Abstract
Background
When problems with compatibility beyond ABO and RhD arise, currently transfusion services search their inventories and perform time-consuming serologic testing to locate antigen-negative blood. These clinically important blood group antigens can be detected reliably by red cell genotyping, which is a technology whereby DNA-based techniques are used to evaluate gene polymorphisms that determine the expression of red cell antigens. We introduced mass-scale genotyping and measured availability of genotyped blood.
Study design and methods
All non-Caucasian donors qualified for genotyping along with Caucasian donors who had a history of repeat donation. Mass-scale red cell genotyping, performed on an electronic interfaced open array platform, was implemented to screen blood donors for 32 SNPs that predicted 42 blood group antigens. Genotype screening results were confirmed by phenotyping, when needed for antigen-negative transfusion, prior to release of the red cell unit.
Results
Approximately 22,000 donors were red cell genotyped within 4 months and a total of 43,066 donors in 4 years. There were 463 discordances (0.52% of 89,596 genotypes with a phenotype). Among the 307 resolved discordances, approximate equal numbers represented historical serological or genotyping discrepancies (n=151 and n=156, respectively). In the final year of the study, an average of 29% of the daily inventory had a genotype.
Conclusions
Red cell genotyping of blood donors using an electronic interface created a large and stable supply of red cell units with historical genotypes. The database served the needs of antigen-negative blood requests for a large regional blood center, and allowed us to abandon screening by serology.
Introduction
Alloimmunized transfusion recipients require antigen-negative blood for safe transfusion. Blood centers are best suited to provide antigen-negative blood because they have the entire regional blood inventory at their disposal. Current practice to provide blood for transfusion recipients relies on phenotyping to confirm that red cell units lack the cognate antigen to which the patient's alloantibody is binding. Hemagglutination is still considered the ‘gold standard’ method for red blood cell (RBC) antigen typing by many serologists1 notwithstanding several important drawbacks; some clinically significant antigens cannot be identified by phenotyping and some variant antigens are known to be consistently mistyped as antigen-negative.
It is now possible to determine a far greater number of antigens by genotyping than by serology,2-4 which if performed on a mass-scale could provide access to a larger supply of antigen-negative blood. Several groups have evaluated red cell genotyping platforms for the potential to genotype large numbers of samples. Testing volumes range from dozens5-7 to several hundred8-15 to thousands of samples.16-22 Because it is considered not economically feasible to genotype all blood donors, selection criteria can be used to maximize the database. Furthermore, blood donors who have been genotyped on a previous donation have historical genotype information that may be utilized without the need for re-testing.
We developed a red cell genotyping process in 200814 and implemented a mass-scale program in 2010.23 The availability of red cell units with a genotype was evaluated over the following three years, which provided genotyped inventory to support blood requests.
Methods
Blood donors
All whole blood donors who declared their ethnicity on the donation questionnaire as Asian, African American, Hispanic, or Native American qualified for red cell genotyping regardless of their ABO/Rh blood group or frequency of donation. In addition, Group O, A, and B whole blood donors qualified if they had a history of at least 3 donations in the previous 3 years, with one donation in the previous 12 months. Occasionally, group AB donors who met the historical donation criteria were included. Blood donors gave informed consent and BloodCenter of Wisconsin's Institutional Review Board approved the retrospective red cell phenotype and genotype review (BCW 12-27).
Red cell genotyping
DNA was extracted from EDTA-anticoagulated whole blood (QIAamp 96 DNA Blood Kit; Qiagen, Valencia, CA). A nanofluidic open array system using real-time PCR-fluorogenic 5’ nuclease TaqMan chemistry (OpenArray Real-Time PCR System; Life Technologies Corporation, Grand Island, NY) was used as described previously.14,23 The analysis was expanded to interrogate 32 single nucleotide polymorphisms (SNPs), which allowed for the prediction of 42 blood group antigens: C, E, c, e, V, hrS,24 VS, hrB,25 Crawford; M, N, S, s, U; Lua, Lub, Lu8, Lu14; K, k, Kpa, Kpb, Jsa, Jsb; Fya, Fyb; Jka, Jkb, Jk3; Dia, Dib; Yta, Ytb; Sc1, Sc2; Doa, Dob, Hy, Joa; Coa, Cob; and Cra. On June 28, 2012, Polynesian/Finn Jk null genotyping was discontinued since no Jk null donors were identified and Uw+ testing was instituted. Genotypes were displayed using ISBT nomenclature allowing the operator to distinguish them from phenotypes (Table 1). Another 14 antigens that we encountered for antigen-negative blood requests were identified by phenotyping (Table 2).
Table 1.
Blood group antigens by common name and ISBT terminology, which were genotyped
| Antigen | ISBT terminology |
Genotypes with a phenotype |
||
|---|---|---|---|---|
| Number | Symbol | All | Discordances | |
| M | 002.001 | MNS1 | 924 | 5 |
| N | 002.002 | MNS2 | 833 | 8 |
| S | 002.003 | MNS3 | 4168 | 72 |
| s | 002.004 | MNS4 | 3104 | 10 |
| U | 002.005 | MNS5 | na | na |
| C | 004.002 | RH2 | 8793 | 126 |
| E | 004.003 | RH3 | 11534 | 36 |
| c | 004.004 | RH4 | 4713 | 3 |
| e | 004.005 | RH5 | 4702 | 30 |
| V | 004.010 | RH10 | 77 | 4 |
| hrS | 004.019 | RH19 | na | na |
| VS | 004.020 | RH20 | na | na |
| hrB | 004.031 | RH31 | na | na |
| Crawford | 004.043 | RH43 | na | na |
| Lua | 005.001 | LU1 | 260 | 8 |
| Lub | 005.002 | LU2 | 3788 | 19 |
| Lu8 | 005.008 | LU8 | 18 | 0 |
| Lu14 | 005.014 | LU14 | na | na |
| K | 006.001 | KEL1 | 12174 | 14 |
| k | 006.002 | KEL2 | 609 | 0 |
| Kpa | 006.003 | KEL3 | 428 | 0 |
| Kpb | 006.004 | KEL4 | 4541 | 0 |
| Jsa | 006.006 | KEL6 | 117 | 5 |
| Jsb | 006.007 | KEL7 | 4314 | 5 |
| Fya | 008.001 | FY1 | 4856 | 19 |
| Fyb | 008.002 | FY2 | 3376 | 25 |
| Jka | 009.001 | JK1 | 4495 | 11 |
| Jkb | 009.002 | JK2 | 4102 | 23 |
| Jk3 | 009.003 | JK3 | na | na |
| Dia | 010.001 | DI1 | 1653 | 7 |
| Dib | 010.002 | DI2 | 102 | 1 |
| Yta | 011.001 | YT1 | 3342 | 15 |
| Ytb | 011.002 | YT2 | 6 | 1 |
| Sc1 | 013.001 | SC1 | 1 | 0 |
| Sc2 | 013.002 | SC2 | 9 | 0 |
| Doa | 014.001 | DO1 | 57 | 11 |
| Dob | 014.002 | DO2 | 49 | 4 |
| Hy | 014.004 | DO4 | 35 | 0 |
| Jo | 014.005 | DO5 | na | na |
| Coa | 015.001 | CO1 | 1820 | 1 |
| Cob | 015.002 | CO2 | 593 | 0 |
| Cra | 021.001 | CROM1 | 3 | 0 |
| Total | na | na | 89,596 | 463 |
na, not applicable/available
Table 2.
Blood group antigens by common name and ISBT terminology, which were phenotyped but not genotyped
| Antigen* | ISBT Terminology |
|
|---|---|---|
| Number | Symbol | |
| He | 002.006 | MNS6 |
| ‘N’ | 002.030 | MNS30 |
| P1 | 003.001 | P1PK1 |
| Pk | 003.003 | P1PK3 |
| f† | 004.006 | RH6 |
| Cw | 004.008 | RH8 |
| Goa | 004.030 | RH30 |
| Lea | 007.001 | LE1 |
| Leb | 007.002 | LE2 |
| Wra | 010.003 | DI3 |
| Xga | 012.001 | XG1 |
| H | 018.001 | H |
| Tca | 021.002 | CROM2 |
| Vel | 034.001 | VEL1 |
All antigens could be genotyped.
Lack of the f antigen expression can be deduced by a negative phenotyping result for the c or e antigens.
Blood sample barcodes were used to create a traceable process from sample selection to results output. Instrument output was reviewed electronically by medical technologists. Control samples of known genotypes were tested in each run except for examples of rare homozygous genotypes. Individual results were excluded by assigning a ‘no call’ when values were below a defined fluorescence or were outside instrument-defined genotype clusters. The no call rate performed on 12,376 samples ranged from 0.28% to 1.60% (mean 0.63%, median 0.60%), with the exception of Yta/Ytb (3.4%), Lua/Lub (3.9%), and Lu8/14 (5.3%). The instrument output file was electronically translated into ISBT genotypes using an electronic ‘rules’ logic table.
Red cell genotype database
Genotype results were electronically transferred to a database, and were displayed alongside historical phenotypes using the blood center computer system (LifeTrak; Mediware Information Systems, Oak Brook, IL). All genotypes were compared to the existing historical phenotypes. The output of the comparison created a report of genotype-phenotype discordances.
Antigen-negative blood requests and phenotyping
A request for antigen-negative blood was considered a single patient ‘encounter’ regardless of the number of red cell units desired. All genotype results were confirmed by serologic phenotyping before the release of a red cell unit, when needed for antigen-negative transfusion.
Results
Donor accrual started on January 2, 2010. Routine red cell phenotype screening was discontinued for non-Caucasian donors (C, U, and Jsb antigens) and Caucasian donors (Lub, Yta, and Coa antigens) on July 17, 2010 because it had been replaced by mass-scale genotype screening.
Red cell genotyping of donors
Among 202,275 individuals who successfully donated a unit of whole blood from 2010 and 2013 inclusive, 79,864 donors matched the criteria for self-declared ethnicity or historical donation data, of which 43,066 (54% of qualified donors) were actually genotyped. A total of 209,540 red cell units had a red cell genotype, representing 32% of all red cell units. These donors provided an average of 2.1 red cell units per year. The database consisted of 24,332 blood donors genotyped in 2010, with 8186, 4498, and 6050 donors genotyped in 2011, 2012 and 2013, respectively (Fig. 1).
Figure 1.
Red cell genotyping of blood donors. The number of blood donors genotyped (black columns) and the cumulative number of donors with a genotype (red line) are shown from 2010 to 2013. DNA extraction started in January 2010 and genetic testing in July 2010. Red cell genotyping is continuing in monthly batches.
We found 463 discordant results (0.52%) among 89,596 genotypes with a historical phenotype (Table 1). The serology performed on a repeat donation sample revealed 151 (32%) data entry or serologic typing errors that included 1 E (RHCE*03.04), 2 Kell modifier (KEL*02M), and 10 Fy(bw+) (FY*02M.01) alleles. The serology was confirmed to be correct for 156 discordant results (34%). Among them, 32 samples (7%) represented variant alleles that included 16 S-silencing (MNS*03N.01/MNS*03N.03), 7 S-deletional (GYPB*01N) and 9 e (RHCE*ceMO) alleles. There were another 156 discordant results (34%) awaiting resolution because the donor had not yet returned for repeat serologic typing.
Availability of red cell units with genotype
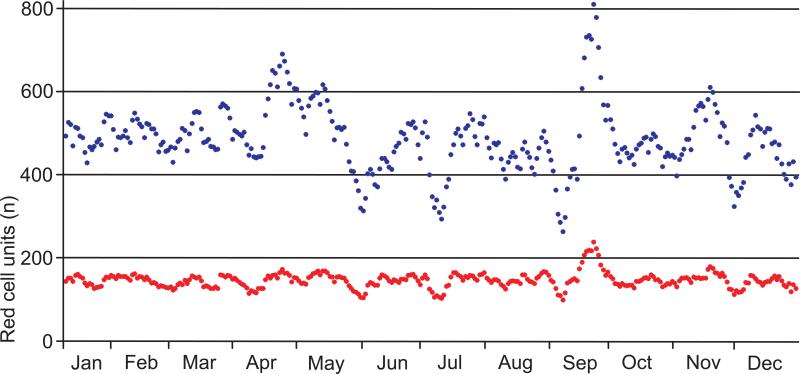
We evaluated genotype information among red cell units in 2013. The blood center collected 151,727 whole blood donations of which 45,681 had a genotype. The monthly number of donations with a red cell genotype remained stable at 29% to 32% (Fig. 2). To meet transfusion recipient needs during 2011-2013, we imported 34 rare red cell units from other blood centers for 11 of 5672 patient encounters.
Figure 2.
Red cell units donated in the blood center in 2013. Blood was collected on 314 days. The collection numbers of a sliding window for 7 collection days are shown (mean, blue dots). The numbers of available red cell units with known genotype are indicated (mean, red dots). Overall, the percentage averaged 29.4% of the inventory throughout 2013.
Discussion
Red cell genotyping of blood donors has been shown to be feasible to screen for both common and rare blood group antigens.19-22,26 Red cell genotyping is a technology whereby DNA-based techniques are used to evaluate genes for the particular single and multiple nucleotide substitutions, deletions, insertions, and gene conversions that determine the expression of red cell antigens. The prime utility for donor testing rests with the large number of samples and the vast number of antigens that can be genotyped in a short period. Evidence of repeat and recent donation has been used in donor screening programs to maximize antigen-negative blood inventory.18,22,27
We performed mass-scale red cell genotyping on over 22,000 samples during the initial 14 weeks of testing and at a lower level thereafter to maintain the number of active donors in our red cell genotype database (Fig. 1). Single determinations using TaqMan chemistry were preferred over the cost of running duplicates or triplicates to gain more accuracy, because our red cell genotyping was designed as screening tool and the phenotype was confirmed before transfusing any red cell unit. Given the low rate of discordances (0.52%), which is consistent with previously published data,6,9,11,14,15,19,21 we had sufficient access to correctly genotyped red cell units to meet the demand for antigen-negative blood. For the 3 years following the implementation in 2010, only 34 red cell units were imported for a blood center that distributes 150,000 red cell units annually to 63 hospitals serving a population of 3.78 million. Recruitment of genotyped donors was not attempted except when we needed to import blood. It will be important to identify the number of future donations by ethnicity and to apply a recruitment strategy to address our gap to provide rare units. The vast majority of patient encounters (5661 equaling 99.8%) were quickly met from our blood center inventory by using the database and confirming the phenotype of genotyped red cell units, if needed, without the delay and expense of shipping red cell units from other blood centers. Our database was applied successfully to serve the needs of transfusion recipients for antigen-negative blood.
The genotyping program provided a steady supply of red cell units with red cell genotype data (Fig. 2). The low rate of genotype-phenotype discordances was caused by roughly equal numbers of phenotypes and genotypes (Table 1). However, a large number of RH2 genotyping errors were observed. These and other low frequency genotyping errors may be a function of the assay chemistry used in combination with the nano-volume of DNA dispensed by the instrument as it is known for limiting dilution assays. The red cell genotyping program incorporated a fully computerized workflow process, from DNA extraction and genotyping electronic worksheets to results review, data handling and error reports. The entire process can be completed within 36 hours, consistent within the turnaround time of infectious disease testing, and the throughput capacity can handle more 700 samples in a single day. Our next step is to validate red cell genotyping as a test-of-record such that the need for serologic confirmation may be eliminated. A network of similarly mass-scale genotyped donors among US blood centers would create a large real-time repository and facilitate rapid access to antigen-negative blood nationwide.
Acknowledgements
We thank Harvey G. Klein, MD for discussing and reviewing the manuscript, and David Allen Stiles, Craig Beczkiewicz and Kathleen Bensing for their computational assistance.
This work was supported by a BloodCenter of Wisconsin Diagnostic Laboratories Strategic Initiative and the Intramural Research Program of the NIH Clinical Center.
Footnotes
Statement of Disclaimer: The views expressed do not necessarily represent the view of the National Institutes of Health (NIH), the Department of Health and Human Services (DHHS), or the U.S. Federal Government.
At the time the red cell genotyping described in this article was implemented, none of the molecular immunohematology assays for red cell genotyping had been approved by the US Food and Drug Administration (FDA).
Conflict of interest disclosure: GAD is the inventor of European patents on red cell genotyping owned by Canadian Blood Services. WAF & JLG do not have a conflict of interest relevant to this article.
Authorship contribution: GAD coordinated red cell genotyping, and collected and summarized the data. JLG contributed to the content of the discussion. GAD and WAF conceptualized the study, analyzed the data, and wrote the manuscript.
Reference List
- 1.Manfroi S, Pagliaro P. Common on “Applying molecular immunohaematology to regularly transfused thalassaemic patients in Thailand”. Blood Transfus. 2015;13:164–5. doi: 10.2450/2014.0158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimring JC, Welniak L, Semple JW, Ness PM, Slichter SJ, Spitalnik SL. Current problems and future directions of transfusion-induced alloimmunization: summary of an NHLBI working group. Transfusion. 2011 Feb;51:435–41. doi: 10.1111/j.1537-2995.2010.03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner FF. Molecular testing in transfusion medicine. Expert Opin Med Diagn. 2010 Sep;4:411–28. doi: 10.1517/17530059.2010.506509. [DOI] [PubMed] [Google Scholar]
- 4.Denomme GA, Flegel WA. Applying molecular immunohematology discoveries to standards of practice in blood banks: now is the time. Transfusion. 2008 Nov;48:2461–75. doi: 10.1111/j.1537-2995.2008.01855.x. [DOI] [PubMed] [Google Scholar]
- 5.Palacajornsuk P, Halter C, Isakova V, Tarnawski M, Farmar J, Reid ME, Chaudhuri A. Detection of blood group genes using multiplex SNaPshot method. Transfusion. 2009 Apr;49:740–9. doi: 10.1111/j.1537-2995.2008.02053.x. [DOI] [PubMed] [Google Scholar]
- 6.St-Louis M, Perreault J, Lemieux R. Extended blood grouping of blood donors with automatable PCR-ELISA genotyping. Transfusion. 2003 Aug;43(8):1126–32. doi: 10.1046/j.1537-2995.2003.00474.x. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka M, Kamada I, Takahashi J, Hirayama F, Tani Y. Evaluation of a blood group genotyping platform (BLOODchip((R)) Reference) in Japanese samples. Transfus Med. 2014 Feb;24:39–44. doi: 10.1111/tme.12085. [DOI] [PubMed] [Google Scholar]
- 8.Polin H, Danzer M, Proll J, Hofer K, Heilinger U, Zopf A, Gabriel C. Introduction of a real-time-based blood-group genotyping approach. Vox Sang. 2008 Aug;95:125–30. doi: 10.1111/j.1423-0410.2008.01067.x. [DOI] [PubMed] [Google Scholar]
- 9.Haer-Wigman L, Ji Y, Loden M, de HM, van der Schoot CE, Veldhuisen B. Comprehensive genotyping for 18 blood group systems using a multiplex ligation-dependent probe amplification assay shows a high degree of accuracy. Transfusion. 2013 Nov;53(11 Suppl 2):2899–909. doi: 10.1111/trf.12410. [DOI] [PubMed] [Google Scholar]
- 10.Le Goff GC, Bres JC, Rigal D, Blum LJ, Marquette CA. Robust, high-throughput solution for blood group genotyping. Anal Chem. 2010 Jul 15;82:6185–92. doi: 10.1021/ac101008d. [DOI] [PubMed] [Google Scholar]
- 11.Latini FR, Gazito D, Arnoni CP, Muniz JG, de Medeiros PR, Carvalho FO, Baleotti W, Jr., Castilho L, Barreto JA. A new strategy to identify rare blood donors: single polymerase chain reaction multiplex SNaPshot reaction for detection of 16 blood group alleles. Blood Transfus. 2014 Jan;12(Suppl 1):s256–s263. doi: 10.2450/2013.0242-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denomme GA, Van OM. High-throughput multiplex single-nucleotide polymorphism analysis for red cell and platelet antigen genotypes. Transfusion. 2005 May;45:660–6. doi: 10.1111/j.1537-2995.2005.04365.x. [DOI] [PubMed] [Google Scholar]
- 13.Hashmi G, Shariff T, Seul M, Vissavajjhala P, Hue-Roye K, Charles-Pierre D, Lomas-Francis C, Chaudhuri A, Reid ME. A flexible array format for large-scale, rapid blood group DNA typing. Transfusion. 2005 May;45:680–8. doi: 10.1111/j.1537-2995.2005.04362.x. [DOI] [PubMed] [Google Scholar]
- 14.Hopp K, Weber K, Bellissimo D, Johnson ST, Pietz B. High-throughput red blood cell antigen genotyping using a nanofluidic real-time polymerase chain reaction platform. Transfusion. 2010 Jan;50(1):40–6. doi: 10.1111/j.1537-2995.2009.02377.x. [DOI] [PubMed] [Google Scholar]
- 15.Paris S, Rigal D, Barlet V, Verdier M, Coudurier N, Bailly P, Bres JC. Flexible automated platform for blood group genotyping on DNA microarrays. J Mol Diagn. 2014 May;16:335–42. doi: 10.1016/j.jmoldx.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Polin H, Danzer M, Hofer K, Gassner W, Gabriel C. Effective molecular RHD typing strategy for blood donations. Transfusion. 2007 Aug;47:1350–5. doi: 10.1111/j.1537-2995.2007.01278.x. [DOI] [PubMed] [Google Scholar]
- 17.Avent ND, Martinez A, Flegel WA, Olsson ML, Scott ML, Nogues N, Pisacka M, Daniels GL, Muniz-Diaz E, Madgett TE, et al. The Bloodgen Project of the European Union, 2003-2009. Transfus Med Hemother. 2009;36:162–7. doi: 10.1159/000218192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner FF, Bittner R, Petershofen EK, Doescher A, Muller TH. Cost-efficient sequence-specific priming-polymerase chain reaction screening for blood donors with rare phenotypes. Transfusion. 2008 Jun;48(6):1169–73. doi: 10.1111/j.1537-2995.2008.01682.x. [DOI] [PubMed] [Google Scholar]
- 19.Meyer S, Vollmert C, Trost N, Bronnimann C, Gottschalk J, Buser A, Frey BM, Gassner C. High-throughput Kell, Kidd, and Duffy matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry-based blood group genotyping of 4000 donors shows close to full concordance with serotyping and detects new alleles. Transfusion. 2014 Dec;54(12):3198–207. doi: 10.1111/trf.12715. [DOI] [PubMed] [Google Scholar]
- 20.Jungbauer C, Hobel CM, Schwartz DW, Mayr WR. High-throughput multiplex PCR genotyping for 35 red blood cell antigens in blood donors. Vox Sang. 2012 Apr;102(3):234–42. doi: 10.1111/j.1423-0410.2011.01542.x. [DOI] [PubMed] [Google Scholar]
- 21.St-Louis M, Perreault J, Lavoie J, Emond J, St-Laurent J, Long A, Richard M. [Genotyping of 21,000 blood donors in Quebec and RHD analysis]. Transfus.Clin.Biol. 2010 Oct;17(4):242–8. doi: 10.1016/j.tracli.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Flegel WA, von Zabern I, Wagner FF. Six years’ experience performing RHD genotyping to confirm D-red blood cell units in Germany for preventing anti-D immunizations. Transfusion. 2009 Mar;49(3):465–71. doi: 10.1111/j.1537-2995.2008.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber KM, Fueger JT, Piefer CL, Leszczynski LA, Kalvelage ML, Sauer DE, Marchan MG, Pugh TM, Denomme GA. Identifying rare blood donors: a study in on-time high-throughput red cell genotyping. Transfusion. 2010;50(S2):174A. [Google Scholar]
- 24.Noizat-Pirenne F, Mouro I, Le Pennec PY, Ansart-Pirenne H, Juszczak G, Patereau C, Verdier M, Babinet J, Roussel M, Rouger P, et al. Two new alleles of the RHCE gene in Black individuals: the RHce allele ceMO and the RHcE allele cEMI. Br J Haematol. 2001 Jun;113(3):672–9. doi: 10.1046/j.1365-2141.2001.02802.x. [DOI] [PubMed] [Google Scholar]
- 25.Daniels GL, Faas BH, Green CA, Smart E, Maaskant-Van Wijk PA, Avent ND, Zondervan HA, von dem Borne AE, van der Schoot CE. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion. 1998 Oct;38(10):951–8. doi: 10.1046/j.1537-2995.1998.381098440860.x. [DOI] [PubMed] [Google Scholar]
- 26.Wagner FF, Bittner R, Petershofen EK, Doescher A, Muller TH. Cost-efficient sequence-specific priming-polymerase chain reaction screening for blood donors with rare phenotypes. Transfusion. 2008 Jun;48:1169–73. doi: 10.1111/j.1537-2995.2008.01682.x. [DOI] [PubMed] [Google Scholar]
- 27.St-Louis M. Molecular blood grouping of donors. Transfus.Apher.Sci. 2014 Apr;50(2):175–82. doi: 10.1016/j.transci.2014.02.012. [DOI] [PubMed] [Google Scholar]