Summary
Septins are essential for the completion of cytokinesis. In budding yeast, Saccharomyces cerevisiae, septins are located at the bud neck during mitosis and are closely connected to the inner plasma membrane. In vitro, yeast septins have been shown to self-assemble into a variety of filamentous structures, including rods, paired filaments, bundles and rings [1–3]. Using electron tomography of freeze-substituted section and cryo-electron tomography of frozen sections, we determined the three dimensional organization of the septin cytoskeleton in dividing budding yeast with molecular resolution [4,5]. Here we describe the detailed procedures used for our characterization of the septin cellular ultrastructure.
Keywords: septin, budding yeast, cytokinesis, cryo-tomography, image processing, cryo-sectionning
1. Introduction
Septins were discovered through a screen for cell division cycle mutants in budding yeast more than forty years ago [6]. Septins are indeed essential for cytokinesis, and play a variety of molecular roles, including the recruitment of proteins like myosin2 [7] or serving as a diffusion barrier for membrane-bound proteins [8] Furthermore, the self-assembly of septin has been shown to be required for cell survival in yeast [9]. From early electron microscopy studies using standard preparation methods [10,11] we know that septins assemble in concentric rings at the bud neck, but several studies point to a variable organization and orientation of septins through the cell cycle, likely regulated by post-translational modifications [12]. In situ FRAP experiments have shown that the assembly of septins at the bud neck is dynamic [12], while fluorescence polarization studies indicate a global reorientation of the septin filaments at the onset of cytokinesis [13]. In agreement with these in vivo observations, we have characterized a variety of septin structures in vitro depending on ionic strength [1], the nature of the septin subunit composition [2] or the phosphorylation state of septins [2]. In high salt (above 200mM) the mitotic septin complex made of Cdc3, Cdc10, Cdc12 and Cdc11 exists as a 32 nm long octameric, symmetric, rod-like structure [1]. At lower ionic strength, these rods self-assemble into long paired filaments resembling railroad tracks, or into bundles of filaments [1]. Remarkably, replacing Cdc11 by Shs1, a less essential and sub-stoichiometric septin, induces the formation of ring-like structure or, for a specific phosphomimetic Shs1 mutation, into gauzes of orthogonal filaments [2]. Hence, the organization of septins is highly variable and plastic. In order to get insight into the organization of septin filaments in situ, it is necessary to use advance electron microscopy methods for sample preparation and visualization that enable the quantitative description under optimized cellular preservation. Using electron tomography we have characterized the three-dimensional organization of septin filaments in dividing budding yeasts [4,5]. This chapter describes the methods we used for sample preparation, data collection and computation.
2. Materials
The methods presented here require specialized equipment for sample preparation and data collection. Below we list the material we have used in our studies, Electron microscopy facilities are often equipped with these or similar tools alternatives.
2.1. Preparation of resin embedded samples for sectioning and EM analysis
Yeast extract peptone glucose, commonly referred to as YPD medium (1% yeast extract, 2% peptone, 2% glucose), autoclaved for 20 minutes at 121°C. To prevent burning the glucose, sterile, filtered glucose can be added after autoclaving. Otherwise, the media will darken and cell growth will not be optimal.
Incubator and shaker (to be set at 30°C) able to contain 2 L cell culture flasks.
Spectrophotometer.
Vacuum filtration device with a pump and a borosilicate glass funnel, equipped with a fritted glass of 25 mm in diameter (Millipore). 0.45 μm polycarbonate filters are used.
High pressure freezing device (EMPACT2-RTS, Leica) and 100 μM deep membrane carriers (Leica). Hexadecene (Fluka) to be used to coat the membrane carriers.
Cryogenic vials (Nalgene) of 2 mL for sample conservation at liquid nitrogen temperature (in a nitrogen tank) or freeze substitution.
Freeze substitution media: 1% osmium tetraoxide, 0.1% uranyl acetate, 5% water in freshly opened dry acetone. The freeze substitution medium can be prepared in advance and stored in liquid nitrogen. We used a Leica AFS2 freeze substitution apparatus.
Epon resin solutions in acetone at increasing concentrations of 30 %, 60 % and 100 %. Epon polymerization molds and oven to be set at 60°C.
Ultamicrotome (Ultracut E, Reichert) equipped with either a homemade glass knife (with a glass “knifemaker”) or a diamond knife of 4.5 mm (Diatome). One of your own eyelash glued (with nailpolish) to a toothpick to be used to handle the sections. Dumont tweezers N7 to be used to hold the grids hexagonal copper grids (mesh size of 100) coated with formvar (0.5%).
Solutions of uranyl acetate (2%) and lead citrate (70%) in methanol, and 10 nm gold beads (Aurion).
A carbon evaporator apparatus.
2.2. Sample cryo-sectioning
YPD medium as described above.
Vacuum filtration device as described above.
High pressure freezing device (EMPACT2-RTS, Leica) and copper tubes (350 μm inner diameter, Leica).
Cryo-ultramicrotome (UC7, Leica) equipped with an anti-contamination “cryosphere” (Leica) and a CRION ionizer (Leica).
Trimming diamong knife (Diatome) and 3.5 mm cryo-immuno diamond knife (Diatome).
Cflat holey carbon grids (Protochips) coated with gold beads (Aurion) and an eyelash glued to a 20 cm long stick.
2.3. Data collection
Data collection for resin embedded samples, at room temperature: A dual axis holder (model 2040, Fischione) enables the collection of two, perpendicular tilt series. For data collection we use an FEG CM200 microscope (FEI) equipped with a 4K×4K CCD camera (Ultrascan 4000, Gatan). We use Digital Micrograph software (Gatan) for automated data collection.
Data collection for cryo-samples: we mount the sample on a 626 cryo-holder (Gatan) and visualize it using a Tecnai 12 electron microscope (FEI) equipped with a 4K×4K Eagle camera (FEI) We use the software suite Xplore 3D (FEI) for automated data collection.
2.3. Data processing
3. Methods
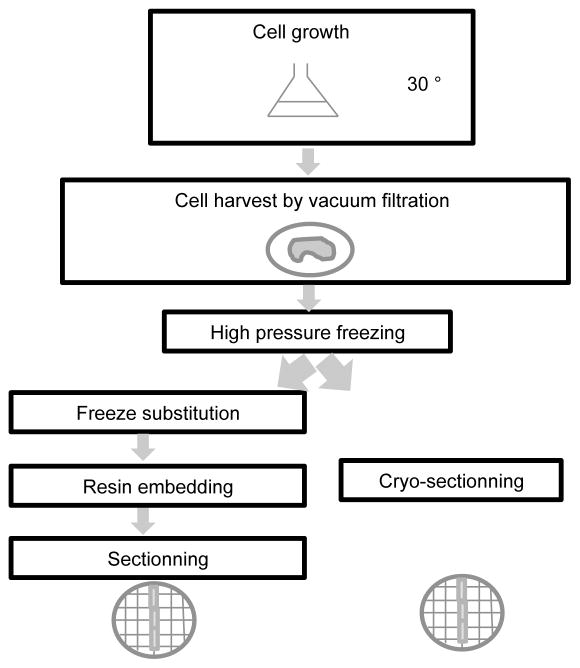
For tomography and cryo-tomography analysis two different strategies can be employed as described in figure 1. Either the harvested cells are directly sectioned after vitrification and then analyzed in a hydrated state or the vitrified cells are gradually freeze substituted in the presence of fixative and staining chemicals, embedded in resin and sectioned at room temperature.
Figure 1. Schematic of the procedure used for sample preparation for electron microscopy analysis.
After cell growth, the cells are harvested by vacuum filtration and vitrified by high pressure freezing. Two strategies can then be employed. Either the vitrified cells are cryo-sectioned right before observation in the scope (or storage) or the cells are freeze substituted, embedded in resin at room temperature and sectioned.
3.1. Sample preparation for resin-embedded cells
Grow yeast cells at 30°C in Erlenmeyer flasks (growing 500 mL of media in 2L flasks should provide cells with sufficient oxygenation). Grow the cells to mid-exponential phase (OD600 nm = 0.4–0.6). Measure the Optical density at 600 nm with a spectrophotometer every 20 minutes.
Harvest the yeast cells by vacuum filtration as described in McDonald (2007) [16] (see note 1). After filtration, a yeast thick paste is isolated on top of the filtration membrane. Transfer the membrane in a wet petri dish (see note 2).
Coat the membrane carriers with hexadecene, which is a cryo-protectant. Use a sterile toothpick to transfer some of the yeast paste to the 100 μm deep membrane carrier already attached to its stand. This step is easier using binoculars. Then transfer the membrane carrier into the high-pressure freezing machine and vitrify the sample at once (see note 3).
Transfer the vitrified samples into cryo vials containing the frozen freeze-substitution media and either store in liquid nitrogen (see note 4) or process further for freeze substitution. During freeze substitution, steadily and accurately raise the temperature from −90°C to −25°C in increments of 2°C per hour. Then raise the temperature to 0°C in 5°C per hour increments (see note 5).
Rinse the samples 3 times for 10 minutes in pure acetone. The samples should be now darker due to the presence of osmium tetraoxide in the media. The samples are most likely to detach from the membrane carrier by themselves during the rinses.
Perform resin embedding by consecutive baths in epon/acetone solutions of increasing epon concentrations: 30, 60 and 100% for one hour or more each (see note 6). During the baths, gently mix the samples on a shaker. To remove any trace of acetone, wash the samples in three consecutive solutions of pure acetone for at least one hour each. Then put the samples into polymerization molds and label them (see note 7) before putting them into an oven at 60°C for 48 hours.
Before sectioning, trim the resin-embedded sample using a homemade glass knife. Then section the resin block using a 4.5 mm diamond knife, using standard procedures. Depending on desired use, choose the proper thickness for sectioning (see note 8). Use the eyelash to move the sections across the air/water interface. Grab the sections with the formvar coated hexagonal grid held with tweezers and coming from below. Then let the grid dry.
Evaporate carefully a thin layer of carbon on top of the grid on the sample side (see note 9). The grids are then ready to be analyzed by electron microscopy.
3.2. Sample preparation of vitrified samples
Grow the yeast cells to mid-log phase as described above (see step 1 in section 3.1) in YPD media containing 20% of sterile filtered dextran (see note 10).
Harvest the cells by vacuum filtration and keep a few milliliters in the funnel without waiting to obtain a yeast paste (see note 11).
Suck some yeast solution into the copper tube mounted on its holder. Make sure the tube is entirely filled in order to ensure good vitrification. Quickly transfer the sample into the high pressure freezing machine for vitrification and operate following standard procedures.
Either store the copper tubes in liquid nitrogen or transfer it into a cryo-ultramicrotome.
Coat some C-flat holey grids with 10 nm gold beads by incubating them upside down on a gold beads solution, followed by several rinses in PBS buffer.
Cool the cryo-ultramicrotome down at −150°C. Attach the copper tube safely into the jaws of the cryo-ultramicrotome and trim one of its ends into a pyramidal shape using the trimming diamond knife (see note 12).
Then use the cryo-immuno knife to section 50 nm thick ribbons. Hold the ribbon with an eyelash attached to a 20 cm long stick. When the ribbon is long enough, hold a pre-cooled grid below the ribbon and invert the charge of the “CRION Ionizer” to make the ribbon stick onto the grid (see note 13). The cryo-sectioning procedure has been described in details by Pierson et al. [17].
Then transfer the grid, kept under liquid nitrogen, into the cryo-holder and either store in liquid nitrogen or transfer to the microscope.
3.3. Data collection for resin-embedded samples
Insert a good grid (see note 14) into the dual axis holder.
Irradiate the area of interest for about 30 minutes to shrink the resin to its final thickness (see note 15). Choose an area of interest in the center of a hexagonal copper hole to prevent a grid bar from coming into the field of view.
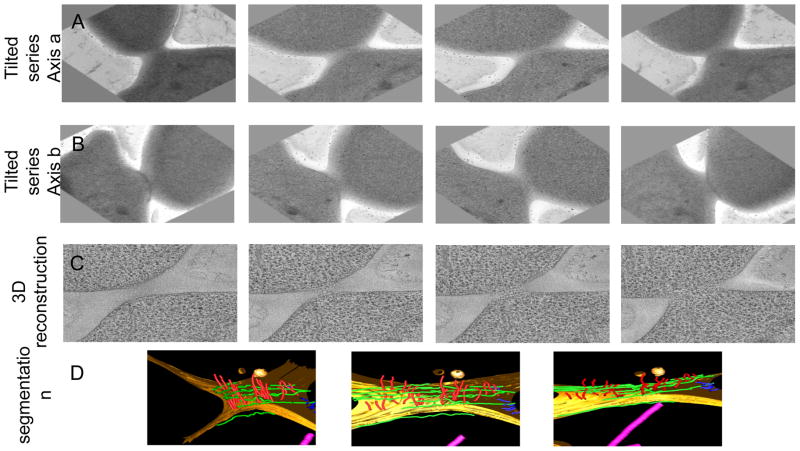
Make sure that the sample is at eucentric height. Tilt the sample from −70° to 70° to make sure a tilt series can be acquired within this range of angles. Collect a tilt series (see note 16) as shown in figure 2.A. On resin sections, tracking and focusing of the sample can be performed on the area of interest. Choose a reasonable magnification to get a resolution that will enable the visualization of filaments within the cell (see note 17).
Rotate the sample by 90° to collect a second tilt serie as shown in figure 2.B (see note 18). Once the sample has been rotated, move the grid have the area of interest in the center of the screen. Collect the second tilt series.
Figure 2. Snapshots from a reconstructed tomogram and its corresponding model.
Panel A. Images from a tilted series collected from a resin-embedded sample. Panel B. After the sample has been rotated by 90°, another tilted series was collected from which a few images are shown. Panel C. Images from different slices of the corresponding 3D reconstruction. Panel D. Model obtained after segmentation of the reconstructed tomogram.
3.4. Data collection for cryo-sections
Place the frozen grid into the cryo-holder, keeping the sample always at liquid nitrogen temperature.
Make sure the ribbon is still vitrified (see note 19). Using low-dose procedures, find a dividing cell in the center of a grid square.
Collect a tilt series from −60 to 60° in two degree increments (~ 60 images), using a total dose of about 50 electrons per Å2. Focus and tracking have to be performed on an area a few microns away from the area of interest to prevent any irradiation and burning of the sample.
3.5. Data processing
Align the tomographic dataset using the fiducial gold beads and software like IMOD [14] (see note 20).
Perform the 3D reconstructions using weighted back projection (for both tilted series in the case of a dual dataset from a resin-embedded sample).
For resin-embedded samples, recombine the 3D reconstruction using the datasets from the two perpendicular tit series. One example is displayed in figure 2.C.
For datasets from cryo-samples, enhance the signal to noise ration trying different types of filtering software (see note 21).
Segment the sample by tracing recognizable features (filaments, vesicles, microtubules, membranes…) through sequential consecutive slices in the depth of the 3D reconstruction (see note 22) as shown in figure 2.D.
Acknowledgments
We thank K. McDonald for training and advice on sample preparation, Manfred Auer and Jeff Triffo for tomography advise, Jason Pierson and Peter Peters for training and hosting A.B. during the cryo-sectioning studies, and Patricia Grob for technical support. This work was funded by a Jane coffin Childs Research post-doctoral fellowship (A.B) and by the Howard Hughes Medical Institute (E.N).
Footnotes
For optimum cell preservation, cells should not be harvested by centrifugation. The mechanical stress induced by centrifugation can disrupt cellular ultrastructural features.
A water soaked filter paper can be set in the petri dish. It prevents the cells from being dried. The yeast paste can be preserved this way for several minutes, the time needed to transfer the sample into the high pressure freezing apparatus.
The transfer needs to be carried out as quickly as possible in order to prevent the cells from drying before being vitrified.
If the freeze substitution device is not readily available, the vitrified samples can be kept in liquid nitrogen for as long as necessary.
It is convenient to start the freeze substitution procedure just before the weekend. The sample will be ready, at 0°C, Monday morning. It is important to make sure that the liquid nitrogen tank connected to the apparatus is full.
The epon resin can be kept at −20°C and defrosted before use. To replace a bath one should use plastic Pasteur pipettes and make sure the samples are not disposed of with the discarded solution. The discarded and rinsing solutions need to be discarded in a dedicated chemical waste container. The preparation of solutions has to be performed under a hood for safety reasons.
To label the samples, you can insert small pieces of paper written with a lead pencil inside the molds.
50 nm sections are fine for 2D imaging. Thicker sections (150 to 200 nm) can and should be used for tomography (irradiation with the electron beam will thin down the sections).
Carbon evaporation prevents charging and subsequent drifting of the sample during data collection. While evaporating carbon, make sure not to produce any sparks, which can damage the formvar film and tear it apart.
Dextran is a cryo-protectant that helps the vitrification process and also makes cryosectionning easier.
Since the media is more viscous because of the dextran, do not wait until a yeast paste is obtained. There will be enough cells when harvesting 2–3 mLs from an initial 500 mL.
Usually the tips of the tubes are not perfectly vitrified, as seen by the presence of holes and cracks in the ice. Trim the first 50 μm of the tube before carving the pyramid. The surface of the vitrified ice, when inspected through the binoculars, should look dark and shiny.
This step is the most delicate and challenging part of the method. Tune the sectioning speed to make it comfortable for yourself. The “CRION ionizer” makes the attachment of the vitrified ribbon onto the grid easier than using a mechanical “stamping” device. The ribbon has to be perfectly attached to the grid and has to lay flat on top of it. Otherwise, cryo-tomography and 3D reconstruction will be difficult to carry out.
It is wise to pre-screen the different grids and samples one has prepared to choose the best one for data collection. The quality of the stain, the density of cells in the sample, and the sample preservation (e.g. the quality of the cellular membranes) are important factors to keep in mind.
Pre-irradiation of the sample is needed to shrink the resin to its final thickness (by about 30%). Otherwise, the resin will shrink during data collection, making it challenging to align the images and to obtain a consistent reconstruction.
Depending on the data collection software use, this procedure will be different (some steps may or not be automated). The quality of the data (tracking and focusing have to be correct) is essential to be able to carry out any further processing.
About 4 Angström per pixels should be enough. We collect our data at a magnification of 50,000 on a 4K×4K camera and bin by 2, resulting in a final resolution of about 4 Å per pixels.
The dual axis holder from Fischione enables a mechanical rotation of the sample gradually so that one can still follow an area of interest (in our case a dividing cell) throughout the rotation. This way the cell of interest is kept in the center of the screen by moving the sample in the x,y directions. Collecting two tilt series at 90° from one another reduces the “missing wedge” in the tomographic data to a “missing pyramid“ and thus minimizes the anisotropy of the tomographic reconstruction.
Check the diffraction pattern to make sure the ice is vitreous. Additionally, the ribbon has to lay perfectly flat onto the grid. Otherwise, this will affect negatively the quality of the reconstruction.
IMOD [14] is a free, downloadable software package (http://bio3d.colorado.edu/imod/). It is powerful, user friendly and widely used in the field. We used IMOD for almost all the processing steps. The IMOD website includes comprehensive instructions and tutorials as well as practical advice and help from the developers. The initial dataset has to be in mrc format.
One can use a nonlinear anisotropic diffusion filter [18] for instance, available in IMOD or TOMOAND [19] (https://sites.google.com/site/3demimageprocessing/tomoand).
For most of our datasets we carry out this task manually using IMOD, but Amira [15] enables some automated rendering in the process.
References
- 1.Bertin A, McMurray MA, Grob P, Park SS, Garcia G, 3rd, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci U S A. 2008;105(24):8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia G, 3rd, Bertin A, Li Z, Song Y, McMurray MA, Thorner J, Nogales E. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J Cell Biol. 2011;195(6):993–1004. doi: 10.1083/jcb.201107123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertin A, McMurray MA, Thai L, Garcia G, 3rd, Votin V, Grob P, Allyn T, Thorner J, Nogales E. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404(4):711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertin A, McMurray MA, Pierson J, Thai L, McDonald KL, Zehr EA, Garcia G, 3rd, Peters P, Thorner J, Nogales E. Three-dimensional ultrastructure of the septin filament network in Saccharomyces cerevisiae. Mol Biol Cell. 2012;23(3):423–432. doi: 10.1091/mbc.E11-10-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertin A, Nogales E. Septin filament organization in Saccharomyces cerevisiae. Commun Integr Biol. 2012;5(5):503–505. doi: 10.4161/cib.21125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwell LH. Genetic control of the cell division cycle in yeast. II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59(1):183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- 7.Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305(5682):393–396. doi: 10.1126/science.1099892. 305/5682/393 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5(5):841–851. doi: 10.1016/s1097-2765(00)80324-x. S1097- 2765(00)80324-X [pii] [DOI] [PubMed] [Google Scholar]
- 9.McMurray MA, Bertin A, Garcia G, 3rd, Lam L, Nogales E, Thorner J. Septin filament formation is essential in budding yeast. Dev Cell. 2011;20(4):540–549. doi: 10.1016/j.devcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69(3):717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soll DR, Mitchell LH. Filament ring formation in the dimorphic yeast Candida albicans. J Cell Biol. 1983;96(2):486–493. doi: 10.1083/jcb.96.2.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4(3):345–357. doi: 10.1016/s1534-5807(03)00061-3. S1534580703000613 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Vrabioiu AM, Mitchison TJ. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature. 2006;443(7110):466–469. doi: 10.1038/nature05109. nature05109 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 15.Stalling D, Westerhoff M, Hege H-C, Hansen C, Johnson C. the visualization handbook. elsevier; New york: 2005. Amira, a highly interactive system for visual analysis; pp. 749–767. [Google Scholar]
- 16.McDonald K. Cryopreparation methods for electron microscopy of selected model systems. Methods Cell Biol. 2007;79:23–56. doi: 10.1016/s0091-679x(06)79002-1. [DOI] [PubMed] [Google Scholar]
- 17.Pierson J, Fernandez JJ, Bos E, Amini S, Gnaegi H, Vos M, Bel B, Adolfsen F, Carrascosa JL, Peters PJ. Improving the technique of vitreous cryo-sectioning for cryo-electron tomography: electrostatic charging for section attachment and implementation of an anti-contamination glove box. J Struct Biol. 2010;169(2):219–225. doi: 10.1016/j.jsb.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Frangakis AS, Hegerl R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol. 2001;135(3):239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez JJ, Li S. An improved algorithm for anisotropic nonlinear diffusion for denoising cryo-tomograms. J Struct Biol. 2003;144(1–2):152–161. doi: 10.1016/j.jsb.2003.09.010. [DOI] [PubMed] [Google Scholar]