Abstract
The Database of Human Gastric Cancer (DBGC) is a comprehensive database that integrates various human gastric cancer-related data resources. Human gastric cancer-related transcriptomics projects, proteomics projects, mutations, biomarkers and drug-sensitive genes from different sources were collected and unified in this database. Moreover, epidemiological statistics of gastric cancer patients in China and clinicopathological information annotated with gastric cancer cases were also integrated into the DBGC. We believe that this database will greatly facilitate research regarding human gastric cancer in many fields. DBGC is freely available at http://bminfor.tongji.edu.cn/dbgc/index.do
Introduction
As one of the most common cancers, gastric cancer has the third highest lethality and fourth highest morbidity of all cancers worldwide [1]. According to the GloboCan statistics in 2012, new gastric cancer cases numbered almost one million (952,000), and more than 700,000 deaths were caused by gastric cancer; almost half of these patients came from China (405,000 new cases and 325,000 deaths) [1, 2]. Although both the lethality and morbidity of gastric cancer have decreased in recent years, the 5-year survival rate remains quite low [3]. Therefore, gastric cancer will remain one of the most difficult challenges for researchers and physicians for a long time [4].
Researchers worldwide have completed many genomics, transcriptomics, proteomics, and epidemiological investigations and clinical trials regarding the pathogenesis and therapies of gastric cancer [5–10]. These investigations have generated vast amounts of data relevant to gastric cancer, and the speed of these investigations is accelerating with the rapid growth of cancer knowledge, decreased costs of detection and computation, and spread of the Internet [11]. These data contain important information for investigating and curing gastric cancer. However, due to the limited background knowledge of clinicians and fundamental researchers, the potential of these data cannot be fully developed. New technologies and research methods still require development; however, low efficiency in managing data is a primary limitation of this development [12]. Due to the long-term accumulation of decentralized research, these data and their formats only satisfy individual needs, lacking integration and standardization and resulting in the diversification, isomerization, and dissection of cancer data [13, 14].
At present, abundant clinical and fundamental studies regarding gastric cancer are planned or in progress. Various types of data are stored in different database systems [13], without sharing or communication. Thus, strongly correlated information remains isolated, in what are called “information islands”. On the one hand, data dissection increases the difficulty of data mining, while on the other hand, it prevents clinicians from making full use of the outcomes of fundamental research to develop clinical trials and applications and keeps fundamental researchers from performing efficient exploratory studies that reference clinically relevant information [15].
In this situation, retrieving comprehensive information on gastric cancer is not an easy task, and portions of these data may disappear in the ocean of the Internet, which would be very unfortunate.
This research took advantage of resources from the Internet and publications from the Chinese Center for Disease Control and Prevention (CDC) and Gastric Cancer Center for Diagnosis and Treatment, Key Laboratory of Gastric Neoplasms in Shanghai. This study systematically collected various types of gastric cancer-related data, integrated these data resources after filtration and standardization, and finally formed the first comprehensive knowledge base for analyzing gastric cancer.
Materials and Methods
Data Resources
The Database of Human Gastric Cancer (DBGC) has integrated the following gastric cancer-related resources:
Epidemiological statistics of gastric cancer patients in China from CDC publications
Clinicopathological information about gastric cancer tissue after surgical resection from patients diagnosed at Shanghai Ruijin Hospital
Molecular biological data on gastric cancer from public online resources (including gastric cancer-related mutations, biomarkers, drug-sensitive genes, transcriptomics projects and corresponding differentially expressed genes, and proteomics projects and corresponding differentially expressed proteins)
Raw research data from the Shanghai Institute of Digestive Surgery and Shanghai Key Laboratory of Gastric Neoplasms
Data Collection
1) Epidemiological statistics of gastric cancer patients in China
The CDC has had an established cancer reporting system for many years and has accumulated abundant epidemiological information on cancer patients in China. The epidemiological statistics of gastric cancer, including case number, death number, incidence rate (crude rate, age-adjusted rate and cumulative rate), mortality rate (crude rate, age-adjusted rate and cumulative rate), and incidence (or mortality) distribution by age group were extracted manually from CDC publications. DBGC 1.0 covers all epidemiological statistics for all typical regions of China from the years 2004 to 2009, and additional statistics will be included in the upgraded version.
2) Clinicopathological information about gastric cancer tissue
Clinicopathological information was provided by Shanghai Ruijin Hospital. The classification and staging methods generally used for gastric cancer diagnosis were annotated using gastric cancer cases diagnosed at Ruijin Hospital. Typical gastric cancer tissues of different stages and types were selected from a gastric cancer biobank that we have maintained for years. All patient information was anonymized and de-identified before our analysis.
3) Molecular biological data on gastric cancer from public online resources
Molecular biological data were extracted and curated from online resources. Transcriptomics data were collected from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) and EBI database (http://www.ebi.ac.uk/). Proteomics data were extracted from the published literature through manual reading and standardization [16, 17]. Mutation data were collected from the dbVar database (http://www.ncbi.nlm.nih.gov/dbvar/), OMIM database (http://www.ncbi.nlm.nih.gov/omim/), HGMD database (http://www.hgmd.org/), and published literature [18, 19]. All biomarker data were extracted from published literature [20, 21]. Drug-related genes were extracted from the PharmGKB database (http://www.pharmgkb.org/), CancerDR database (http://crdd.osdd.net/raghava/cancerdr/) and published literature [22, 23]. We designed detailed extraction standards for each type of molecular biological data resource, and every data collection procedure had to follow these standards to ensure data coherency. The detailed collection procedure is provided below:
Transcriptomics data:
Search the GEO database using the following keywords:(“stomach neoplasms” [MeSH Terms] OR “stomach cancer” [All Fields]) AND "Homo sapiens" [porgn].
Filter the results manually, and select the publications related to human gastric cancer for subsequent information extraction.
Classify the programs by sample size and sample type.
Extract publication information (title, publication time, experiment type, tissue type, sample quantity, sample description, sample of experiment, sample of control, platform, GSE ID, GSM IDs, download links, and literature citation) manually, referring to MIAME (Minimum Information About a Microarray Experiment).
Pre-process raw data (series matrix files in the GEO database) using Perl to eliminate the differences from diverse platforms.
Extract differentially expressed genes using R language.
Proteomics data:
Search raw data in PubMed using the following keywords: (“proteomics” [MeSH Terms] OR “proteomics” [All Fields]) AND (“stomach neoplasms” [MeSH Terms] OR (“stomach” [All Fields] AND “neoplasms” [All Fields]) OR “stomach neoplasms” [All Fields] OR (“gastric” [All Fields] AND “cancer” [All Fields]) OR “gastric cancer” [All Fields]).
Filter the results manually, and select the proteomics publications related to human gastric cancer for subsequent information extraction.
Use these papers as seed literature and filter the references again.
Classify the publications by sample size and sample type.
Manually read papers and extract publication information (title, publication time, sample quantity, sample experiment, sample control, sample description, technology method used, fold change, up-regulated protein quantity, down-regulated protein quantity, and reference) and corresponding up-regulated proteins and down-regulated proteins (based on conclusions of the authors).
Mutation data:
Search in the OMIM, HGMD and dbVar databases using the keywords “gastric cancer” and extract mutation information (gene, mutation type, description of cDNA, description of full AA, description of AA, and reference).
Search in PubMed using the following keywords: (“mutation” [MeSH Terms] OR “mutation” [All Fields]) AND (“stomach neoplasms” [MeSH Terms] OR (“stomach” [All Fields] AND “neoplasms” [All Fields]) OR “stomach neoplasms” [All Fields] OR (“gastric” [All Fields] AND “cancer” [All Fields]) OR “gastric cancer” [All Fields]).
Filter the results manually, and select the papers related to human gastric cancer for subsequent information extraction.
Take these papers as seed literature and filter the references again.
Read these papers and extract mutation information manually (gene, mutation type, description of cDNA, description of full AA, description of AA, and reference).
Remove duplicate data from the four sources.
Biomarker data:
-
Search in PubMed using the following keywords:
(“biological markers” [MeSH Terms] OR (“biological” [All Fields] AND “markers” [All Fields]) OR “biological markers” [All Fields] OR “biomarker” [All Fields]) AND (“stomach neoplasms” [MeSH Terms] OR (“stomach” [All Fields] AND “neoplasms” [All Fields]) OR “stomach neoplasms” [All Fields] OR (“gastric” [All Fields] AND “cancer” [All Fields]) OR “gastric cancer” [All Fields]).
Filter the results manually, and select the papers related to human gastric cancer for subsequent information extraction.
Take these papers as seed literature and filter the references again.
Read these papers and extract mutation information manually (biomarker name, full name, type, stage, description, mechanism, sensitivity, specificity, and reference).
Classify the biomarkers by biomarker type, stage, specificity and sensitivity.
Drug-sensitive data:
Search in PharmGKB using the keywords “gastric cancer” and manually extract drug-sensitive information (drug name, gene name, gene type, mechanism, and reference).
Search in PubMed using the following keywords:“resistance” [All Fields] AND (“stomach neoplasms” [MeSH Terms] OR (“stomach” [All Fields] AND “neoplasms” [All Fields]) OR “stomach neoplasms” [All Fields] OR (“gastric” [All Fields] AND “cancer” [All Fields]) OR “gastric cancer” [All Fields])
Filter the results manually, and select the papers related to human gastric cancer drug resistance for subsequent information extraction.
Take these papers as seed literature and filter the references again.
-
Summarize the 19 drugs generally used for the clinical treatment of gastric cancer (5-fluorouridine, camptothecin, carboplatin, cisplatin, docetaxel, doxorubicin, doxorubicin hydrochloride, epirubicin, etoposide, fluorouracil, irinotecan, leucovorin, mitomycin c, oxaliplatin, paclitaxel, tamoxifen, trastuzumab, vinblastine, and vincristine).
Taking “cisplatin” as an example, search in PubMed using keywords:
(“cisplatin” [MeSH Terms] OR “cisplatin” [All Fields]) AND “resistance” [All Fields] AND (“stomach neoplasms” [MeSH Terms] OR (“stomach” [All Fields] AND “neoplasms” [All Fields]) OR “stomach neoplasms” [All Fields] OR (“gastric” [All Fields] AND “cancer” [All Fields]) OR “gastric cancer” [All Fields]).
Filter the results manually, and select the papers related to human gastric cancer drug resistance for subsequent information extraction.
Take these papers as seed literature and filter the references again.
Read these papers and extract drug-sensitive information manually (drug name, gene name, gene type, mechanism, and reference).
We annotated all genes and drugs in this database to help users better understand and use these data resources. The genes are annotated according to NCBI (http://www.ncbi.nlm.nih.gov), HGNC (http://www.genenames.org/), Ensembl (http://feb2014.archive.ensembl.org/) and Gene Cards (http://www.genecards.org/). The drugs are annotated according to DrugBank (http://www.drugbank.ca/).
Moreover, mutations detected in the TCGA project were also included to annotate genes in the DBGC. Users can find all mutations of a certain gene detected in the TCGA project. These mutations were processed by ICGC (https://dcc.icgc.org) based on TCGA data and referenced by each mutation in the DBGC.
In addition, several gastric cancer-related fundamental research projects have been conducted by our research team. Project descriptions and raw data are provided in the DBGC for download and further analysis.
Database Construction
The DBGC is a relational database with a MySQL data layer. A user-friendly interface was designed to organize and display data resources using HTML and JavaScript. The interaction between the data layer and the web interface was completed using the Java EE Platform.
Results and Discussion
Database Description
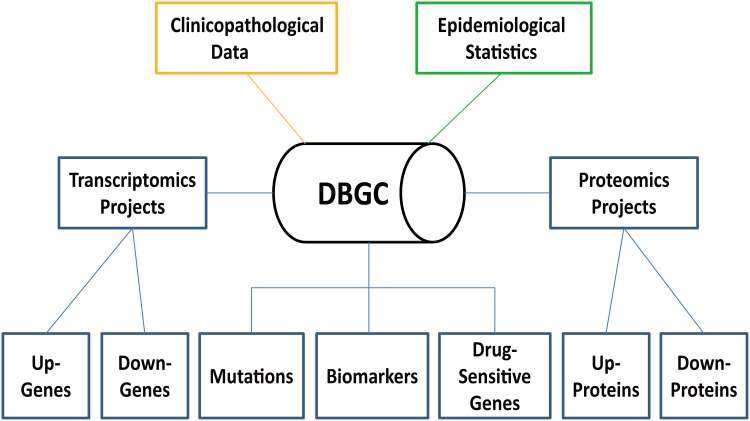
This database consists primarily of three longitudinal data systems, epidemiological, clinicopathological and molecular biological data (Fig 1). The molecular biological data consist of gastric cancer-related transcriptomics, proteomics, mutation, biomarker and drug-sensitive gene data. The overall statistics of these data are listed in Table 1. Along with the epidemiological statistics of gastric cancer patients in China and the clinicopathological information annotated with gastric cancer cases, all these data were extracted from public databases, publications and published literature.
Fig 1. Schematic architecture of the DBGC.
Table 1. Primary statistics of molecular biological data in the DBGC.
| Data resource type | Description | Statistics |
|---|---|---|
| Transcriptomics data | ||
| Transcriptomics projects | 121 | |
| Samples | 5629 | |
| Up-regulated genes | 3463 | |
| Down-regulated genes | 3380 | |
| Proteomics data | ||
| Proteomics projects | 47 | |
| Up-regulated proteins | 1513 | |
| Down-regulated proteins | 808 | |
| Mutation data | ||
| Mutations | 850 | |
| Genes | 97 | |
| TCGA Mutations | 1049822 | |
| Drug-sensitive data | ||
| Genes | 132 | |
| Drugs | 19 | |
| Biomarker data | ||
| Biomarkers | 221 |
Database Interfaces
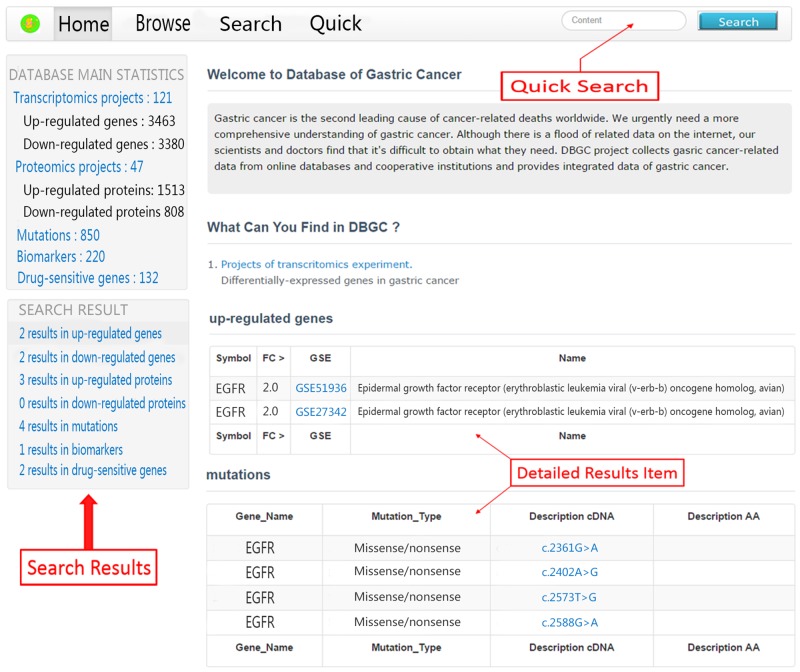
1) Quick Search (Fig 2). The quick search function module makes identifying the role of a gene or protein in gastric cancer possible via inputting keywords into the search box located at the navigation bar. The search result will tell you whether the gene or protein is differentially expressed in any transcriptomics projects or proteomics projects and whether it has been identified as a biomarker for gastric cancer or a drug-sensitive gene. Moreover, if the gene has any mutation that is related to gastric cancer, a detailed list will be displayed on the results page. For example, using “EGFR” as a keyword, we can conclude that it was identified as an up-regulated gene in GSE51936 and GSE27342 and as a down-regulated gene in GSE29630. The corresponding protein of the gene EGFR was identified as an up-regulated protein in 3 proteomics projects (PubMed Ids: 23161554, 24263233 and 24722433). EGFR has been reported as a prognosis factor of gastric cancer and is related to drug resistance to irinotecan, which is a commonly used drug for the treatment of gastric cancer. Four mutations of EGFR related to gastric cancer have been reported (c.2361G>A, c.2402A>G, c.2573T>G, c.2588G>A).
Fig 2. Quick search function module.
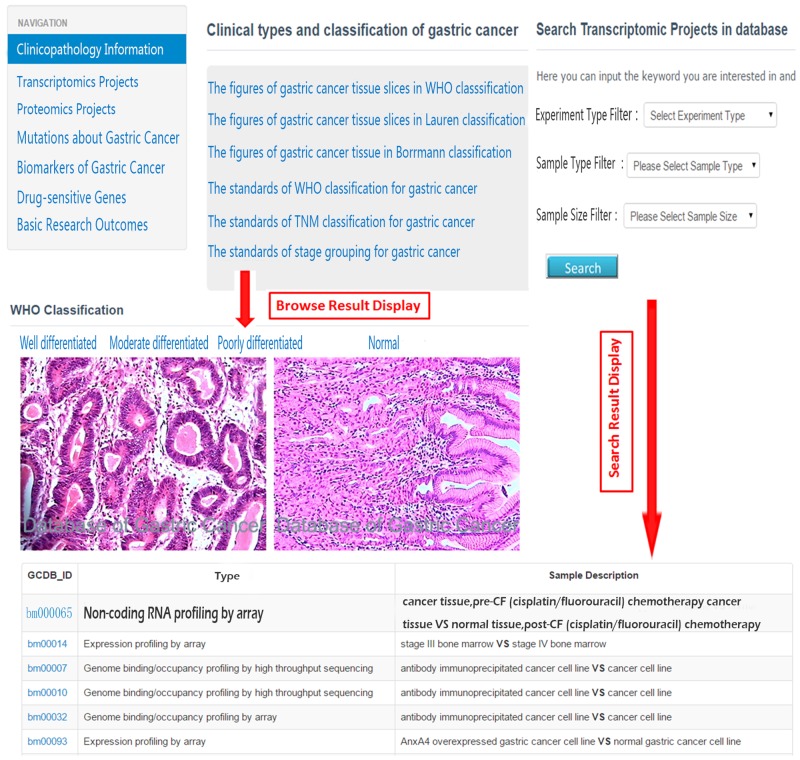
2) Browse and Search (Fig 3). Using the navigation, users can click corresponding items to browse the data resources provided in the DBGC. Detailed information will be listed below. We have also established several search criteria for each type of data resource through which all data items that fulfill the conditions will be displayed.
Fig 3. Browse function module and search function module.
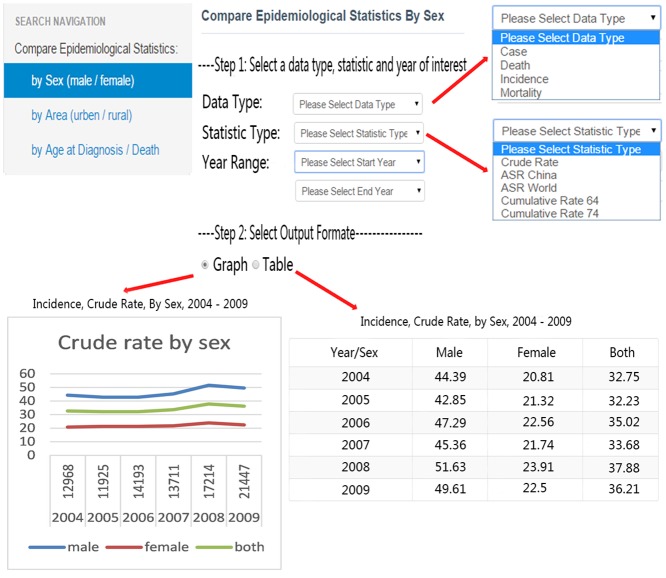
3) Our database differs from other online resources because of the inclusion of epidemiological statistics of gastric cancer patients in China. Users can compare statistics by sex (male and female), area (urban and rural), and age at diagnosis or death. Case number, death number, incidence rate, and mortality rate in a selected year range can be displayed in both graph and table format (Fig 4).
Fig 4. Statistical function module for epidemiological data.
Discussion
Gastric cancer is a leading cancer worldwide in both mortality and morbidity. Higher incidence and mortality of gastric cancer are observed in Asian regions, particularly in China. The epidemiological statistical data of gastric cancer in this database were obtained primarily from publications of the CDC, which has been engaged in malignant tumor studies for several decades and has established comprehensive archives of malignant tumor patients in China. These data have played an important role in promoting cancer prevention and health policymaking in China [24–26]. Through retrieving the gastric cancer epidemiology data in this database, researchers and clinicians can quickly determine the epidemiological trends of gastric cancer in China.
Gastric cancer-related mutations, biomarkers, drug-sensitive genes, transcriptomics projects and corresponding differentially expressed genes, and proteomics experiments and corresponding differentially expressed proteins were manually collected from online databases and published literature. The quick search function provided by the DBGC enables researchers to identify the role of a gene or protein in gastric cancer. These differentially expressed genes and proteins contain abundant important information about gastric cancer, and many analytical studies could be conducted using them.
Our research team has been engaged in gastric cancer research for many years and has accumulated considerable experience in tumor epidemiological research, gastric cancer clinicopathology and biomarker research, biobank building, molecular biological mechanism research, bioinformatics analysis and large-scale database construction [27–30]. To provide inquiry and analysis tools that are more convenient and practical for gastric cancer researchers, we constructed this database. The current version is 1.0. Because the large amounts of data generated by different experimental platforms in different fields are greatly dispersed and heterogeneous, some useful information may have been missed in our process of data collection. We will continue retrieving these data and updating the latest data for a long time to ensure the timeliness and completeness of the data. In the next version, we intend to cover the newest human gastric cancer-related mutations, biomarkers and drug-sensitive genes. Transcriptomics data will be the emphasis of the next version, in which all transcriptomics projects will be re-analyzed to extract differentially expressed genes at different fold change values. Thus, users could query whether a certain gene is differentially expressed through configuring the sample type and fold change value.
Conclusion
The database described in this article, the DBGC, is a comprehensive and web-accessible database of human gastric cancer. This database has integrated a variety of data resources related to gastric cancer and provided several easy-to-use web-based functional modules. We believe that the DBGC will be an important tool for gastric cancer clinicians, tumor fundamental research scientists, cancer genome researchers, government health policymakers, and gastric cancer patients.
Data Availability
Data are availabe via Datadryad (https://datadryad.org). The unique accession number is: doi:10.5061/dryad.271dk.
Funding Statement
This work was partially supported by grants from the National Natural Science Foundation of China (81172329, 31571363, 81372644, 81372645 and 8157111077), Chinese National High Tech Program (2012AA02A504 and 2012AA02A203), International Cooperative Project from Shanghai Science and Technology Commission (12410706400), Innovation Foundation of Translational Medicine of Shanghai Jiao Tong University School of Medicine (15ZH1002 and 15ZH3001), Fong Shu Fook Tong Foundation and Gastrointestinal Carcinoma Biobank Project of Shanghai Jiao Tong University School of Medicine. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://globocan.iarc.fr, accessed on 15/1/2015.
- 2. Bray F, Ren JS, Masuyer E et al. Estimates of global cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013; 132(5): 1133–45. 10.1002/ijc.27711 [DOI] [PubMed] [Google Scholar]
- 3.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al. SEER Cancer Statistics Review, 1975–2011, National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2011/, based on November 2013 SEER data submission, posted to the SEER web site, April 2014.
- 4. Penon D, Cito L, Giordano A. Novel findings about management of gastric cancer: A summary from 10th IGCC. World J Gastroenterol. 2014; 20(27): 8986–8992. 10.3748/wjg.v20.i27.8986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sulahian R, Casey F, Shen J, Qian ZR, Shin H, Ogino S, et al. An integrative analysis reveals functional targets of GATA6 transcriptional regulation in gastric cancer. Oncogene. 2014; 33(49): 5637–5648. 10.1038/onc.2013.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang X, Lu Y, Yang J, Shi Y, Lan M, Liu Z, et al. Identification of triosephosphate isomerase as an anti-drug resistance agent in human gastric cancer cells using functional proteomic analysis. J Cancer Res Clin Oncol. 2008; 34(9): 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ju H, Lim B, Kim M, Noh SM, Han DS, Yu HJ, et al. Genetic variants A1826H and D2937Y in GAG-beta domain of versican influence susceptibility to intestinal-type gastric cancer. J Cancer Res Clin Oncol. 2010; 36(2): 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang F, Sun GP, Zou YF, Hao JQ, Zhong F, Ren WJ. MicroRNAs as promising biomarkers for gastric cancer.Cancer Biomark. 2012; 11(6): 259–267. 10.3233/CBM-2012-00284 [DOI] [PubMed] [Google Scholar]
- 9. Tanaka M, Kataoka H, Yano S, Ohi H, Kawamoto K, Shibahara T, et al. Anti-cancer effects of newly developed chemotherapeutic agent, glycoconjugated palladium (II) complex, against cisplatin-resistant gastric cancer cells. BMC Cancer. 2013; 13:237 10.1186/1471-2407-13-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matsuda T, Saika K. The 5-year relative survival rate of stomach cancer in the USA, Europe and Japan.Jpn J Clin Oncol. 2013; 43(11): 1157–1158. 10.1093/jjco/hyt166 [DOI] [PubMed] [Google Scholar]
- 11. von Eschenbach AC, Buetow K. Cancer Informatics Vision:caBIG. Cancer Inform. 2007; 2: 22–24. [PMC free article] [PubMed] [Google Scholar]
- 12. Gadaleta E, Lemoine NR, Chelala C. Online resources of cancer data: barriers, benefits and lessons. Brief Bioinform. 2011; 12(1), 52–63. 10.1093/bib/bbq010 [DOI] [PubMed] [Google Scholar]
- 13. Buetow KH. Cyberinfrastructure: empowering a ‘‘thirdway” in biomedical research. Science. 2005; 308: 821–824. [DOI] [PubMed] [Google Scholar]
- 14. Saltz J, Oster S, Hastings S, Langella S, Kurc T, Sanchez W, et al. caGrid: design and implementationof the core architecture of the cancer biomedical informatics grid. Bioinformatics. 2006; 22: 1910–1916. [DOI] [PubMed] [Google Scholar]
- 15. Butler D. Translational research: crossing the valley of death. Nature. 2008; 453(7197): 840–842. 10.1038/453840a [DOI] [PubMed] [Google Scholar]
- 16. Nishigaki R, Osaki M, Hiratsuka M, Toda T, Murakami K, Jeang KT, et al. Proteomic identification of differentially-expressed genes in human gastric carcinomas. Proteomics. 2005; 5(12): 3205–3213. [DOI] [PubMed] [Google Scholar]
- 17. Yang YX, Xiao ZQ, Chen ZC, Zhang GY, Yi H, Zhang PF, et al. Proteome analysis of multidrug resistance in vincristine-resistant human gastric cancer cell line SGC7901/VCR. Proteomics. 2006; 6(6): 2009–2021. [DOI] [PubMed] [Google Scholar]
- 18. van Grieken NC, Aoyama T, Chambers PA, Bottomley D, Ward LC, Inam I, et al. KRAS and BRAF mutations are rare and related to DNA mismatch repair deficiency in gastric cancer from the East and the West: results from a large international multicentre study. Br J Cancer. 2013; 108(7): 1495–1501. 10.1038/bjc.2013.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei ZH, Guo WH, Wu J, Suo WH, Fu GH. A nonsense mutation in the Xeroderma pigmentosum complementation group F (XPF) gene is associated with gastric carcinogenesis. Gene. 2014; 537(2): 238–244. 10.1016/j.gene.2013.12.061 [DOI] [PubMed] [Google Scholar]
- 20. Huang CH, Chiou SH. Clinical proteomics identifies potential biomarkers in Helicobacter pylori for gastrointestinal diseases. World J Gastroenterol. 2014; 20(6): 1529–1536. 10.3748/wjg.v20.i6.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Raza Y, Khan A, Farooqui A, Mubarak M, Facista A, Akhtar SS, et al. Oxidative DNA Damage as a Potential Early Biomarker of Helicobacter pylori Associated Carcinogenesis. Pathol Oncol Res. 2014; 20(4): 839–846. 10.1007/s12253-014-9762-1 [DOI] [PubMed] [Google Scholar]
- 22. Kwon OH, Park JL, Baek SJ, Noh SM, Song KS, Kim SY, et al. Aberrant upregulation of ASCL2 by promoter demethylation promotes the growth and resistance to 5-fluorouracil of gastric cancer cells. Cancer Sci. 2013; 104(3): 391–397. 10.1111/cas.12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xian XS, Park H, Choi MG, Park JM. Cannabinoid receptor agonist as an alternative drug in 5-fluorouracil-resistant gastric cancer cells. Anticancer Res. 2013; 33(6): 2541–2547. [PubMed] [Google Scholar]
- 24. Gao Ling, Chen Xiaohua, Ma Zenghai, Li Fengsheng. STYK1/NOK—A Potential Radiotherapeutic Target and Biomarker for Gastric Cancer and Cervical Cancer. Journal of Cancer Therapy. 2013; 4(2): 575–577. [Google Scholar]
- 25. Du P, Zhang W, Wang H, Chen C. Comparative genomic analysis of Escherichia coli O104:H4 stx2 prophage reveals a potential new method to identify virulence factors. Genome. 2012; 55(9): 697–700. 10.1139/g2012-053 [DOI] [PubMed] [Google Scholar]
- 26. Guo Y, Wang H, Li Y, Song Y, Chen C, Liao Y, et al. Genome of Helicobacter pylori strain XZ274, an isolate from a tibetan patient with gastric cancer in China. J Bacteriol. 2012; 194(15): 4146–4147. 10.1128/JB.00804-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hao Y, Yu YY, Wang LS. IPO-38 is identified as a novel serum biomarker of gastric cancer based on clinical proteomics technology. J Proteome Res. 2008; 7(9): 3668–3677. 10.1021/pr700638k [DOI] [PubMed] [Google Scholar]
- 28. Jiang J, Liu W, Guo X. IRX1 influences peritoneal spreading and metastasis via inhibiting BDKRB2-dependent neovascularization on gastric cancer. Oncogene. 2011; 30(44): 4498–4508. 10.1038/onc.2011.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Chen-fei, Ma Tao, Su Yang. UGT1A1 Gene Polymorphisms and the Toxicities of FOLFIRI in Chinese Han Patients with Gastrointestinal Cancer. Anti-Cancer Agents in Medicinal Chemistry. 2013; 13(2): 235–241. [DOI] [PubMed] [Google Scholar]
- 30. Zhao X, Liu Q, Cai Q, Li Y, Xu C, Li Y, et al. Dr.VIS: a database of human disease-related viral integration sites. Nucleic Acids Res. 2012; 40: D1041–D1046. 10.1093/nar/gkr1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are availabe via Datadryad (https://datadryad.org). The unique accession number is: doi:10.5061/dryad.271dk.