Abstract
Cyclooxygenases (COXs) are enzymes that play a vital role in the inflammatory cascade through the generation of prostaglandins. Their over-expression has been implicated in numerous diseases. In particular, over-expression of COX-2 has been shown to be a predictive biomarker for progression of pre-malignant lesions towards invasive cancer in various tissues. This makes the early detection of COX-2 expressing lesions of high clinical relevance. Herein we describe the development of the first self-immolating trigger which targets COXs. We incorporated our trigger design into 2 activatable fluorogenic probe and demonstrated COX-specific activation in vitro. Experimental data revealed probe activation was likely caused by solvent-exposed amino acids on the surface of the COXs. Overall, the probes reported here mark the first step towards developing self-immolating imaging/therapeutic agents targeted to specific COXs.
Introduction
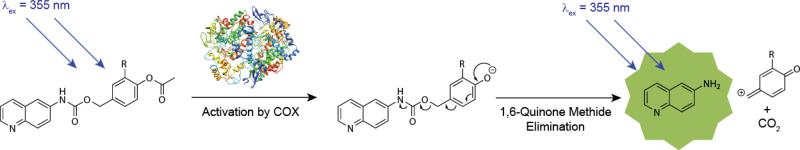
Self-immolating molecules hold tremendous promise for the imaging and treatment of diseases. They are designed to be selectively cleaved by a biological target, leading to spontaneous degradation and the release of a previously inactive fluorophore or drug.1 In this way imaging probes and cytotoxic drugs can be delivered to cells which over-express the target enzyme. The development over the last 30 years of self-immolating macromolecules has allowed for a single cleavage event to result in the release of numerous active agents, greatly increasing the efficacy of this approach.1,2 However, for an enzyme to be targeted in this manner, a self-immolating trigger specific to that enzyme must be found. Herein we describe our initial efforts towards developing the first COX-2 specific self-immolating triggers. We incorporated our self-immolating trigger design into activatable fluorophores (Figure 1) for 2 reasons: the release of a fluorescent compound provides a convenient signal to assess the efficacy of the trigger and COX-specific imaging agents are themselves of considerable interest. COX-2 targeted fluorescent imaging agents have been reported previously,3–5 however to the best of our knowledge none have been based on a self-immolating structure.
Figure 1.
Molecular Design of COX-Sensitive Fluorogenic Probes. (A) Overall Design Strategy. (B) Structure of CP-1 and CP-2.
The COX enzymes6–8 sit at a crucial node point in the inflammatory cascade, catalyzing the conversion of arachidonic acid into prostaglandin H2, the precursor of several members of the prostaglandin family, including prostaglandin E2 (PGE2). There are 2 COX isoforms, COX-1 and COX-2, which are both homo-dimers of approximately 70 kDa. They share 60% of their amino acid sequence and their 3-D structures are virtually super-imposable. COX-1 is constitutively expressed in a wide range of tissues (kidney, stomach, lung, small intestine and colon) and is thought to regulate baseline prostaglandin synthesis. In contrast, COX-2 is normally produced only in a few tissues, including specific regions of the kidney, central nervous system and seminal vesicles. However, COX-2 expression may increase dramatically in response to inflammatory stimuli. The link between chronic inflammation and carcinogenesis is well established,9 and the COX-2/PGE2 pathway is now well recognized as an instrumental signaling pathway in cancer progression. Over-expression of COX-2 has been reported not only in primary tumors but also in morphologically normal pre-malignant lesions and is linked to poor prognosis and disease progression,10,11 making this enzyme attractive for both early detection imaging and targeted drug delivery applications.
The design of our COX-targeted self-immolating triggers is outlined in Figure 1. The core is based on the structure of aspirin, which is known to selectively acetylate both COX-1 and COX-2 at the equivalent serine residue (Ser-530 in COX-1 and Ser-516 in COX-2).6 Deacetylation of the probes by COXs reveals a phenol moiety, provoking a 1,6-quinone-methide elimination to release a previously masked fluorophore (Figure 2). Activatable fluorescent probes have been widely used in biological imaging due to the improved signal-to-background ratios compared to their “always on” counterparts. They have been used to study a plethora of analytes, including metal ions, reactive oxygen species, reactive nitrogen species and thiols (see recent reviews).12–14 With respect to enzymes, several general strategies to achieve enzyme-specific fluorescent enhancement have been developed.13,15 One of the most common is to link a compatible fluorophore/quencher pair via an enzymatically cleavable linker. The resulting probe is optically silent due to the Förster resonance energy transfer (FRET) interaction between the fluorophore/quencher pair, but upon cleavage these moieties are separated and the signal from the fluorophore is restored. This probe design has found widespread use in the field, especially for the detection of proteases. Compounds based on a coumarin motif (e.g. fluorescein, rhodamine, umbelliferone) fluoresce due to intramolecular charge transfer (ICT), a process which can be altered via conversion of the 7-hydroxy/amino moieties to the analogous esters/amides. The resulting change in fluorescent properties can be used to create fluorogenic probes in which ester/amide cleavage causes fluorescent enhancement at carefully selected wavelengths. The activity of numerous enzymes, including proteases, phosphatases and esterases, has been interrogated using this approach. The detection of other enzymes, such as dehydrogenases and monoamine oxidase, can be realized using enzymatic activity to convert non-fluorescent molecules into optically active reporters. For our studies we selected 6-aminoquinoline, an ICT fluorophore, as an optical reporter on the basis of molecular modeling studies (vide infra), which suggested that this sterically undemanding moiety would not inhibit the binding of our probes to COXs.
Figure 2.
Mechanism of Probe Activation by COXs. The probe is optically silent until deacetylation by a COX reveals a phenol group, resulting in a 1,6-quinone-methide elimination and the release of fluorescent 6-aminoquinoline.
Aspirin is known to be 10-100 fold more active against COX-1 than COX-2. Because of the larger binding pocket in COX-2, the orientation of the acetyl functionality of aspirin towards its target serine residue is sub-optimal.8 Given the high relevance of COX-2 to diseases, such as cancer, we set out to design probes based on aspirin but with greater activation efficacies for COX-2 compared to COX-1. Our strategy was inspired by previous studies which showed that the glyceryl ester derivative of aspirin retained its anti-inflammatory properties while by-passing gastric irritation via the lipid metabolic pathway,16 this association of activities being characteristic of selective COX-2 inhibitors. Hence, taking a similar synthetic approach, we synthesized CP-1. The carboxylate group on aspirin binds ionically to Arg-120 in the cyclooxygenase active site, positioning it for transacetylation.6 Seminal work by Kalgutkar et al. showed that replacing this carboxylate with an alkyl sulfide moiety leads to the generation of selective COX-2 inhibitors.17,18 Based on this work we synthesized CP-2, with a thioocy-2-ynyl group in place of the carboxylate. In both cases, we predicted that the replacement of the carboxylate would lead to COX-2 selectivity.
Results and Discussion
Synthetic Chemistry
The syntheses of CP-1 and CP-2 are outlined in Scheme 1. The synthesis of CP-1 began with the esterification of 5-formylsalicylic acid (1) under Mitsunobu conditions with a di-protected glycerol derivative (2). Purification over silica afforded amixture of 3 and small amounts of di-iso-propyl azodicarboxylate derived impurities. We anticipated that these impurities would not interfere with the next step and hence proceeded to treat 3 with NaBH4 at 0 °C to selectively reduce the aldehyde moiety. Selective acetylation of the phenolic alcohol, using K2CO3 to form the reactive phenolic anion, furnished us with 5.
Scheme 1.
Synthesis of CP-1 and CP-2. ia) PPh3, DIAD, THF; 0 °C to r.t.; 16 h b) NaBH4, MeOH; 0 °C to r.t.; 3 h; 28% over 2 steps ii) K2CO3, Ac2O, DMF; 0 °C; 2 h; 58% iiia) 6-aminoquinoline, pyridine, triphosgene, DCM/toluene; 0 °C to reflux; 16 h b) TBAF, THF, acetic acid; r.t.; 15 h; 34% over 2 steps iv) HSO3Cl; r.t.; 15 h; 53% va) PPh3, THF/toluene b) H2SO4, MeOH; reflux; 2 days c) LiAlH4, THF; 0 °C to reflux; 15 h d) 1-chloro-2-octyne, KHCO3, DMF; r.t.; 15 h; 39% over 4 steps vi) K2CO3, Ac2O, DMF; 0 °C; 1 h; 79% viia) Na2CO3, triphosgene, toluene; 0 °C to r.t.; 4 h b) 6-aminoquinoline, THF; r.t.; 1 h; 26% over 2 steps. DIAD = diisopropyl azodicarboxylate; TBAF = tetrabutylammonium fluoride; TBDMS = tert-butyldimethylsilyl.
6-Aminoquinoline was converted to the isocyanate with triphosgene, which was then coupled with 5 at reflux in toluene. The crude material from this step was treated with TBAF in THF to remove the TBDMS protecting groups. Chromatographic purification of this compound proved challenging due to its instability in the presence of silica and the polar solvents required to elute it. However, CP-1 could be crystallized from a mixture of H2O, acetonitrile and acetic acid. HPLC analysis of CP-1 (Figure S1) revealed that it was 95% pure with two minor impurities, one of which was 6-aminoquinoline (~1%). Given the instability of CP-1 in aqueous solutions (vide infra), it is probable that the presence of 6-aminoquinoline was the result of CP-1 degradation during the HPLC analysis. Hence we judged that the material was of sufficient purity to carry forth for in vitro testing. 4-Hydroxybenzoic acid was the starting material for the synthesis of CP-2. Following a published procedure19, an aromatic sulfonyl chloride was installed ortho to the phenolic alcohol. Without the need for chromatographic purification of any of the intermediates, the sulfonyl chloride was reduced to a thiol using PPh3, the carboxylic acid was esterified and then reduced to a benzylic alcohol with LiAlH4. Thiophenol 8 was then alkylated with 1-chloro-2-octyne to generate 9. Selective acetylation was achieved as described above. However, for reasons which are unclear, we were not able to obtain the carbamate using the methodology used to synthesize CP-1. Instead, 10 was treated with triphosgene to form the analogous chloroformate, which was subsequently coupled in situ with 6-aminoquinoline to generate CP-2. The lower polarity of CP-2 allowed us to purify it via silica chromatography. The purity of CP-2 was over 98% by HPLC, again sufficient for in vitro testing. No 6-aminoquinoline release was observed by HPLC, likely a result of the greater stability of CP-2 against hydrolysis (vide infra).
Spectroscopy
The spectral properties of CP-1 and CP-2 were investigated via UV-visible and emission spectroscopy. Three solvents were used: water, water and methanol (1:1, v:v) and a mixture of saturated NaHCO3, water and methanol (1:1:2, v:v:v). The last solvent system was chosen to deprotect the phenolic acetates,20 hence allowing us to confirm the release of 6-aminoquinoline via our proposed mechanism. Solutions of either probe in DMSO (10 mgmL−1) were diluted to a final concentration of 0.1 mgmL−1 prior to the measurement of absorption and emission (λex = 355 nm) spectra. In all cases the spectra in water and the mixture of water and methanol were virtually identical and for clarity only the spectra in water are shown. Comparison of the spectra in water to those in the deprotection solution showed an increase in absorbance from 300 to 360 nm for both CP-1 and CP-2, with the maxima being blue-shifted by approximately 10-15 nm (Figure S2). As shown in Figure 3, the changes in the emission spectra were more profound. A new peak, centered atapproximately 540 nm, appeared for both CP-1 and CP-2, an observation which is consistent with the release of 6-aminoquinoline under these conditions.21,22 Minor changes were observed at lower wavelengths. In the case of CP-1, the emission at 430 nm was red-shifted by approximately 20 nm and was reduced in intensity whereas there was a gain in intensity of the analogous peak for CP-2. The cause of these minor differences is unclear. Nonetheless, the release of 6-aminoquinoline was established unambiguously in both cases, as documented by the emergence of a new peak at 540 nm.
Figure 3.
Emission spectra for (A) CP-1 and (B) CP-2. Spectra were measured at 0.1 mgmL−1 in water and deprotection solution (2:1:1, v:v:v, methanol:water:saturated NaHCO3). Release of 6-aminoquinoline via removal of an acetyl moiety by the deprotection solution is clearly documented by the appearance of a new peak around 540 nm.
Probe Stability
COXs are reported to display catalytic activity between pH 6.0 and 9.023, hence we measured the stability of our probes at pH 6.0 (100 mM citrate-PBS), 7.4 (PBS), 8.0 (100 mM Tris) and 9.0 (100 mM glycine) (Figure S3). Following incubation at 37 °C for 30 mins, the release of 6-aminoquinoline was measured via fluorescence spectroscopy (λex = 355 nm, λem = 535 nm). CP-2 was essentially stable at every pH analyzed, with only a small increase in fluorescence (1.1 a.u.) at pH 9.0 and none in physiological conditions (PBS buffer). In contrast, CP-1 was highly unstable at pH 9.0 (increase of 77.4 a.u.) and significant release of 6-aminoquinoline was measured at pH 7.4 (14 a.u.) and pH 6 (7.9 a.u.).
The greater susceptibility of CP-1 towards hydrolysis was presumably due to the electron-withdrawing effects of the ester functionality that resulted in the stabilization of the incipient phenolate anion.
In Vitro Testing
To evaluate the efficacy of CP-1 and CP-2, we then tested their activation in the presence of commercial purified ovine COX-1 and recombinant human COX-2. Solutions containing 100 μM of either CP-1 or CP-2 were incubated with 0.125 mgmL−1 of COX-1 or COX-2 at 37 °C for 180 minutes in 80 mM Tris buffer (pH 8.0) and the emission at 535 nm (λex = 355 nm) was measured at regular intervals (Figure 4). Activation of each probe by both isozymes was recorded over the entire time-course, but was most rapid over the initial 60 minutes. Hence 60 minutes was chosen as a suitable time-point for subsequent studies.
Figure 4.
Activation of A) CP-1 B) CP-2 by COX-1 and COX-2 over a time-course of 180 mins. Purified COX-1 and COX-2 (0.125 mgmL−1) were incubated with either CP-1 or CP-2 (100 μM) in Tris buffer (80 mM, pH = 8.0, 0.1% Tween 20, 300 μM DDC) at 37 °C for 180 mins and fluorescent readings (λex = 355 nm, λem = 535 nm) were taken at 0, 30, 60, 120 and 180 mins.
Our pH studies had demonstrated that probe hydrolysis was significant at pH 8.0, especially for CP-1. To control for this and ensure that only probe activation by COXs was measured, we also incubated CP-1 and CP-2 in blank buffer solutions containing no enzyme. We then calculated the relative increase in fluorescence for the COX samples compared to the blank (Figure 5). Despite CP-1 exhibiting higher absolute increases in fluorescence compared to CP-2, its hydrolytic instability resulted in lower relative values. Both probes were more reactive towards COX-1 than COX-2, although this effect was orders of magnitude less than reported previously for aspirin, documenting the successful generation of a probe with similar detection potential for COX-1 and COX-2. We also measured probe activation by an equimolar amount (compared to the COXs) of porcine liver esterase under identical conditions. Relative increases in fluorescence similar to those observed for the COXs were measured, indicating that CP-1 and CP-2 were also substrates for this esterase.
Figure 5.
Activation of CP-1 and CP-2 by Purified COX and Porcine Liver Esterase Enzymes in Vitro. Either COX-1, COX-2, albumin, carbonic anhydrase (all 0.125 mgmL−1) or porcine liver esterase (0.309 mgmL−1) were incubated with either CP-1 or CP-2 (100 μM) in Tris buffer (80 mM, pH = 8.0, 0.1% Tween 20, 300 μM DDC) at 37 °C for 60 minutes. Pre-treatment with the COX-2 inhibitor celecoxib (100 mM) was performed at 0 °C for 10 minutes prior to addition of probe. Albumin and carbonic anhydrase were tested as non-enzymatic protein controls. Fluorescence readings (λex = 355 nm, λem = 535 nm) were then taken. Results are reported as fold increase in fluorescent signal compared to a blank solution with no protein.
Pre-incubation of COX-2 with the COX-2 inhibitor celecoxib (100 mM) did not completely ablate the activation of either probe, although a significant reduction (5.5- to 2.5-fold relative to blank) was observed for CP-2 (Figure 5). These results were counter to our expectations, as celecoxib should sterically prevent the probes from reacting with the Ser-516 residue in COX-2. This prompted us to further investigate the mechanism by which the probes were activated. To ensure that the activation was COX-specific, we tested both probes with 2 control proteins for which they have no affinity: albumin and carbonic anhydrase. As expected, no evidence for activation by either control protein was observed (Figure 5). Next, we conducted molecular modelling studies to estimate the relative affinity of the probes for COX-2 compared to celecoxib. AutoDock Vina24 was used to dock the probes into the active site of COX-225 and binding affinities of −8.2, −8.4 and −11.8 kcal/mol were calculated for CP-1, CP-2 and celecoxib respectively. These numbers translate into 300-400 fold weaker binding affinities for our probes compared to celecoxib, suggesting that their binding affinities are approximately 600-800 nM.
The lack of celecoxib-induced inhibition led us to hypothesize that our probes were being activated by other nucleophilic amino acids on COX-2. To examine this, we analyzed a sample of CP-1 treated COX-2 by LC/MS/MS (following tryptic digestion) to identify sites of acetylation (Figure S4). We unexpectedly found 18 putative lysine acetylation sites and 7 serine sites, none of which were Ser-516. This strongly implied that our probes were being activated by solvent exposed nucleophilic amino acids. In light of this data, we postulate that both CP-1 and CP-2 bind to COXs with an orientation which prevents activation by Ser-516 and are subsequently activated by random surface solvent-exposed nucleophilic amino acids. This mechanism of activation may be advantageous as several probes can be activated by a single COX enzyme, increasing the sensitivity of the probes.
In Cellulo Testing
Encouraged by our in vitro data we tested CP-1 and CP-2 for their ability to detect upregulation of COX-2 in cellulo. We chose a murine macrophage cell line, RAW264.7, which is known to upregulate COX-2 upon stimulation with lipopolysaccharide (LPS) and interferon-γ.17 We incubated both stimulated and non-stimulated RAW 264.7 cells with either CP-1 or CP-2 at various concentrations (0 μM, 50 μM, 100 μM and 250 μM) at 37 °C for 1 hour, followed by a thorough PBS wash and live cell imaging via multispectral fluorescence microscopy. Post-processing of the acquired multispectral image cube allowed us to isolate the fluorescence signal at 550 nm. A comparison with data acquired from non-treated cells demonstrated that this signal was not caused by cellular autofluorescence. However, only weak signals were observed at all concentrations for both probes, with no evidence of COX-2 specific fluorescence from stimulated RAW264.7s.
Discussion
Our in vitro experiments clearly demonstrate that self-immolating probes based on an aspirin motif can be activated by the COX proteins. CP-2 proved to be the most effective probe, exhibiting greater relative increases in fluorescence in the presence of COXs than CP-1. A key determinant of its efficacy was its greater stability against hydrolysis. Neither albumin nor carbonic anhydrase reacted with either probe, demonstrating that affinity for COXs is vital for probe activation. Molecular modeling studies calculated binding affinities of 600-800 nM for our probes, consistent with their in vitro activation by COXs. The inability of celecoxib to completely inhibit probe activation, combined with LC/MS/MS data, strongly supports the hypothesis that probe activation is the result of deacetylation by random surface exposed amino acids rather than Ser-516 specifically. Interestingly, pre-incubation with celecoxib did reduce the activation of CP-2 by COX-2, consistent with the hypothesis that CP-2 binds non-covalently to the COX active site prior to activation and that the avidity of this interaction could be reduced by celecoxib. Overall, given this and its vastly superior hydrolytic stability, CP-2 is our optimal probe to date. Both probes were activated by porcine liver esterase, and at present they are not capable of discriminating between esterases and COXs. Prior literature suggests that stability against esterases can be improved by increasing the steric bulk of the alkyl portion of an ester.26 We are currently investigating whether this is a viable approach to improve the selectivity of our probes. Disappointingly, neither probe exhibited any evidence of COX-specific activation in stimulated RAW264.7 cells. Our hypothesis is that their affinity for the COXs is too low to allow them to function effectively in cellulo. We are currently investigating synthetic approaches to improve their affinity, guided by molecular modeling studies.
Conclusions
We have synthesized and tested 2 self-immolating fluorogenic probes for COXs. To the best of our knowledge, these are the first self-immolating molecules specific for the COXs. Both probes are activated by COX-1 and COX-2 in vitro and molecular modelling calculations suggest that they are submicromolar binders of these enzymes. The greater hydrolytic stability of CP-2 leads to greater relative increases in fluorescence compared to blank controls, making it our optimal probe to date. Our original molecular design envisaged activation by a specific serine residue in the COX active site, however the inability of celecoxib to completely ablate probe activation, along with analysis of acetylation sites on COX-2 by LC/MS/MS, strongly implied activation by numerous other solvent-exposed amino acids. Neither probe showed evidence of COX-specific activation in cellulo. Future work will focus on improving the selectivity of the probe for specific COX isozymes, improving their stability against esterases and developing probes which can detect COXs in a cellular setting.
Experimental
Synthetic chemistry
1,3-Di(tert-butyldimethylsilyl)propan-2-yl 2-hydroxy-5-(hydroxymethyl)benzoate (4)
1 (3.13 g, 18.8 mmol) was dissolved in anhydrous THF (60 mL) and the resulting solution was cooled to 0 °C over an ice-bath. DIAD (3.71 mL, 18.8 mmol) was added, followed by PPh3 (4.94 g, 18.8 mmol) in four separate portions over 20 mins. 2 (2.01 g, 6.28 mmol) was dissolved in anhydrous THF (10 mL) and added to the reaction, which was subsequently stirred at 0 °C for 1 h. The temperature of the reaction was then allowed to rise to ambient and it was stirred for a further 15 h. The reaction mixture was concentrated under reduced pressure and then purified over silica (eluent = 19:1 hexane:EtOAc) to give a crude sample of 3 as a colorless oil (1.258 g). Rf = 0.40 (eluent = 19:1 hexane:EtOAc). This crude material was then dissolved in anhydrous MeOH (40 mL) and cooled to 0 °C over an ice-bath. NaBH4 (102 mg, 2.68 mmol, “1 eq” based on 3 being pure) was added, the ice-bath was removed and the reaction was stirred for 3 h. The reaction was diluted with EtOAc (50 mL) and extracted with ddH2O (25 mL). The aqueous fraction was washed with EtOAc (2 × 25 mL) and the combined organic fractions were dried (MgSO4), filtered and concentrated to give the crude product which was subsequently purified over silica (eluent = 9:1-7:3 hexane:EtOAc) to give 4 as a waxy white solid (828 mg, 1.76 mmol, 28% over 2 steps). Rf = 0.12 (eluent = 9:1 hexane:EtOAc). 1H NMR (CDCl3, 400 MHz): δH = 0.05-0.08 (m, 12H, SiCH3), 0.89 (s, 18H, SiC(CH3)3), 3.80-3.95 (m, 4H, CH(CH2OTBDMS)2), 4.62 (s, 2H, ArCH2OH), 5.19 (quint, J = 5 Hz, 1H, CO(O)CH), 6.99 (d, J = 8.5 Hz, 1H, ArH), 7.49 (dd, J = 8.5 Hz, 1.5 Hz, 1H, ArH), 7.87 (d, J = 1 Hz, 1H, ArH), 10.73 (s, 1H, ArOH). 13C NMR (CDCl3, 400 MHz): δC = −5.5, −5.4, 18.2, 25.8, 61.1, 64.7, 75.9, 112.4, 117.9, 128.8, 131.6, 134.8, 161.3, 169.3. HR-MS (m/z, ESI): Calculated (C23H42O6Si2.H+) 471.2598; Found 471.2593.
1,3-Di(tert-butyldimethylsilyl)propan-2-yl 2-(acetyloxy)-5-(hydroxymethyl)benzoate (5)
4 (828 mg, 1.76 mmol) was dissolved in anhydrous DMF (40 mL) and the resulting solution was cooled to 0 °C over an ice-bath. K2CO3 (486 mg, 3.52 mmol) was added followed by acetic anhydride (183 μL, 1.94 mmol) and the resulting suspension was stirred at 0 °C for 2 h. The reaction was then diluted with EtOAc (25 mL) and extracted with ddH2O (3 × 25 mL). The combined aqueous extracts were washed with EtOAc (1 × 25 mL) and this single organic fraction was then washed with ddH2O (1 × 25 mL). The combined organic extracts were dried (MgSO4), filtered and concentrated under reduced pressure to give the crude product, which was subsequently purified over silica (eluent = 9:1-7:3 hexane:EtOAc) to give 5 as a colorless oil (523 mg, 1.02 mmol, 58%). 1H NMR (CDCl3, 400 MHz): δH = 0.06 (s, 12H, SiCH3), 0.89 (s, 18H, SiC(CH3)3), 2.35 (s, 3H, ArOC(O)CH3), 3.75-3.95 (m, 4H, CH(CH2OTBDMS)2), 4.72 (s, 2H, ArCH2OH), 5.07 (quint, J = 5 Hz, 1H, CO(O)CH), 7.11 (d, J = 8.5 Hz, 1H, ArH), 7.58 (dd, J = 8 Hz, 2.5 Hz, 1H, ArH), 8.01 (d, J = 2.5 Hz, 1H, ArH). 13C NMR (CDCl3, 400 MHz): δC = −5.5, −5.4, 18.3, 21.0, 25.8, 61.0, 64.3, 75.4, 123.4, 124.0, 130.1, 132.1, 138.7, 150.1, 163.6, 169.7. HR-MS (m/z, ESI): Calculated (C25H44O7Si2.Na+) 535.2523; Found 535.2518.
1,3-Dihydroxypropan-2-yl 2-(acetyloxy)-5-({[(quinolin-6-yl)carbamoyl]oxy}methyl) Benzoate (CP-1)
6-Aminoquinoline (28 mg, 0.19 mmol) and pyridine (0.05 mL, 0.7 mmol) were dissolved in anhydrous DCM and the resulting pale orange solution was cooled to 0 °C over an ice-bath. Triphosgene (19 mg, 0.065 mmol) was added, at which point the orange color of the solution intensified, and the reaction was stirred at 0 °C for 1 h. 5 (100 mg, 0.195 mmol) was dissolved in anhydrous toluene (2.5 mL) and added to the reaction, which was then heated at reflux for 15 h. The resulting suspension was cooled to ambient temperature, filtered and concentrated under reduced pressure. The residue was then dissolved in anhydrous THF (10 mL) and acetic acid (3 mL). TBAF (1M in THF, 975 μL, 0.975 mmol) was added and the reaction was stirred for 15 h. The reaction was diluted with toluene (5 mL) and then concentrated under reduced pressure. The residue was re-dissolved in a mixture of acetonitrile:H2O:acetic acid (4:1:0.1) and cooled to −20 °C until crystallization was observed (1-2 days). The solid was filtered, washed with cold acetonitrile (−20 °C) and Et2O and dried under reduced pressure to give CP-1 as an off-white solid (33 mg, 0.064 mmol, 34%). 1H NMR (d6-DMSO, 400 MHz): δH = 2.29 (s, 3H, ArOC(O)CH3), 3.60 (m, 4H, C(O)CH(CH2OH)2), 4.87 (t, J = 5.5 Hz, 2H, CH2OH), 4.95 (quint, J = 5.5 Hz, 1H C(O)CH(CH2OH)2), 5.26 (s, 2H, ArCH2OC(O)NH), 7.27 (d, J = 8.0 Hz, 1H, ArH), 7.46 (dd, J = 8.0 Hz, 4.5 Hz, 1H, ArH), 7.75 (m, 2H, ArH), 7.94 (d, J = 9.0 Hz, 1H, ArH), 8.08 (s, 1H ArH), 8.15 (s, 1H, ArH), 8.25 (d, J = 8.0 Hz, 1H, ArH), 8.75 (d, J = 4.5 Hz, 1H, ArH), 10.20 (br s, 1H, C(O)NH). 13C NMR (CDCl3, 400 MHz): δC = 20.7, 25.1, 59.7, 65.1, 67.0, 76.8, 121.8, 122.7, 123.7, 124.3, 128.4, 129.6, 131.3, 133.8, 134.5, 135.3, 137.0, 144.4, 148.8, 149.7, 153.3, 163.7, 169.1. HR-MS (m/z, ESI): Calculated (C23H22N2O8.H+) 455.1454; Found 455.1449. The purity of CP-1 was measured via HPLC using the following gradient of MeCN (+0.1% TFA) in H2O (+0.1% TFA): 2-20%, 0-10 mins; 20-90%, 10-25 mins. Under these conditions 6-aminoquinoline eluted at 10.2 mins (see supporting information for trace).
3-(Chlorosulfonyl)-4-hydroxybenzoic acid (7)19
4-Hydrobenzoic acid (5.50 g, 39.8 mmol) was added in small portions to HSO3Cl (31.1 mL, 468 mmol). The resulting solution was stirred for 15 h, before being poured onto crushed ice (~100 g). The resulting precipitate was filtered and then re-dissolved in EtOAc (50 mL). The resulting solution was washed with brine (1 × 25 mL), dried (MgSO4), filtered and concentrated under reduced pressure to give 7 as a white solid (5.56 g, 21.1 mmol, 53%). 1H NMR (d6-DMSO, 400 MHz): δH = 6.86 (dd, J = 8.5 Hz , 0.5 Hz, 1H, ArH), 7.78 (ddd, J = 8.5 Hz, 2 Hz, 1 Hz, 1H, ArH), 8.07 (dd, J = 2 Hz, 0.5 Hz, 1H, ArH).
4-(Hydroxymethyl)-2-(oct-2-yn-1-ylsulfanyl)phenol (9)
7 (2.00 g, 7.59 mmol) was dissolved in a mixture of anhydrous toluene and anhydrous THF (1:1, v:v, 60 mL). PPh3 (7.96 g, 30.3 mmol) was added to the reaction in 4 portions, an exothermic process which caused the formation of a white precipitate, and the resulting suspension was stirred for 15 h. The reaction was then diluted with toluene (50 mL) and extracted with 10 % NaOH (aq) (3 × 50 mL). The combined aqueous extracts were washed with toluene (1 × 50 mL), acidified to ~pH = 1 with 1M HCl and extracted with EtOAc (3 × 50 mL). The combined organic extracts were dried (MgSO4), filtered and concentrated under reduced pressure to give the crude thiophenol (1.411 g), which was subsequently dissolved in methanol (25 mL). H2SO4 (1 mL) was added to the resulting solution, which was then heated at reflux for 2 days, before being allowed to cool to ambient temperature. The reaction was then poured onto crushed ice (20 g) and the resulting solution was extracted with EtOAc (3 × 50 mL). The combined organic extracts were dried (MgSO4), filtered and concentrated to give the crude methyl ester (1.179 g), which was then dissolved in anhydrous THF (50 mL) under N2 and vigorous stirring. The resulting solution was cooled to 0 °C and 1M LiAlH4 in THF (25.6 mL, 25.6 mmol, 4 eq. assuming that crude methyl ester is 100 % pure) was added dropwise. The reaction was then heated at reflux for 15 h, before being allowed to cool to ambient temperature. H2O (10 mL) was added dropwise to the reaction, resulting in vigorous effervescence, to quench the remaining LiAlH4, followed by 1M HCl (10 mL). Following 15 mins of stirring the reaction was diluted with a further portion of 1M HCl (50 mL) and extracted with EtOAc (3 × 50 mL). The combined organic extracts were dried (MgSO4), filtered and concentrated to give the crude benzyl alcohol 8 (665 mg, 4.26 mmol, 56%) as a white solid, which was used in the next step without any further purification. 1H NMR (CDCl3, 400 MHz): δH = 4.40 (s, 2H, ArCH2OH), 6.72 (m, 1H, ArH), 6.92 (m, 1H, ArH), 7.16 (m, 1H, ArH). 8 (415 mg, 2.66 mmol) was dissolved in anhydrous DMF (50 mL) under N2 and vigorous stirring. KHCO3 (320 mg, 3.19 mmol) and 1-chloro-2-octyne (413 μL, 2.66 mmol) were added and the resulting suspension was stirred for 15 h. The reaction was then diluted with H2O (50 mL) and extracted with EtOAc (3 × 50 mL). The combined organic extracts were dried (MgSO4), filtered and concentrated under reduced pressure to give the crude product which was subsequently purified over silica (eluent = 4:1-1:1 hexane:EtOAc) to give 9 as a colorless oil (482 mg, 1.82 mmol, 69 %). Rf = 0.35 (eluent = 1:1 hexane:EtOAc). 1H NMR (CDCl3, 400 MHz): δH = 0.89 (m, 3H, CH2CH3), 1.29 (m, 4H, CH2CH2CH3), 1.45 (m, 2H, CH2CH2CH2CH3), 2.14 (m, 2H, CCCH2), 3.41 (s, 2H, SCH2CC), 4.61 (s, 2H, ArCH2OH), 7.00 (d, J = 8.0 Hz, 1H, ArH), 7.31 (dd, J = 8.0 Hz, 2.0 Hz, 1H, ArH), 7.55 (d, J = 2.0 Hz, 1H, ArH). 13C NMR (CDCl3, 100 MHz): δc = 13.9, 18.7, 22.2, 25.6, 28.2, 31.0, 64.7, 75.2, 85.4, 115.1, 118.1, 130.9, 133.2, 135.4, 157.0. HR-MS (m/z, ESI): Calculated (C15H20O2S.Na+) 287.1082; Found 287.1087.
4-(Hydroxymethyl)-2-(oct-2-yn-1-ylsulfanyl)phenyl acetate (10)
9 (420 mg, 1.59 mmol) was dissolved in anhydrous DMF (30 mL) under N2 and vigorous stirring. The resulting solution was cooled to 0 °C and K2CO3 (439 mg, 3.18 mmol) and then acetic anhydride (165 μL, 1.75 mmol) were added and the resulting suspension was stirred at 0 °C for 1 h, at which point TLC analysis (eluent = 7:3 hexane:EtOAc) revealed the complete consumption of 10. The reaction mixture was diluted with H2O (50 mL) and extracted with EtOAc (3 × 50 mL). The combined organic extracts were dried (MgSO4), filtered and concentrated under reduced pressure to give the crude product, which was subsequently purified over silica (eluent = 7:3 hexane:EtOAc) to give 10 as a colorless oil (385 mg, 1.26 mmol, 79 %). Rf = 0.20 (eluent = 7:3 hexane:EtOAc). 1H NMR (CDCl3, 400 MHz): δH = 0.88 (m, 3H, CH2CH3), 1.30 (m, 4H, CH2CH2CH3), 1.46 (m, 2H, CH2CH2CH2CH3), 2.15 (m, 2H, CCCH2), 2.35 (s, 3H, ArOC(O)CH3), 3.60 (m, 2H, SCH2CC), 4.67 (s, 2H, ArCH2OH), 7.06 (d, J = 8.0 Hz, 1H, ArH), 7.26 (dd, J = 8.0 Hz, 2.0 Hz, 1H, ArH), 7.54 (d, J = 2.0 Hz, 1H, ArH). 13C NMR (CDCl3, 100 MHz): δc = 13.9, 18.8, 20.8, 22.2, 22.4, 28.3, 31.0, 64.6, 74.9, 84.7, 122.6, 126.3, 129.0, 129.5, 139.3, 148.8, 169.1. HR-MS (m/z, ESI): Calculated (C17H22O3S.Na+) 329.1187; Found 329.1200.
2-(Oct-2-yn-1-ylsulfanyl)-4-({[(quinolin-6-yl)carbamoyl]oxy}methyl)phenyl acetate (CP-2)
Na2CO3 (103 mg, 0.969 mmol) was flamed-dried under reduced pressure. Anhydrous toluene (2 mL) was added and the resulting suspension was cooled to 0 °C. Triphosgene (64 mg, 0.22 mmol) was added and the reaction was stirred vigorously for 30 mins. 10 (33 mg, 0.11 mmol) was dissolved in anhydrous toluene (2 mL) was added drop-wise to the reaction over the course of 20 mins. The reaction was then allowed to warm to ambient temperature and was stirred vigorously for a further 3 h, after which time TLC analysis (eluent = 7:3 hexane:EtOAc) revealed the complete consumption of 10. The reaction mixture was concentrated under reduced pressure for 2 h to ensure the removal of any remaining triphosgene before being re-dissolved in anhydrous THF (2 mL). 6-Aminoquinoline (39 mg, 0.27 mmol) was added, at which point a precipitate was formed, and the reaction was stirred vigorously for a further 1 h. The reaction was filtered and the filtrate was concentrated under reduced pressure to give the crude product, which was subsequently purified over silica (eluent = 1:1 to 2:3 hexane:EtOAc) to give CP-2 as a yellow oil (20 mg, 0.0042 mmol, 38%). Rf = 0.28 (7:3 hexane:EtOAc). 1H NMR (CDCl3, 400 MHz): δH = 0.85 (m, 3H, CH2CH3), 1.20-1.30 (m, 4H, CH2CH2CH3), 1.42 (m, 2H, CH2CH2CH2CH3), 2.13 (m, 2H, CCCH2CH2), 2.36 (s, 3H, ArOC(O)CH3), 3.61 (s, 2H, ArSCH2), 5.23 (s, 2H, ArCH2O), 7.11 (d, J = 8.0 Hz, 1H, ArH), 7.32 (m, 1H, ArH), 7.34 (m, 1H, C(O)NH), 7.41 (dd, J = 8.5Hz, 4.5 Hz, 1H, ArH), 7.53 (dd, J = 9.0 Hz, 2.5 Hz, 1H, ArH), 7.60 (d, J = 2.0 Hz, 1H, ArH), 8.09 (d, J = 9.0 Hz, 1H, ArH), 8.11-8.17 (m, 2H, ArH), 8.83 (dd, J = 4.0 Hz, 1.0 Hz, 1H, ArH). 13C NMR (CDCl3, 100 MHz): δc = 13.9, 18.7, 20.8, 22.1, 22.4, 28.3, 31.0, 66.4, 74.7, 84.8, 114.3, 121.7, 122.8, 122.8, 126.3, 127.8, 128.9, 129.4, 129.9, 131.0, 134.3, 135.9, 136.1, 148.7, 149.5, 153.1, 169.0. HR-MS (m/z, ES): Calculated (C27H29N2O4S) 477.1848; Found 477.1838. The purity of CP-2 was measured via HPLC using the following gradient of MeCN (+0.1% TFA) in H2O (+0.1% TFA): 40-60%, 0-20 mins; 60-100%, 20-25 mins. Under these conditions 6-aminoquinline eluted at 8.8 mins (see supporting information for trace).
Measurement of Absorption and Emission Spectra
Solutions of either CP-1 or CP-1 in DMSO (10 mgmL−1) were diluted to a final concentration of 0.1 mgmL−1 in water or a mixture of methanol, water and saturated NaHCO3 (2:1:1, v:v:v). Spectra in water were measured immediately, those in methanol, water and saturated NaHCO3 were measured after a 30 minute incubation at ambient temperature. Emission spectra were measured using an excitation wavelength of 355 nm.
Testing of probes with purified proteins
Unless otherwise stated 80 mM Tris (pH 8.0, 0.1% Tween 20, 300 μM diethyldithiocarbamate) was used throughout these experiments. All stock solutions were stored at −20 °C when not in use. Commercially available solutions of COX enzymes were used as stock solutions for these enzymes. Porcine liver esterase, albumin and carbonic anhydrase were dissolved at 1.07 mgmL−1 in buffer. Hematin was dissolved at 0.1 mgmL−1 in 9:1 v:v buffer:DMSO. CP-1 and CP-2 were dissolved at 10 mgmL−1 in DMSO. Celecoxib was dissolved at 1 mgmL−1 in DMSO.
Over ice, solutions comprised of 1 protein, hematin (1 μM) and DMSO (7.6 μL) were mixed in buffer. COX-1, COX-2, albumin and carbonic anhydrase were all used at a final concentration of 0.125 mgmL−1. Porcine liver esterase was used at a final concentration of 0.309 mgmL−1 (equimolar concentration to COX-1 and COX-2). In the case of celecoxib inhibitor experiments, celecoxib in DMSO (7.6 μL) was used in place of DMSO and the solution was incubated for 10 mins over ice. CP-1 or CP-2 was added to give a final solution of 200 μL containing the probe at a concentration of 100 μM. The solution was transferred to a black-sided 96 well plate and incubated in a fluorimeter at 37 °C. At various time-points fluorescence readings (λex = 355 nm, λem = 535 nm) was measured to give the data presented in figure 4. Blank solutions compromising hematin (1 μM), DMSO (7.6 μL) and CP-1 or CP-2 (100 μM) were also mixed. Fluorescence readings (λex = 355 nm, λem = 535 nm) were taken at identical points.
To obtain the data presented in figure 5, the increase in fluorescence for the COX samples was divided by the increase in fluorescence for the blank sample over the same time period (60 mins).
Molecular modeling
CP-1 and CP-2 were docked into the COX-2 active site using AutoDock Vina to predict their binding modes and affinities. Specifically, the crystal structure of celecoxib-bound COX-2 (PDB code: 3LN1) with celecoxib or other ligands removed was treated rigid during docking; whereas either compound was treated flexible with 13 rotatable bonds each and further allowed to rotate or translate externally within a 20×20×20 Å3 cubical region whose center is located at the center of the bound celecoxib. The rationale of treating COX-2 as rigid is that no significant conformational differences for the backbone or even many active-site side chains were observed among over 30 ligand-bound COX-2 crystal structures deposited in the Protein Data Bank. For each compound, 100 independent docking jobs (each of which has 10 independent minimization runs) were performed using AutoDock Vina starting from random conformations and the model with the lowest predicted energy value was retained. According to AutoDock Vina, the binding energy values were predicted to be −8.2 and −8.4 kcal/mol for CP-1 and CP-2 respectively, compared with −11.8 kcal/mol predicted for celecoxib.
Mass spectrometry analysis of CP-1 treated COX-2
COX-2 was incubated with CP-1 as described in the section “testing of probes with purified proteins”. The sample was analyzed for sites of acetylation by MS Bioworks (Ann Arbor, MI). Briefly, COX-2 was purified by SDS-PAGE and digested with trypsin, chymotrysin and elastase. The gel digest was subsequently analyzed by LC/MS/MS using a Waters NanoAcquity HPLC system interfaced to a ThermoFisher Orbitrap Velos Pro. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column at 350nL/min; both columns were packed with Jupiter Proteo resin (Phenomenex). The mass spectrometer was operated in data-dependent mode, with MS performed in the Orbitrap at 60,000 FWHM resolution and MS/MS performed in the LTQ. The fifteen most abundant ions were selected for MS/MS. See supporting information for details on MS data analysis, a summary of acetylation sites and a representative mass spectrum.
In cellulo testing
RAW264.7 cells were stimulated to express COX-2 as previously described.17 Solutions of either CP-1 or CP-2 in media (1 mL + 30 μL DMSO) at a range of concentrations (0 μM, 50 μM, 100 μM and 250 μM) were incubated at 37 °C on either stimulated or non-stimulated cells for 1 h. The media was removed and the cells were washed twice with PBS and PBS (1 mL) was added for the imaging.
Fluorescence image cubes were measured with an inverted Nikon Eclipse Ti microscope using DAPI 365/10x excitation filter, FITC 510LP emission filter and DAPI dichroics. The fluorescence image at λem = 550 nm was isolated from the image cube. ImageJ software was used to pseudocolorize these images and quantify the pixel intensity.
Supplementary Material
Acknowledgements
This work was supported by National Cancer Institute grant CA135626 to TDT and EFJ, National Cancer Institute grants CA58207, CA097214, CA107584 and CA143803, Avon Research Foundation grants 07-2007-074 and 02-2011-110, California Breast Cancer Research Program grant 14OB-0165 and Cancer League grant to TDT and by Award K12GM081266 from the National Institute of General Medical Sciences/National Institutes of Health to LES. National Science Foundation award CCF-1546278 to YS
Footnotes
Electronic Supplementary Information (ESI) available: [Experimental details, HPLC traces, UV-Vis spectra and NMR spectra]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Wong A, DeWit M, Gillies E. Adv. Drug Deliv. Rev. 2012;64:1031–1045. doi: 10.1016/j.addr.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe CA, Russell AT, Greco F, Hayes W, Thornthwaite DW. Polym. Chem. 2011;2:773. [Google Scholar]
- 3.Uddin MJ, Crews BC, Blobaum AL, Kingsley PJ, Gorden DL, McIntyre JO, Matrisian LM, Subbaramaiah K, Dannenberg AJ, Piston DW, Marnett LJ, Uddin J. Cancer Res. 2010;70:3618–27. doi: 10.1158/0008-5472.CAN-09-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Fan J, Wang J, Zhang S, Dou B, Peng X. J. Am. Chem. Soc. 2013;135:11663–9. doi: 10.1021/ja4056905. [DOI] [PubMed] [Google Scholar]
- 5.Uddin MJ, Crews BC, Ghebreselasie K, Marnett LJ. Bioconjug. Chem. 2013;24:712–23. doi: 10.1021/bc300693w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouzer CA, Marnett LJ. Chem. Rev. 2003;103:2239–304. doi: 10.1021/cr000068x. [DOI] [PubMed] [Google Scholar]
- 7.Rouzer CA, Marnett LJ. Biochem. Biophys. Res. Commun. 2005;338:34–44. doi: 10.1016/j.bbrc.2005.07.198. [DOI] [PubMed] [Google Scholar]
- 8.Simmons DL, Botting RM, Hla T. Pharmacol. Rev. 2004;56:387–437. doi: 10.1124/pr.56.3.3. [DOI] [PubMed] [Google Scholar]
- 9.Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Biochem. Pharmacol. 2006;72:1605–21. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 10.Greenhough A, Smartt HJM, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A, Moore E. Carcinogenesis. 2009;30:377–86. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 11.Subbaramaiah K, Dannenberg AJ. Trends Pharmacol. Sci. 2003;24:96–102. doi: 10.1016/S0165-6147(02)00043-3. [DOI] [PubMed] [Google Scholar]
- 12.Tang Y, Lee D, Wang J, Li G, Yu J, Lin W, Yoon J. Chem. Soc. Rev. 2015;44:5003–5015. doi: 10.1039/c5cs00103j. [DOI] [PubMed] [Google Scholar]
- 13.Guo Z, Park S, Yoon J, Shin I. Chem. Soc. Rev. 2014;43:16–29. doi: 10.1039/c3cs60271k. [DOI] [PubMed] [Google Scholar]
- 14.Ding Y, Tang Y, Zhu W, Xie Y. Chem. Soc. Rev. 2015;44:1101–12. doi: 10.1039/c4cs00436a. [DOI] [PubMed] [Google Scholar]
- 15.Drake CR, Miller DC, Jones EF. Curr. Org. Synth. 2011;8:498–520. doi: 10.2174/157017911796117232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paris GY, Garmaise DL, Cimon DG, Swett L, Carter GW, Young P. J. Med. Chem. 1979;22:683–7. doi: 10.1021/jm00192a014. [DOI] [PubMed] [Google Scholar]
- 17.Kalgutkar AS, Kozak KR, Crews BC, Hochgesang GP, Marnett LJ. J. Med. Chem. 1998;41:4800–18. doi: 10.1021/jm980303s. [DOI] [PubMed] [Google Scholar]
- 18.Kalgutkar AS, Crews BC, Rowlinson SW, Garner C, Seibert K, Marnett LJ. Science (80−. ) 1998;280:1268–1270. doi: 10.1126/science.280.5367.1268. [DOI] [PubMed] [Google Scholar]
- 19.1985 4,548,951.
- 20.Wuts PGM, Greene TW. Protective Groups in Organic Synthesis. 4th edn. Wiley; 2006. [Google Scholar]
- 21.Brynes PJ, Bevilacqua P, Green A. Anal. Biochem. 1981;413:408–413. doi: 10.1016/0003-2697(81)90381-x. [DOI] [PubMed] [Google Scholar]
- 22.Huang W, Hicks SN, Sondek J, Zhang Q. ACS Chem. Biol. 2011;6:223–8. doi: 10.1021/cb100308n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshimoto A, Ito H, Tomita K. J. Biochem. 1970;68:487–499. doi: 10.1093/oxfordjournals.jbchem.a129379. [DOI] [PubMed] [Google Scholar]
- 24.Trott O, Olson AJ. J. Comput. Chem. 2009;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang JL, Limburg D, Graneto MJ, Springer J, Hamper JRB, Liao S, Pawlitz JL, Kurumbail RG, Maziasz T, Talley JJ, Kiefer JR, Carter J. Bioorg. Med. Chem. Lett. 2010;20:7159–63. doi: 10.1016/j.bmcl.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 26.Tian L, Yang Y, Wysocki LM, Arnold AC, Hu A, Ravichandran B, Sternson SM, Looger LL, Lavis LD. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4756–4761. doi: 10.1073/pnas.1111943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.