FIG. 4.
Histones become hyperacetylated in RBBP4 KD oocytes. Full-grown, GV-intact oocytes were injected with siRNA and morpholino cocktail, followed by IVM. H3K4ac (A), H4K8ac (B), H4K12ac (C), and H4K16ac (D) were detected in Met II eggs. The experiments were performed three times, and at least 20 oocytes were analyzed for each sample. Representative images are shown. The graphs to the right are quantifications of the fluorescence intensity. The data are expressed as the mean ± SEM; Student t-test was used to analyze the data. *P < 0.05, **P < 0.01. Bar = 10 μm.
RBBP4 Regulates AURKC-CPC Localization and Activity in Mouse Oocytes
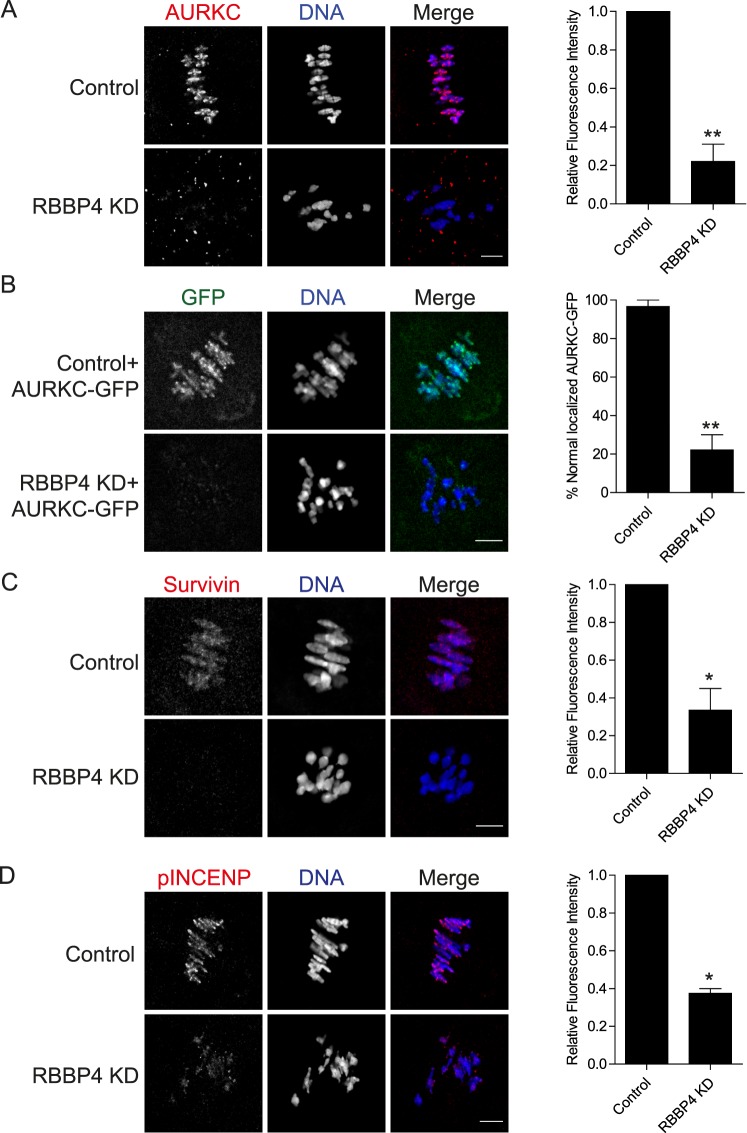
In mitosis, AURKB, the catalytic subunit of the CPC, regulates bipolar spindle formation, chromosome alignment, spindle assembly checkpoint, and cytokinesis [25–28]. In mammalian oocytes, AURKC, an AURKB homolog, appears to be the primary CPC kinase that carries out many of these cellular functions [29–31]. In mitosis, AURKB activity is positively regulated by histone hypoacetylation [32]. Moreover, we previously demonstrated that the maturation-associated decrease of histone acetylation is required for AURKC-CPC localization and activity in mouse oocytes [30]. Given the essential role of AURKB in bipolar spindle assembly in mitosis, it is possible that RBBP4-mediated histone deacetylation regulates bipolar spindle assembly in oocyte meiosis by promoting AURKC-CPC function. To investigate this hypothesis, we first assessed AURKC-CPC localization in RBBP4 KD oocytes. As shown in Figure 5A, depleting RBBP4 resulted in loss of kinetochore and interchromatid axis localization of AURKC in Met I oocytes. To confirm this finding, we expressed an AURKC-GFP fusion. Consistent with the immunocytochemistry results, exogenous AURKC localization was perturbed in RBBP4 KD oocytes when compared to injected controls (Fig. 5B). These results indicate that RBBP4-mediated histone deacetylation is required for AURKC localization.
FIG. 5.
RBBP4 KD perturbs CPC localization and activity during oocyte maturation. Full-grown, GV-intact oocytes were injected with the siRNA and morpholino cocktail, followed by IVM. Met I oocytes were fixed and immunostained with an anti-AURKC antibody (red in merge; A), anti-survivin antibody (red in merge; C), and anti-phospho-INCENP antibody (red in merge; D). DNA was detected with 4′,6-diamidino-2-phenylindole (blue in merge). In B, 100 ng/μl of Aurkc-Gfp (green in merge) cRNA was injected with the siRNA/morpholino mixture. The graphs to the right show the quantifications. The experiments were performed at least twice, and at least 15 oocytes were analyzed for each sample. Representative images are shown. The data are expressed as the mean ± SEM; Student t-test was used to analyze the data. *P < 0.05, **P < 0.01. Bars = 10 μm.
To investigate whether depleting RBBP4 affects just AURKC localization or the entire CPC, we examined the localization of survivin, another member of the CPC. Depletion of RBBP4 caused abnormal localization of survivin similar to its effect on AURKC in Met I oocytes (Fig. 5C). Phosphorylation of INCENP, a specific marker for CPC activity [30], also was significantly decreased when compared to control oocytes (Fig. 5D). To confirm that RBBP4 KD perturbs AURKC localization through histone hyperacetylation, we incubated oocytes with TSA. The TSA treatment perturbed AURKC localization at Met I when compared to control oocytes, and the effect appeared identical to that observed for RBBP4-depleted oocytes (Supplemental Fig. S2). These data indicate that RBBP4-mediated histone deacetylation is critical not only for AURKC-CPC localization but also for its activity.
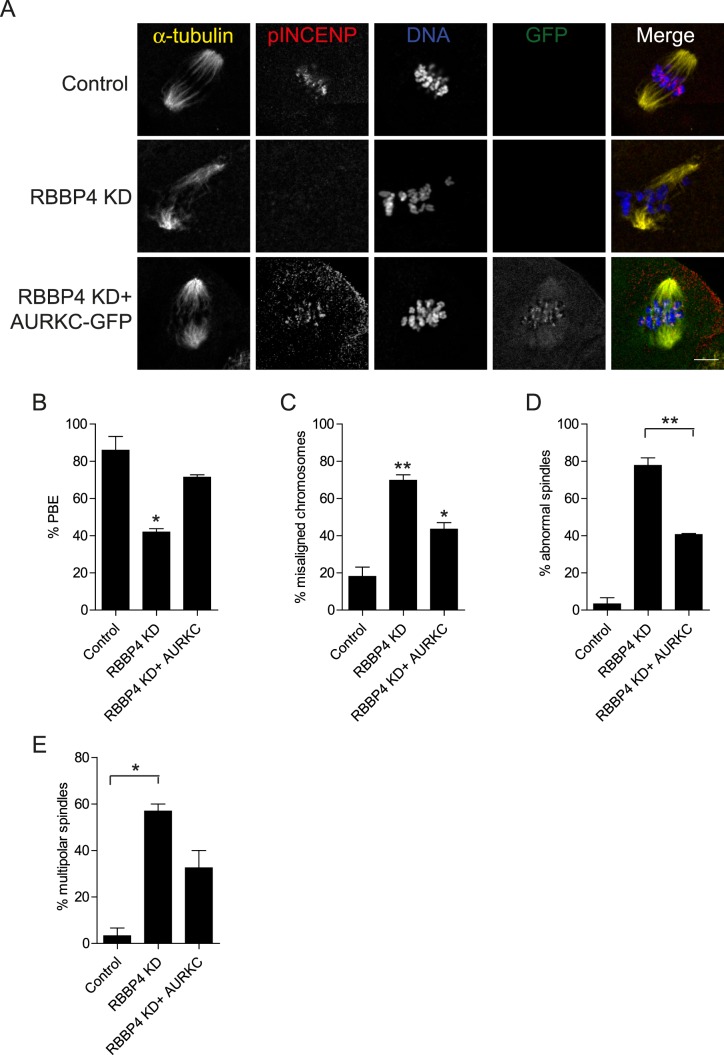
To our knowledge, no evidence connects histone deacetylation and AURKC activity to regulation of meiotic bipolar spindle assembly. To ascertain whether such a linkage exists, we determined if AURKC overexpression rescues the abnormal spindle phenotype in RBBP4-depleted oocytes. Overexpressing AURKC-GFP partially rescued the meiotic spindle defects as well as the percentage of PBE and chromosome misalignment in RBBP4-depleted eggs (Fig. 6). Taken together, these data suggest that RBBP4-mediated histone deacetylation regulates bipolar spindle assembly and meiotic progression by promoting localized AURKC function.
FIG. 6.
Overexpression of AURKC partially rescues the RBBP4 KD phenotypes. Full-grown oocytes were injected with the indicated materials, followed by IVM. A) After 18 h, meiotic progression was evaluated, followed by fixation and staining of spindles with an anti-α-tubulin antibody (yellow in merge), phospho-INCENP (red in merge), and DNA with 4′,6-diamidino-2-phenylindole (blue in merge). Representative confocal microscopy images are shown. B–D) Quantification of the percentage of oocytes in A that extruded a polar body (PBE; B) had misaligned chromosomes (C), contained abnormal spindles (D), and displayed multipolar spindles (E). The experiments were performed three times, and at least 15 oocytes were analyzed for each sample. The data are expressed as the mean ± SEM, and one-way ANOVA was used to analyze the data. *P < 0.05, **P < 0.01. Bar = 10 μm.
RBBP4 and RBBP7 KD Leads to Defects in Meiotic Progression, Spindle Formation, and Chromosome Segregation During Oocyte Maturation
Our observations suggest that in addition to RBBP7 [7], RBBP4 is another HDAC regulator that is required for oocyte meiotic maturation. We next asked whether these proteins have overlapping functions. Both RBBP4 and RBBP7 were targeted with an siRNA and morpholino cocktail, and the oocytes were then matured in vitro. We found that the incidence of chromosome misalignment in the double KD did not increase compared to the RBBP4 (Figs. 2C and 7A) or RBBP7 single KDs [7]. The incidence of aneuploidy, however, was significantly increased in the double KD oocytes compared to RBBP4-depleted oocytes (compare Figs. 2D and 7B). This incidence is similar to that observed when RBBP7 was depleted. Importantly, compared to the single KDs [7], the phenotype was additive when both proteins were knocked down because the incidence of PBE was significantly lower (Fig. 7C). These data suggest that RBBP4 and RBBP7 function in separate HDAC complexes to control MI through different mechanisms that impinge on the ability to complete MI.
FIG. 7.
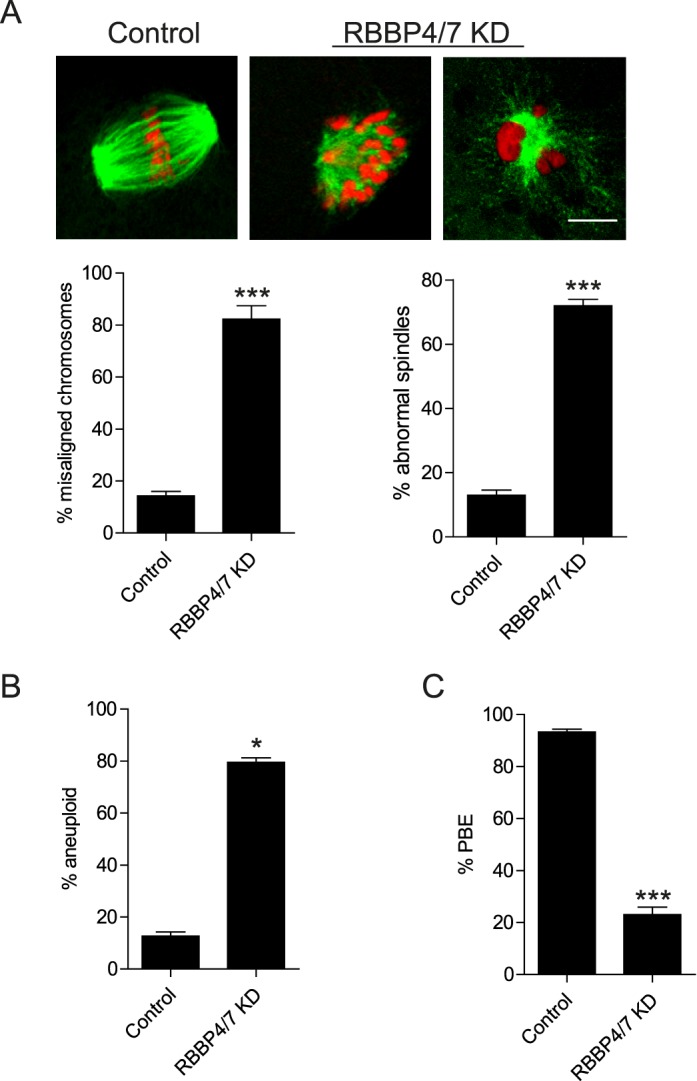
Knockdown of RBBP4 and RBBP7 perturbs meiotic progression and chromosome segregation. Full-grown, GV-intact oocytes were injected with combination of siRNAs and morpholinos, followed by IVM for 18 h. A) Representative confocal microscopy images of the Met II spindle detected with anti-β-tubulin antibody (green) and DNA with TOPRO-3 (red) are shown. The percentage of misaligned chromosomes and abnormal spindle formation was assessed from these images. B) Met II eggs were examined for aneuploidy using an in situ chromosome spread. Kinetochores were stained with CREST anti-serum, and DNA was detected by TOPRO-3 staining. C) First PBE was assessed to evaluate meiotic progression. The experiments were carried out at least three times, and at least 20 oocytes were examined for each sample. The data are expressed as the mean ± SEM, and Student t-test was used to analyze the data. *P < 0.05, ***P < 0.001. Bar = 10 μm.
DISCUSSION
We show here, to our knowledge for the first time, that RBBP4 is a regulator of histone deacetylation at H3K4, H4K8, H4K12, and H4K16 during MI. In addition, we find that depleting RBBP4 leads to meiotic progression defects, chromosome misalignment, multipolar spindles, and aneuploidy, which phenocopies oocytes in which HDAC2 or RBBP7 is depleted [6, 7] or when HDACs are inhibited with TSA [24]. Given that RBBP4 regulates HDACs in mitosis, these data indicate that RBBP4 is also involved in regulating histone deacetylation during meiosis in females. In further support of this proposal is that immunoprecipitation experiments conducted with mitotic cells indicate that RBBP4 and RBBP7 bind HDAC complexes [9–11, 33]. The multipolar spindle phenotype observed in RBBP4 KD oocytes is, at least partially, through impaired AURKC function because overexpression of AURKC rescues this phenotype. Given that either RBBP4 or RBBP7 [7] alone cannot fully support HDAC activity, it is possible that these RBBPs work in separate pathways to regulate HDAC activity. Although our experiments were conducted using denuded oocytes isolated from superovulated mice, this in vitro method is the best approach currently available until an Rbbp4 knockout mouse is created for in vivo studies. Taken together, these results identify RBBP4 as a regulator of histone deacetylation in mouse oocytes, and they provide new insight regarding the underlying role of histone deacetylation in regulating bipolar spindle formation during MI. How RBBP4 regulates HDAC activity during oocyte meiosis is still unknown.
The HDACs lack histone-binding domains and execute their functions by being components of repressor complexes [34]. In mitosis, RBBP4 is a subunit of HDAC complexes and has the ability to bind directly to histones [8]. Our observation that RBBP4 localizes to chromatin in mouse oocytes (Supplemental Fig. S1) suggests a model in which RBBP4 regulates histone deacetylation by providing a docking site for HDAC complexes. It is still possible, however, that the cytoplasmic pools of RBBP4 are essential to regulate histone deacetylation [35], and further studies are required to answer this question.
The analyses of RBBP4 and RBBP7 double depletion during MI reveal nonoverlapping functions for the two proteins. The percentage of aneuploid eggs is higher in the double KD than in the RBBP4 single KD. This finding can be explained by our observation that one-half of the RBBP4-depleted oocytes were arrested at Met I, and possibly the remaining oocytes were healthier and progressed to Met II. In contrast, the percentage of oocytes with misaligned chromosomes and abnormal spindles is similar in the double and RBBP4 single KDs. Therefore, our data suggest that RBBP4 is the main HDAC regulator of bipolar spindle formation in mouse oocytes. Because the incidence of PBE is lower in the double-depletion experiments than in either of the single depletions, these results imply that perturbing the function(s) of each regulator at the same time ultimately impairs completion of MI.
Abnormal bipolar spindle assembly is a pronounced phenotype when either RBBP4 or histone deacetylation is perturbed in oocytes [6, 7]. In mitosis, HDACs suppress centrosome overduplication [36]. Therefore, inhibiting histone deacetylation is accompanied by multipolar spindles and mitotic defects [37]. We also observe an increase in multipolar spindles in RBBP4-depleted oocytes. However, at the time when we knock down RBBP4, the acentriolar centrosomes (MTOCs) have already undergone duplication, ruling out this function as causing the multipolar spindle phenotype. Spindle assembly in oocytes depends on self-organization of numerous (upward of 80) MTOCs into two poles [38]. In mitosis, centrosomal proteins, including γ-tubulin, undergo acetylation and deacetylation to control their functions [36]. Deacetylation of γ-tubulin could be required for MTOC congression into two poles because an increase in the number of γ-tubulin foci occurs in RBBP4-depleted oocytes.
We previously demonstrated that histone deacetylation is required for localized AURKC function in mouse oocytes [7]. Importantly, our observation that overexpression of AURKC rescues the meiotic spindle defects in RBBP4-depleted cells indicates that the abnormal spindle is, at least partially, a result of perturbing AURKC function. In mitosis, AURKB activity is critical for bipolar spindle formation, likely by regulating MCAK activity [25]. Although the mechanism of AURKC's role in meiotic spindle formation is not fully understood, one model is that perturbing AURKC function induces merotelic and syntelic kinetochore-microtubule attachments [30]. These abnormal attachments subsequently disturb chromosome forces and spindle pole integrity, resulting in a multipolar spindle [39, 40]. Whether AURKC has a specific, direct function at meiotic spindle poles is still unknown.
In conclusion, we define a role for RBBP4 to ensure the maturation-associated decrease of histone acetylation during mouse oocyte meiosis, and we provide additional evidence that such deacetylation is required for proper chromosome segregation. Further understanding of how histone deacetylation is regulated will be critical to determine how MI is regulated in female meiosis. Furthermore, these data shed new light on underlying causes of oocyte aneuploidy that could help address the majority of clinical aneuploidies in females.
ACKNOWLEDGMENT
The authors thank Michael Lampson for the phospho-INCENP antibody and Pengpeng Ma, Richard Jimenez, Amanda Gentilello, Sergey Medvedev, Jun Ma, and Olga Davydenko for their valuable discussions and help.
Footnotes
This research was supported by grants from the National Institutes of Health (HD022681 to R.M.S. and HD061657 to K.S.) and Rutgers University Institutional start-up funds to K.S. A.Z.B. was supported by a fellowship funded from Science and Technology Development Fund (U.S.-Egypt Joint Science and Technology Board).
REFERENCES
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Jones KT. Meiosis in oocytes: predisposition to aneuploidy and its increased incidence with age. Hum Reprod Update. 2008;14:143–158. doi: 10.1093/humupd/dmm043. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Nagata M, Aoki F. Inadequate histone deacetylation during oocyte meiosis causes aneuploidy and embryo death in mice. Proc Natl Acad Sci U S A. 2006;103:7339–7344. doi: 10.1073/pnas.0510946103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Yan LY, Lei ZL, Miao YL, Shi LH, Yang JW, Wang Q, Ouyang YC, Sun QY, Chen DY. Changes in histone acetylation during postovulatory aging of mouse oocyte. Biol Reprod. 2007;77:666–670. doi: 10.1095/biolreprod.107.062703. [DOI] [PubMed] [Google Scholar]
- Kim JM, Liu H, Tazaki M, Nagata M, Aoki F. Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol. 2003;162:37–46. doi: 10.1083/jcb.200303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Schultz RM. Histone deacetylase 2 (HDAC2) regulates chromosome segregation and kinetochore function via H4K16 deacetylation during oocyte maturation in mouse. PLoS Genet. 2013;9:e1003377. doi: 10.1371/journal.pgen.1003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboula AZ, Stein P, Schultz RM, Schindler K. Knockdown of RBBP7 unveils a requirement of histone deacetylation for CPC function in mouse oocytes. Cell Cycle. 2014;13:600–611. doi: 10.4161/cc.27410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol Cell Biol. 1999;19:5847–5860. doi: 10.1128/mcb.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun ZW, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- Shuda K, Schindler K, Ma J, Schultz RM, Donovan PJ. Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev. 2009;76:1094–1105. doi: 10.1002/mrd.21075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM, Montgomery RR, Belanoff JR. Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol. 1983;97:264–273. doi: 10.1016/0012-1606(83)90085-4. [DOI] [PubMed] [Google Scholar]
- Tsafriri A, Chun SY, Zhang R, Hsueh AJ, Conti M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol. 1996;178:393–402. doi: 10.1006/dbio.1996.0226. [DOI] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil. 1989;86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2005;7:93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- Duncan FE, Chiang T, Schultz RM, Lampson MA. Evidence that a defective spindle assembly checkpoint is not the primary cause of maternal age-associated aneuploidy in mouse eggs. Biol Reprod. 2009;81:768–776. doi: 10.1095/biolreprod.109.077909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimian KJ, Ballister ER, Smoak EM, Wood S, Panchenko T, Lampson MA, Black BE. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr Biol. 21:1158–1165. doi: 10.1016/j.cub.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen KM, Forer A, Yao C, Girton J, Johansen J. Do nuclear envelope and intranuclear proteins reorganize during mitosis to form an elastic, hydrogel-like spindle matrix? Chromosome Res. 2011;19:345–365. doi: 10.1007/s10577-011-9187-6. [DOI] [PubMed] [Google Scholar]
- Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- Yang F, Baumann C, Viveiros MM, De La Fuente R. Histone hyperacetylation during meiosis interferes with large-scale chromatin remodeling, axial chromatid condensation and sister chromatid separation in the mammalian oocyte. Int J Dev Biol. 2012;56:889–899. doi: 10.1387/ijdb.120246rd. [DOI] [PubMed] [Google Scholar]
- Jambhekar A, Emerman AB, Schweidenback CT, Blower MD. RNA stimulates Aurora B kinase activity during mitosis. PLoS One. 2014;9:e100748. doi: 10.1371/journal.pone.0100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori M, Fumoto K, Fukuta Y, Iwasaki T, Kikuchi A, Tatsuka M, Hosoya H. Myosin II regulatory light chain as a novel substrate for AIM-1, an Aurora/Ipl1p-related kinase from rat. J Biochem. 2000;128:903–907. doi: 10.1093/oxfordjournals.jbchem.a022840. [DOI] [PubMed] [Google Scholar]
- Vigneron S, Prieto S, Bernis C, Labbe JC, Castro A, Lorca T. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N, Brenkman AB, van den Broek NJ, Cruijsen CW, van Osch MH, Lens SM, Medema RH, Kops GJ. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Schindler K, Schultz RM. CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol Reprod. 2009;80:795–803. doi: 10.1095/biolreprod.108.074906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboula AZ, Schindler K. Selective disruption of Aurora C kinase reveals distinct functions from Aurora B kinase during meiosis in mouse oocytes. PLoS Genet. 2014;10:e1004194. doi: 10.1371/journal.pgen.1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AL, Gentilello AS, Balboula AZ, Shrivastava V, Ohring J, Schindler K. Phosphorylation of threonine 3 on histone H3 by haspin kinase is required for meiosis I in mouse oocytes. J Cell Sci. 127:5066–5078. doi: 10.1242/jcs.158840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20:2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- Endo T, Kano K, Naito K. Nuclear histone deacetylases are not required for global histone deacetylation during meiotic maturation in porcine oocytes. Biol Reprod. 2008;78:1073–1080. doi: 10.1095/biolreprod.107.067397. [DOI] [PubMed] [Google Scholar]
- Ling H, Peng L, Seto E, Fukasawa K. Suppression of centrosome duplication and amplification by deacetylases. Cell Cycle. 2012;11:3779–3791. doi: 10.4161/cc.21985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, Bernard L, Draetta GF, Alcalay M, Seiser C, Chiocca S. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Maiato H, Logarinho E. Mitotic spindle multipolarity without centrosome amplification. Nat Cell Biol. 2014;16:386–394. doi: 10.1038/ncb2958. [DOI] [PubMed] [Google Scholar]
- Mattiuzzo M, Vargiu G, Totta P, Fiore M, Ciferri C, Musacchio A, Degrassi F. Abnormal kinetochore-generated pulling forces from expressing a N-terminally modified Hec1. PLoS One. 2011;6:e16307. doi: 10.1371/journal.pone.0016307. [DOI] [PMC free article] [PubMed] [Google Scholar]