Abstract
Solid cancer cells commonly enter the blood and disseminate systemically but are highly inefficient at forming distant metastases for poorly understood reasons. We studied human melanomas that differed in their metastasis histories in patients and in their capacity to metastasize in NSG mice. All melanomas had high frequencies of cells that formed subcutaneous tumours, but much lower percentages of cells that formed tumours after intravenous or intrasplenic transplantation, particularly among inefficient metastasizers. Melanoma cells in the blood and visceral organs experienced oxidative stress not observed in established subcutaneous tumours. Successfully metastasizing melanomas underwent reversible metabolic changes during metastasis that increased their capacity to withstand oxidative stress, including increased dependence upon NADPH-generating enzymes in the folate pathway. Anti-oxidants promoted distant metastasis in NSG mice. Folate pathway inhibition using low-dose methotrexate, ALDH1L2 knockdown, or MTHFD1 knockdown inhibited distant metastasis without significantly affecting the growth of subcutaneous tumors in the same mice. Oxidative stress thus limits distant metastasis by melanoma cells in vivo.
Circulating cancer cells are commonly observed in the blood of patients and mice with various cancers1–4. However, metastasis is a very inefficient process5 in which few disseminated cancer cells survive and even fewer proliferate6–8. Some patients can have circulating cancer cells in their blood without evidence of metastasis or worse outcomes9–11.
Epithelial cells undergo cell death when they detach from extracellular matrix in culture as a result of reduced glucose uptake, ATP depletion, and oxidative stress 12,13. Oncogenic signaling can promote their survival by increasing glucose uptake and flux through the pentose phosphate pathway, which generates NADPH and regenerates glutathione, a buffer against oxidative stress14. Glutathione is necessary for the initiation of some cancers and anti-oxidants can promote cancer initiation and progression15–18. Cancer cells thus undergo genetic changes within primary tumours that increase their capacity to withstand oxidative stress, raising the question of whether additional adaptations are required during metastasis. Breast and lung cancer cell lines undergo metabolic changes during invasion in culture and metastasis in vivo that would be expected to reduce the generation of reactive oxygen species (ROS) 18–23. Nonetheless, it is unknown whether ROS levels change in metastasizing cells in vivo or whether this limits distant metastasis. In fact, anti-oxidants inhibit the metastasis of some cancer cell lines, raising the possibility that ROS promotes metastasis in certain contexts 24–26.
We addressed these issues by studying melanomas from multiple patients that were xenografted into NOD/SCID IL2Rγnull (NSG) mice. Melanoma metastasis in this assay is predictive of clinical outcome in patients: stage III melanomas that metastasize efficiently in NSG mice go on to form distant metastases in patients, despite surgical resection, whereas melanomas that metastasize inefficiently in mice are usually cured by surgery in patients27.
Blood and viscera are hostile to metastasis
We obtained four efficiently (UT10, M481, M405, and M514) and four inefficiently (M597, M528, M610, and M498) metastasizing melanomas from patients. All expressed melanoma markers (Extended data figure 1). The efficiently metastasizing melanomas formed distant metastases in patients and in NSG mice after subcutaneous injection (Extended data figure 2a).
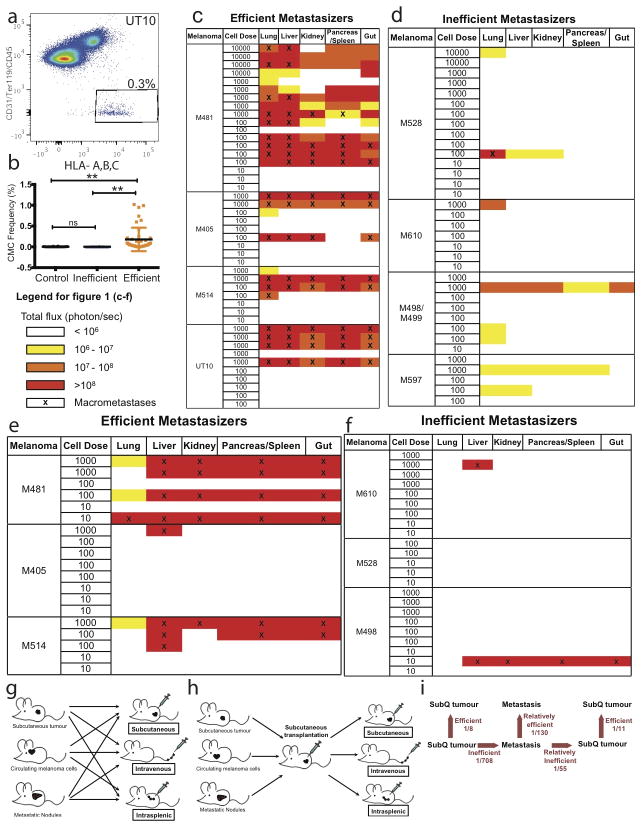
The inefficiently metastasizing melanomas did not form distant metastases in patients or macrometastases in NSG mice (Extended data figure 2a). The efficient and inefficient metastasizers did not significantly differ with regard to the frequency of cells that formed tumours after subcutaneous injection in NSG mice (Table 1) or the rate at which these tumours grew (Extended data figure 2b). One in 8 cells from efficient metastasizers and 1 in 11 cells from inefficient metastasizers formed tumours upon subcutaneous injection (Table 1). We often detected circulating melanoma cells (CMC) by flow cytometry in the blood of mice with efficiently metastasizing, but not inefficiently metastasizing, melanomas (Extended data figure 3a and b).
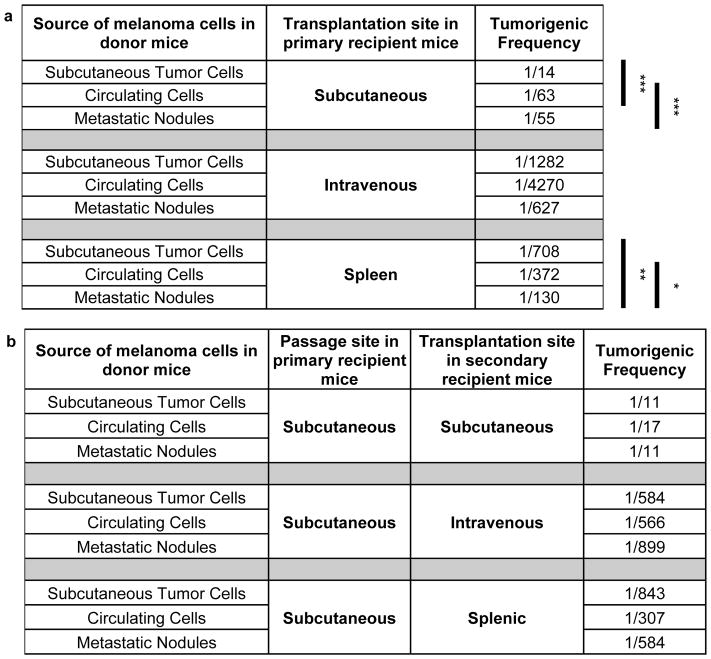
Table 1. Melanoma cells more readily formed tumors upon subcutaneous injection as compared to intravenous or intrasplenic injection.
Limit dilution analysis of data in Extended data figure 3 to infer the minimum frequency of tumor-forming melanoma cells after subcutaneous, intravenous, or intrasplenic transplantation into NSG mice. Each mean value reflects 5 (subcutaneous), 10 (intravenous), or 4 (intrasplenic) independent experiments using melanomas obtained from 8 different patients (see details in Extended data figure 3). Statistical significance was assessed by a chi-square test using the ELDA software46
| Melanoma | Subcutaneous | Intravenous | Intrasplenic | |||
|---|---|---|---|---|---|---|
| Inefficient | 1/11 | 1/2540 |
# |
1/3677 |
# |
|
| Efficient | 1/8 | 1/235 | 1/173 | |||
p<0.0001.
To test if the main difference between efficient and inefficient metastasizers is the ability to enter circulation, we intravenously injected 10, 100, 1000, or 10,000 cells from efficiently and inefficiently metastasizing melanomas into NSG mice. The melanoma cells were marked by luciferase expression, allowing us to confirm micro and macrometastases by bioluminescence imaging. Efficiently metastasizing melanomas from all four patients formed macrometastases in multiple visceral organs (Extended data figure 3c). Limiting dilution analysis indicated that at least 1 in 235 cells formed tumours upon intravenous injection (Table 1). In contrast, only 1 in 2,540 cells from inefficiently metastasizing melanomas formed tumours after intravenous injection (Table 1 and Extended data figure 3d). Therefore the ability to enter circulation is not the only factor that limits distant metastasis.
These data also demonstrate that even efficiently metastasizing melanomas more readily formed tumours after subcutaneous injection (1 in 8 cells) as compared to intravenous injection (1 in 235 cells, Table 1). This was true even when efficiently metastasizing melanoma cells were injected subcutaneously without Matrigel (1 in 60 cells formed tumors; p<0.001). This suggests the blood was more hostile to melanoma cells than the subcutaneous environment.
If distant metastasis is limited mainly by survival in the blood, direct injection into a visceral organ should increase tumour formation. To test this we injected efficiently and inefficiently metastasizing melanomas into the spleens of NSG mice. Efficiently metastasizing melanomas from two of three patients formed macrometastases in multiple visceral organs in most mice (Extended data figure 3e and Table 1; at least 1 in 173 cells formed tumours upon intrasplenic injection). In contrast, only 1 in 3,677 cells from inefficiently metastasizing melanomas formed tumours after intrasplenic injection (Extended data figure 3f and Table 1). The ability to survive in circulation is therefore not the only factor that limits distant metastasis. The spleen is also a relatively hostile environment for melanoma cells.
Reversible tropic changes during metastasis
To test whether melanoma cells undergo changes in properties during metastasis, we obtained efficiently metastasizing melanoma cells from 12 donor mice that had been grafted with melanomas from 3 patients (M481, M405 and UT10). We compared the capacity of melanoma cells from subcutaneous tumours versus the blood versus etastatic liver nodules (2–5 mm in diameter) in the same donor mice to form tumours upon subcutaneous, intravenous, or intrasplenic injection in recipient mice (Table 2a; see Extended data figure 3g for experimental design). Upon subcutaneous injection, melanoma cells from subcutaneous tumors were significantly better at forming tumours (1 in 14 cells formed tumours) as compared to melanoma cells from the blood (1 in 63 cells) or metastatic nodules (1 in 55 cells; Table 2a). In contrast, upon intrasplenic injection, melanoma cells from metastatic nodules were significantly better at forming tumours (1 in 130 cells) as compared to melanoma cells from the blood (1 in 372 cells) or subcutaneous tumours (1 in 708 cells; Table 2a). This suggests melanoma cells adapt to sites of metastasis as they metastasize.
Table 2. Melanoma cells undergo reversible changes in tropism during metastasis.
a) Schematic and limiting dilution analysis of the frequency of melanoma cells from subcutaneous tumours, the blood (circulating cells), or metastatic nodules that formed tumours after subcutaneous, intravenous, or intrasplenic transplantation. These data reflect three independent experiments performed with efficiently metastasizing melanomas (M405, M481 and UT10; n=10 mice/melanoma/melanoma cell source/transplantation site for a total of 270 mice). b) Schematic and limiting dilution analysis of the frequency of melanoma cells from subcutaneous tumours, the blood (circulating cells), or metastatic nodules that formed tumours after being passaged subcutaneously in primary recipient mice and then transplanted subcutaneously, intravenously, or intrasplenically into secondary recipient mice. These data reflect one experiment performed with efficiently metastasizing melanoma cells (M481; n=8–10 mice/melanoma cell source/transplantation site for a total of 85 mice). Statistical significance was assessed by a chi-square test using ELDA software46
p<0.05;
p<0.005;
p<0.0005.
To test whether the changes were irreversible (i.e. genetic) or reversible (i.e. epigenetic or metabolic), we tested whether small numbers of metastatic melanoma cells would reacquire subcutaneous properties after being passaged subcutaneously for a short period of time. We subcutaneously transplanted 100 cells from subcutaneous tumours, the blood, or metastatic liver nodules of the same donor mice into primary recipient mice and allowed them to form tumours for up to 12 weeks (Table 2b; all injections formed tumours). Then we retransplanted melanoma cells from the subcutaneous tumours into subcutaneous, intravenous, or intrasplenic sites in secondary recipient mice (Extended data figure 3h shows experimental design). Melanoma cells that originated in all sites formed subcutaneous tumours with high efficiency in secondary recipient mice after being passaged subcutaneously (Table 2b). Melanoma cells that originated in all sites formed metastatic tumours with low efficiency after intravenous or intrasplenic injections in secondary recipient mice after being passaged subcutaneously (Table 2b). The changes in tumourigenic tropism during metastasis are thus reversible. Since these reversible changes were observed very consistently, even in small numbers of melanoma cells, these changes cannot reflect selection for rare genetic events during metastasis followed by reversion mutations after subcutaneous re-transplantation.
Oxidative stress limits distant metastasis
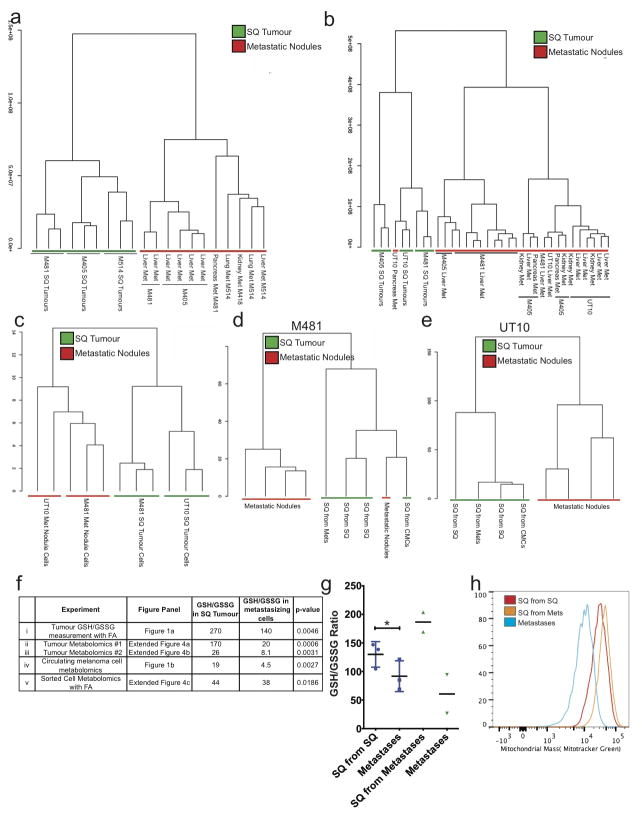
We performed LC-MS/MS metabolomics on subcutaneous tumours and visceral metastatic nodules from the same NSG mice transplanted with efficiently metastasizing melanomas derived from 4 patients. In two independent experiments, unsupervised clustering of metabolomics data showed that metastatic nodules obtained from the liver, pancreas, and kidney almost always clustered together, distinct from subcutaneous tumours, irrespective of xenograft line (Extended data figure 4a and b; complete metabolomics data are shown in Extended data tables 1 and 2). Among subcutaneous tumours and among metastatic nodules, samples clustered by patient. We obtained similar results when melanoma cells were isolated by flow cytometry, excluding mouse cells (Extended data figure 4c). The metabolic differences between subcutaneous tumours and visceral metastases appeared to be largely reversible as metabolomics analysis of tumours from Table 2b showed that subcutaneous tumours clustered together irrespective of whether they arose from the transplantation of subcutaneous, circulating, or metastatic cells (Extended data figure 4d and e).
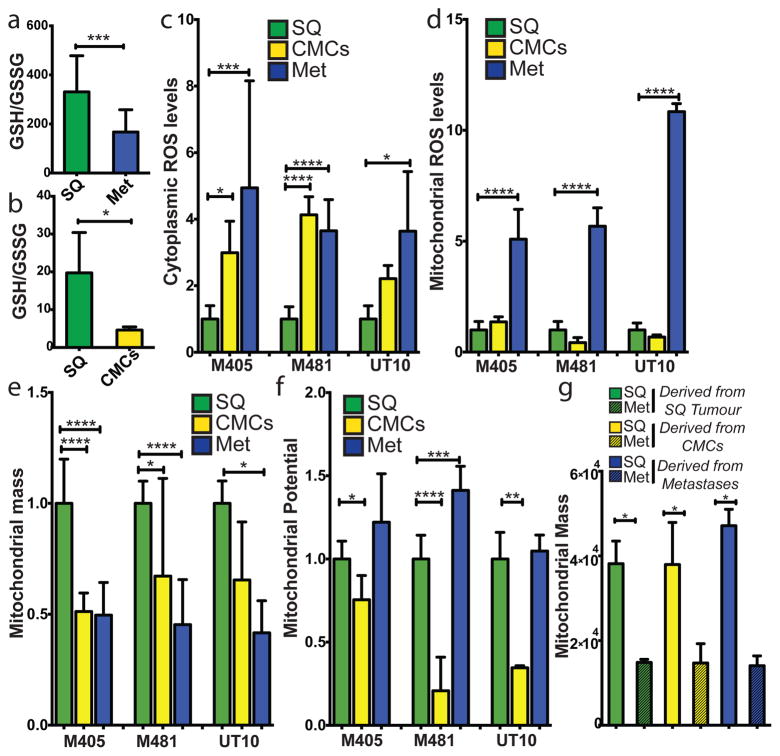
In a total of six independent experiments that involved four different technical approaches, the glutathione (GSH) to oxidized glutathione (GSSG) ratio was always significantly higher in subcutaneous tumours as compared to metastatic nodules or circulating melanoma cells (Figure 1a and 1b and Extended data figure 4f). This was true irrespective of whether melanoma cells were isolated by dissection or by flow cytometry (to eliminate stromal cells). The lower GSH/GSSG ratio in circulating melanoma cells and metastatic nodules suggested that metastasizing cells experienced oxidative stress not observed in established subcutaneous tumours, and they consumed GSH in an effort to maintain redox homeostasis.
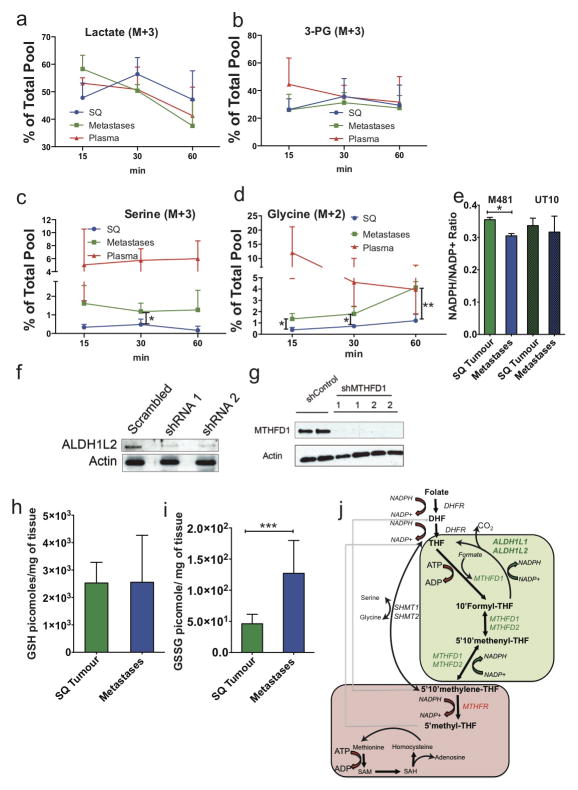
Figure 1. Metastasizing melanoma cells experience high levels of oxidative stress.
a) GSH/GSSG ratio in subcutaneous tumours as compared to metastatic nodules (n=15 mice from 2 independent experiment with 3 melanomas, M481, M405, UT10; note that extractions were performed with 0.1% formic acid to prevent spontaneous oxidation47). Total amounts of GSH and GSSG are shown in Extended data Figure 5h and 5i. b) GSH/GSSG ratio in subcutaneous tumours as compared to circulating melanoma cells (n=7 mice from 3 independent experiments with 2 melanomas, M405 and UT10; these were different experiments than those in panel a, performed under different technical conditions). c, d) cytoplasmic (c) and mitochondrial (d) ROS levels in dissociated melanoma cells from subcutaneous tumours, the blood, and metastatic nodules obtained from the same mice (n=9 mice from 3 independent experiments using 3 different melanomas). e, f) Mitochondrial mass (e) and mitochondrial membrane potential (f) in dissociated melanoma cells from subcutaneous tumours, the blood, and metastatic nodules obtained from the same mice (n=6 mice from 2 independent experiments using 3 different melanomas). g) Melanoma cells underwent reversible changes in mitochondrial mass during metastasis: mitochondrial mass in dissociated melanoma cells from subcutaneous tumours versus metastatic nodules obtained from the same mice transplanted with subcutaneous, circulating, or metastatic melanoma cells. All data represent mean±sd. Statistical significance was assessed using two-tailed Student’s t-tests (a and b) and one-way analyses of variance (ANOVAs) followed by Dunnett’s tests for multiple comparisons (c–g; *, p<0.05; ***, p< 0.0005; ****, p<0.00005).
Consistent with this, cytoplasmic ROS levels were significantly higher in circulating melanoma cells and visceral metastatic nodules as compared to subcutaneous tumours (Figure 1c). Mitochondrial ROS levels were significantly higher in visceral metastatic nodules as compared to circulating melanoma cells and subcutaneous tumours (Figure 1d).
Mitochondrial respiration is one of the main sources of ROS. Mitochondrial mass declined significantly in circulating melanoma cells and metastatic nodules as compared to subcutaneous tumours (Figure 1e). Mitochondrial membrane potential also declined significantly in circulating melanoma cells as compared to subcutaneous tumours but not in metastatic nodules (Figure 1f). These data raised the possibility that mitochondrial function is reduced in circulating melanoma cells in an effort to reduce ROS generation.
These changes in redox status between subcutaneous and metastasizing cells appeared to be reversible. Even though the GSH/GSSG ratio was always higher in subcutaneous tumours as compared to metastatic nodules, the GSH/GSSG ratio in subcutaneous tumours derived from the transplantation of metastatic cells was even higher than the GSH/GSSG ratio in subcutaneous tumours derived from the transplantation of subcutaneous cells (Extended data figure 4g).
Changes in mitochondrial mass between subcutaneous and metastatic melanoma cells were also reversible. Subcutaneous tumours always exhibited significantly higher mitochondrial mass as compared to metastatic nodules, irrespective of whether the subcutaneous tumours arose from the transplantation of subcutaneous tumour cells, circulating cells, or metastatic cells (Figure 1g and Extended data figure 4h).
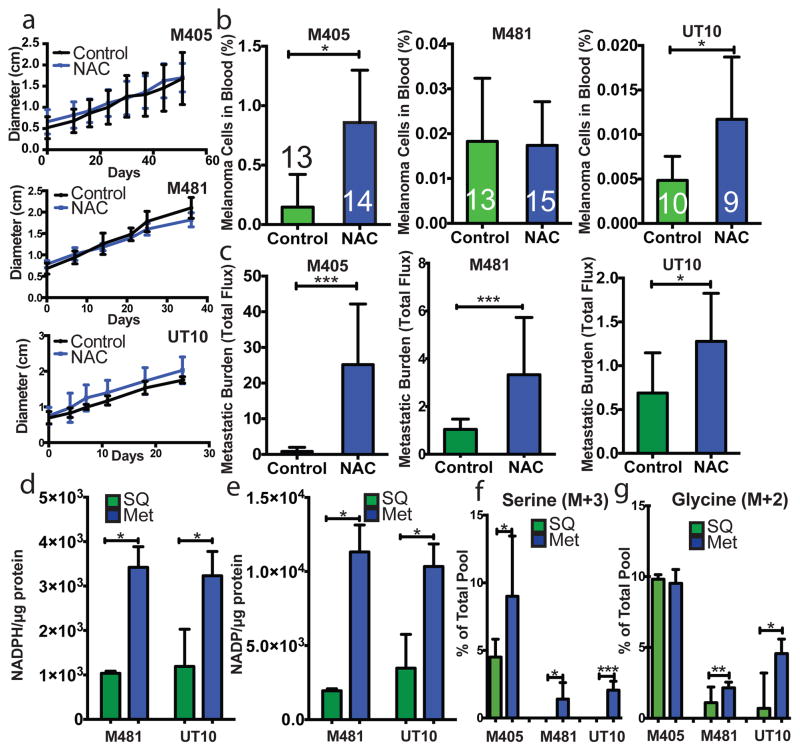
To test if oxidative stress limits melanoma metastasis we subcutaneously transplanted efficiently metastasizing melanoma cells derived from three patients into NSG mice and treated the mice with daily subcutaneous injections of the anti-oxidant N-acetyl-cysteine (NAC; 200 mg/kg/day). In no case did NAC treatment significantly affect the growth of established subcutaneous tumours (Figure 2a) but it significantly increased the frequency of melanoma cells in the blood of mice transplanted with M405 and UT10 (Figure 2b) and significantly increased metastatic disease burden in mice with all three melanomas (Figure 2c). Oxidative stress therefore limits the metastasis of melanoma cells in vivo.
Figure 2. Melanoma cell metastasis, but not subcutaneous tumour growth, is promoted by anti-oxidants in vivo.
a–c) Growth of established subcutaneous tumours in NSG mice treated with either PBS (Control) or N-acetyl-cysteine (NAC) by daily subcutaneous injection. Tumor diameter source data are shown in Supplementary Figure 1. Frequency of circulating melanoma cells in the blood (b) and metastatic disease burden (c) assessed based on total bioluminescence signal from the visceral organs of the same mice. Data in panels a–c represent 8 independent experiments with total replicates/treatment shown in the bars of panel b. Panel a shows a single representative experiment per melanoma due to the difficulty of reflecting tumour growth measurements from independent experiments in the same graph. No statistically significant differences among treatments were observed in subcutaneous tumor growth in any experiment. d and e) Levels of NADPH (d) and NADP (e) in subcutaneous tumours versus metastatic nodules (n=4 mice from 2 independent experiments with M481 and UT10). f and g) In vivo isotope tracing of uniformly 13C-labelled glucose into serine (f), and glycine (g) in subcutaneous tumours versus metastatic nodules from the same mice (n=6 mice in 2 independent experiments for M405; n=3 mice in one experiment for each of M481 and UT10). The fragments for uniformly labeled serine (M+3) and glycine (M+2), which come from labeled glucose via de novo serine synthesis, are shown. All data represent mean±sd. Statistical significance was assessed using two-tailed Student’s t-tests (d–f, h and i), or the Mann-Whitney test (g, due to unequal variance), and repeated-measures two-way analyses of variance (ANOVAs) (a–c; *. p<0.05; **, p<0.005; ***, p< 0.0005, ****, p<0.00005).
Among efficiently metastasizing melanomas from three patients, NAC pre-treatment of cells and administration to mice increased tumour formation after intravenous injection by 10-fold (p<0.0001; Extended data table 3a). Among inefficiently metastasizing melanomas from two patients, tumours only arose from cells treated with NAC (Extended data table 3b). Oxidative stress thus limits tumorigenesis by circulating melanoma cells.
Metabolic adaptations during metastasis
We hypothesized that successfully metastasizing cells undergo reversible metabolic changes that increase their capacity to withstand oxidative stress. One such adaptation that could promote survival would be increased GSH regeneration28–30. NADPH is needed to convert GSSG into GSH and increased production of NADPH promotes the regeneration of GSH28–30. In fact, we observed significantly higher levels of NADPH and NADP in metastatic cells as compared to subcutaneous tumours (Figure 2d and e). The NADPH/NADP ratio was significantly reduced in metastases as compared to subcutaneous tumours for one melanoma (M481) but not for a second (UT10; Extended data Figure 5e). The higher levels of NADPH in metastases raised the possibility that metastasizing cells generate more NADPH to increase their capacity to regenerate GSH. The oxidative stress in these cells would be predicted to consume more NADPH, potentially explaining why the NADPH/NADP ratio did not necessarily change despite increases in the amounts of NADPH and NADP.
Metastatic nodules had high levels of purine intermediates relative to subcutaneous tumors in the same mice (Extended data tables 1 and 2). This could reflect changed folate pathway activity. Moreover, folate metabolism is a major source of NADPH for oxidative stress management29–31, raising the question of whether the folate pathway promotes distant metastasis. NADPH regeneration by the folate pathway involves the conversion of serine to glycine and the donation of a methyl group to tetrahydrofolate (see Extended data figure 5j). Serine can be imported into cells as well as produced de novo from glucose via the glycolytic intermediate 3-phosphoglycerate (3-PG). Elevated de novo serine synthesis promotes the growth of some melanomas and breast cancers32,33. To test for alterations of these pathways during metastasis we administered uniformly 13C labeled glucose to NSG mice bearing melanomas derived from three patients. We observed significantly increased 13C labeling of serine and glycine in metastatic tumours as compared to subcutaneous tumours (Figure 3f and 3g and Extended data figure 5c and 5d). However, we observed no differences between subcutaneous tumours and metastases in terms of the fractional enrichments of uniformly labeled lactate or 3-PG, the precursor used for de novo serine synthesis (Extended data figure 5a and 5b). Metastatic tumours thus display enhanced contribution of glucose carbon to tissue serine and glycine levels, suggesting increased flux through the folate pathway.
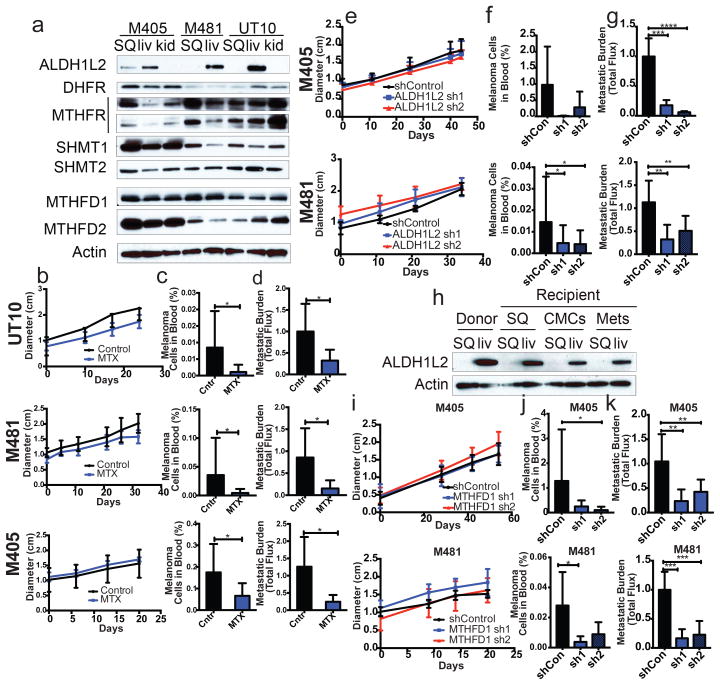
Figure 3. During metastasis, some melanoma cells reversibly increase their expression of folate pathway enzymes that generate NADPH and folate pathway inhibition selectively impairs metastasis.
a) Western blot analysis of folate pathway enzymes in subcutaneous tumours versus metastatic liver and kidney nodules from NSG mice transplanted with three different melanomas. b–d) Growth of subcutaneous tumours in mice bearing three different melanomas (M405, M481, UT10) treated with DMSO (control) or methotrexate (n=5 mice/treatment). The frequency of circulating melanoma cells in the blood (c) and metastatic disease burden (d) in the same mice (n=10 mice/treatment for each melanoma except n=8 for M405). Data in panels b–d reflect 6 independent experiments but only one representative experiment per melanoma is shown in panel b. e–g) Growth of subcutaneous tumours in mice transplanted with two different melanomas expressing scrambled control shRNA versus two shRNAs against ALDH1L2. The frequency of circulating melanoma cells (f) and metastatic disease burden in visceral organs based on total bioluminescence signal (g). The data in panels e–g reflect 6 independent experiments (n=10 mice per shRNA for M405 and n=19 mice per shRNA for M481) but only one representative experiment per melanoma is shown in panel e. h) Western blot analysis of ALDH1L2 expression in subcutaneous tumours versus metastatic liver nodules from a donor mouse or from recipient mice subcutaneously transplanted with subcutaneous, circulating, or metastatic melanoma cells from the donor mouse. The increase in ALDH1L2 expression in metastatic liver nodules was reversible upon subcutaneous transplantation. Data in panels a and h are from two independent experiments. i–k) Growth of subcutaneous tumours in mice transplanted with cells from two melanomas expressing either scrambled control shRNAs or two shRNAs against MTFHD1. Frequency of circulating melanoma cells (j) and metastatic disease burden in visceral organs (k) from the same mice. Data in panels i–k reflect four independent experiments with a total of 9 mice per control shRNA and 10 mice per shRNA against MTFHD1 for each melanoma. Panel i shows one representative experiment per melanoma. All error bars represent standard deviation. Statistical significance was assessed using one-way analyses of variance (ANOVAs) followed by Dunnett’s tests for multiple comparisons (f, g, j and k), two-tailed Student’s t-tests (c and d) and repeated measures two-way ANOVAs (b, e and i) (*, p<0.05; **, p,0.005, ***, p< 0.0005; ****, p<0.00005). Tumor diameter and western blot source data are in Supplementary Figure 1.
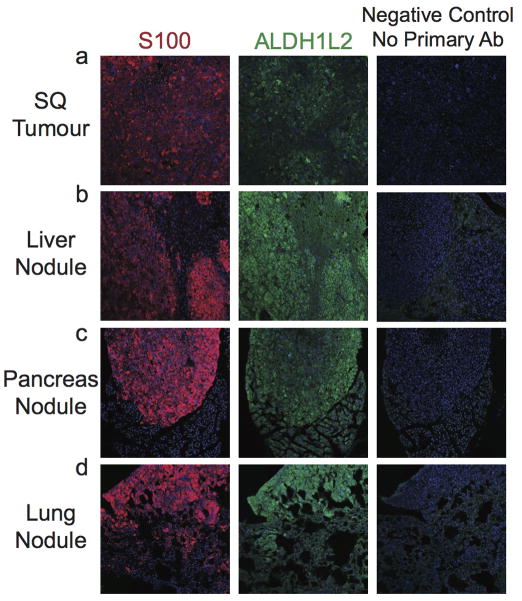
Western blotting and immunofluorescence analysis of key enzymes in the folate pathway showed a striking increase in the expression of the NADPH-regenerating enzyme ALDH1L2 in liver, pancreas, and lung metastases compared to subcutaneous tumours, though not in kidney metastases (Figure 3a; Extended data figure 6). Some metastases also showed reduced expression of an NADPH-consuming folate metabolism enzyme, MTHFR (Figure 3a). The expression of other folate metabolism enzymes did not consistently change during metastasis, nor did a variety of other enzymes in anti-oxidant pathways (Figure 3a; Extended data table 4).
We compared ALDH1L2 levels in subcutaneous tumours versus metastatic nodules from mice transplanted with subcutaneous, circulating, or metastatic melanoma cells. ALDH1L2 was much more highly expressed in liver nodules than subcutaneous tumours in both donor and recipient mice, irrespective of whether the subcutaneous tumours arose from subcutaneous, circulating, or metastatic melanoma cells (Figure 3h). The changes in ALDH1L2 expression during metastasis were thus reversible.
To test whether metastasizing cells are more dependent than subcutaneous cells on the folate pathway we transplanted melanoma cells from three patients subcutaneously in NSG mice and treated with low dose of methotrexate (1.25 mg/kg/day), an inhibitor of dihydrofolate reductase (Extended data figure 5j). We simultaneously provided thymidine (3 mg/kg/day) and hypoxanthine (750 μg/mg/day) to the mice to ameliorate the effects of folate pathway inhibition on nucleotide metabolism. Methotrexate treatment in these conditions had no significant effect on the growth of subcutaneous tumours (Figure 3b); however, the frequency of circulating melanoma cells in the blood of the same mice was significantly reduced (Figure 3c), as was metastatic disease burden (Figure 3d), in mice bearing all three melanomas. Metastasizing melanoma cells are therefore particularly sensitive to folate pathway inhibition.
Depletion of ALDH1L2 can decrease the GSH/GSSG ratio in vitro30. To test whether ALDH1L2 is required for metastasis we identified two shRNAs that knocked down ALDH1L2 expression in melanoma cells (Extended data figure 5f). We infected melanoma cells derived from three patients with either of these shRNAs or a scrambled control shRNA then injected the cells subcutaneously in NSG mice. Neither shRNA against ALDH1L2 significantly affected the growth of subcutaneous tumours (Figure 3e) but both significantly reduced the frequency of circulating melanoma cells in the blood of mice bearing 1 of 2 melanomas (Figure 3f) and overall metastatic disease burden in both melanomas (Figure 3g). ALDH1L2 thus promotes melanoma metastasis in vivo.
We performed a similar experiment on another NADPH-regenerating enzyme in the folate pathway, MTHFD1 (Extended data figure 5j). We identified two shRNAs that knocked down MTHFD1 expression in melanoma cells (Extended data figure 5g). Neither shRNA against MTHFD1 significantly affected the growth of subcutaneous tumours (Figure 3i) but they reduced the frequencies of circulating melanoma cells in the blood (Figure 3j) and metastatic disease burden in mice bearing both melanomas (Figure 3k). MTHFD1 thus promotes melanoma metastasis in vivo.
Our results suggest that few circulating cancer cells survive and proliferate6–8 because of oxidative stress. Our results further suggest that metabolic pathways that regenerate NAPDH and buffer oxidative stress represent therapeutic targets to impede distant metastasis. Metastasizing melanoma cells are more dependent on certain NADPH-regenerating folate pathway enzymes than subcutaneous melanoma cells. However, these enzymes cannot account for the increase in NADP in metastatic nodules as compared to subcutaneous tumours (Figure 2d and 2e). Therefore, metastasizing cells likely also undergo additional metabolic changes that promote the generation of NADP and NADPH.
Changes in the expression of individual folate pathway enzymes are unlikely to be sufficient to drive metastasis. Since the key changes driving distant metastasis were reversible adaptations rather than stochastic genetic changes, there is no reason why these changes would need to be driven by a single sufficient driver. Adaptive changes might involve coordinated changes in the expression of multiple enzymes to increase flux through metabolic pathways. Other metabolic pathways also contribute to the synthesis and regeneration of NADPH, including the pentose phosphate12,13 and malate34 pathways. These pathways could potentially also contribute to the ability of melanoma cells to survive during metastasis.
Our experiments showed that oxidative stress increases in metastasizing cells as compared to established subcutaneous tumors and limits distant metastasis. However, we did not test whether oxidative stress also limits the initiation and early growth of primary cutaneous melanomas. Thus, transient oxidative stress could occur during the formation of primary tumors15–18 in addition to limiting distant metastasis at a later stage of cancer progression.
ROS can cause oncogenic mutations and activate oncogenic pathways, raising the possibility that treatment with anti-oxidants could suppress the initiation or progression of some cancers28,35. Anti-oxidants or anti-oxidant enzymes suppress cancer initiation in some contexts36–38 while increasing cancer initiation in other contexts 15–17,39. Increasing dietary antioxidants has generally not reduced cancer incidence in clinical trials40. Dietary supplementation with anti-oxidants actually increased incidence and death from lung and prostate cancer41–43. Dietary supplementation with folate promotes the development and progression of breast cancer44,45. Our results suggest that anti-oxidants promote disease progression, at least in melanoma, by promoting metastasis.
METHODS
Obtaining melanomas and enzymatic dissociation
Melanoma specimens were obtained with informed consent from all patients according to protocols approved by the Institutional Review Boards of the University of Michigan Medical School (IRBMED approvals HUM00050754 and HUM00050085; see ref27) and the University of Texas Southwestern Medical Center. Tumours were dissociated in Sterile Closed System Tissue Grinders (SKS Science) in enzymatic digestion medium containing 200 U/ml collagenase IV (Worthington) for 20 minutes at 37°C. DNase (50–100 U/ml) was added to reduce clumping of cells during digestion. Cells were filtered with a 40 μm cell strainer to obtain a single-cell suspensions.
Cell labeling and sorting
All melanomas used in this study stably expressed DsRed and luciferase so that the melanoma cells could be unambiguously distinguished from mouse cells by flow cytometry and by bioluminescence imaging. When isolated by flow cytometry, cells were also stained with antibodies against mouse CD45 (30-F11-APC, eBiosciences), mouse CD31 (390-APC, Biolegend), Ter119 (TER-119-APC, eBiosciences) and human HLA-A,B,C (G46-2.6-FITC, BD Biosciences) in order to select live human melanoma cells and to exclude contaminating mouse endothelial and haematopoietic cells. Live human melanoma cells were thus isolated by flow cytometry by sorting cells that were positive for DsRed and HLA and negative for mouse CD45, Ter119, and CD31. All antibody labeling was performed for 20 minutes on ice, followed by washing and centrifugation. Prior to flow cytometric analysis, cells were re-suspended in staining medium (L15 medium containing bovine serum albumin (1 mg/ml), 1% penicillin/streptomycin, and 10 mM Hepes (pH 7.4)) containing 4′,6-diamidino-2-phenylindole (DAPI; 5 μg/ml; Sigma) to eliminate dead cells from sorts and analyses. Sorts and analyses were performed using a FACSAria flow cytometer (Becton Dickinson). After sorting, an aliquot of sorted melanoma cells was always reanalysed to check for purity, which was usually greater than 95%. For analysis of circulating melanoma cells, blood was collected from each mouse by cardiac puncture with a syringe pretreated with citrate-dextrose solution (Sigma). Red blood cells were precipitated by Ficoll sedimentation according to the manufacturer’s instructions (Ficoll Paque Plus, GE Healthcare). Remaining cells were washed with Hanks’ balanced salt solution (Invitrogen) before antibody staining and flow cytometric analysis. For limiting dilution analysis, cells for each mouse were sorted into individual wells of 96-well V-bottomed plates containing staining medium and loaded into syringes directly from the well (one well into one syringe into one mouse).
Transplantation of melanoma cells
After sorting, cells were counted and resuspended in staining medium with 25% high-protein Matrigel (product 354248; BD Biosciences). Subcutaneous injections were performed into the right flank of NOD.CB17-Prkdcscid Il2rgtm1Wjl/SzJ (NOD/SCID IL2Rγnull or NSG) mice (Jackson Laboratory) in a final volume of 50 μl. Each mouse was transplanted with 100 melanoma cells unless otherwise specified. Tumour formation was evaluated regularly by palpation of the injection site, and the subcutaneous tumours were measured every 10 days until any tumour in the mouse cohort reached 2.5 cm in its largest diameter. Mice were monitored daily for signs of distress and euthanized according to a standard body condition score or when their tumours reached 2.5 cm in largest diameter, whichever came first. We adhered to this limit in all experiments. Organs were analyzed visually and by bioluminescence imaging (see details below) for presence of macrometastases and micrometastases. These experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center (protocol 2011-0118). Intravenous injections were done by injecting cells into the tail vein of NSG mice in 100 μl of staining medium. For intrasplenic injections the mice were anesthetized with isoflourane, then the left flank was shaved and disinfected with an ethanol wipe and iodine swab. An incision was made into the intraperitoneal cavity. The spleen was exposed with forceps and cells were injected slowly in a 40μl volume of staining medium. The peritoneum was then sutured and skin was closed with clips. Mice were injected with buprenex before surgery and then again 12 hours after surgery.
Lentiviral transduction of human melanoma cells
A bicistronic lentiviral construct carrying dsRed2 and luciferase (dsRed2-P2A-Luc) was generated (for bioluminescence imaging) and cloned into the FUW lentivrial expression construct. The primers that were used for generating this construct were: dsRed2 forward, 5′-CGACTCTAGAGGATCCatggatagcactgagaacgtc-3′ (capital letters indicate homology to FUW backbone); dsRed2 reverse, 5′-TCCACGTCTCCAGCCTGCTTCAGCAGGCTGAAGTTAGTAGCTCCGCTTCCctggaacaggtggtg gc-3′ (capital letters indicate P2A sequences); luciferase forward, 5′-GCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTGGATCCatggaagacgcc aaaaacataaag-3′ (capital letters indicate P2A sequences) and luciferase reverse, 5′-GCTTGATATCGAATTCttacacggcgatctttccgc-3′ (capital letters indicate homology to FUW backbone). All constructs were generated using the In-Fusion HD cloning system (Clontech) and sequence verified.
For virus production, 0.9 μg of the appropriate plasmid and 1 μg of helper plasmids (0.4 μg pMD2G and 0.6 μg of psPAX2) were transfected into 293T cells using polyjet (Signagen) according to the manufacturers instructions. Replication incompetent viral supernatants were collected 48 hours after transfection and filtered through a 0.45 μm filter. 300,000 freshly dissociated melanoma cells were infected with viral supernatants supplemented with 10 μg/ml poybrene (Sigma) for 4 hours. Cells were then washed twice with staining medium, and about 25,000 cells (a mixture of infected and noninfected cells) were suspended in staining medium with 25% high-protein Matrigel (product 354248; BD Biosciences) then injected subcutaneously into NSG mice. After growing to 1 to 2 cm in diameter, tumours were excised and dissociated into single-cell suspensions, and luciferase-dsRed+ or GFP+ cells were collected by flow cytometry for injection into secondary recipients. Metastasis was monitored by bioluminescence imaging in secondary recipients. All shRNAs were expressed from a pGIPZ miRNA-based construct with TurboGFP from GE Dharmacon (Lafayette, Colorado). For ALDH1L2, the following GE Dharmacon shRNA clones were used: V2LHS_30207, V2LHS_30209. For MTHFD1 the following GE Dharmacon shRNA clones were used: V2LHS_216208 and V2LHS_196832.
Bioluminescence imaging
Mice were injected with 100 luciferase-dsRed+ cells on the right flank and monitored until tumour diameters approached 2.5 cm, at which point they were imaged along with an uninjected control mouse using an IVIS Imaging System 200 Series (Caliper Life Sciences) with Living Image software. Mice were injected intraperitoneally with 100 μl of PBS containing D-luciferin monopotassium salt (40 μg/ml) (Biosynth) 5 minutes before imaging, followed by general anesthesia 2 minutes before imaging. After imaging of the whole mouse, the mice were euthanized and individual organs were surgically removed and quickly imaged. The exposure time of images ranged from 10 to 60 seconds depending on signal intensity. The bioluminescence signal was quantified with “region of interest” measurement tools in Living Image (Perkin Elmer) software. After imaging, tumours and organs were fixed in 10% neutral-buffered formalin for histopathology. For live imaging, mice were imaged once a month, and whole body bioluminescence was quantified using Living Image Software (Perkin Elmer).
LC-MS/MS metabolomic analysis
Mice were euthanized by cervical dislocation. Subcutaneous tumours and metastatic nodules were dissected, immediately homogenized in 80% methanol chilled with dry ice (Honeywell), vortexed vigorously, and metabolites were extracted overnight at −80°C. The following day, samples were centrifuged at 13,000xg for 15 minutes at 4°C, the supernatant was collected, and metabolites from the pellet were re-extracted with 80% methanol at −80°C for 4 hours. After centrifugation, both supernatants were pooled and lyophilized using a SpeedVac (Thermo). To inhibit spontaneous oxidation, samples were extracted with 80% methanol containing 0.1% formic acid in some experiments47. Dried metabolites were reconstituted in 0.03% formic acid in water, vortexed and centrifuged, then the supernatant was analysed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). A Nexera Ultra High Performance Liquid Chromatograph (UHPLC) system (Shimadzu) was used for LC, with a Polar-RP HPLC column (150 × 2 mm, 4 μm, 80 Å, Phenomenex) and the following gradient: 0–3 min 100% mobile phase A; 3–15 min 100%–0% A; 15–17 min 0% A; 17–18 min 0%–100% A; 18–23 min 100% A. Mobile Phase A was 0.03% formic acid in water. Mobile Phase B was 0.03% formic acid in acetonitrile. The flow rate was 0.5 ml/min and the column temperature 35°C. A triple quadrupole mass spectrometer (AB Sciex QTRAP 5500) was used for metabolite detection as previously described48. Chromatogram peak areas were integrated using Multiquant (AB Sciex). To measure GSH and GSSG levels, some metabolite extractions were performed with 0.1% formic acid in 80% methanol, to inhibit spontaneous GSH oxidation. To calculate GSH and GSSG amounts, a standard curve was prepared by adding known quantities of GSH and GSSG to tumour metabolite extracts.
Isotope tracing with [U-13C]glucose
Mice were injected intraperitoneally with 2g/kg body mass of [U-13C]Glucose (Cambridge Isotopes) and were analyzed 15, 30, and 60 minutes later. Mice were fasted for 14 hours prior to the injection. In most experiments, subcutaneous tumours and metastatic nodules were surgically excised and homogenized in ice cold 50% methanol for GC-MS and in 80% dry ice cold methanol for LC-MS analysis. Metabolites were extracted with three freeze-thaw cycles in liquid nitrogen. Supernatant was collected after a 15 minute centrifugation at 13,000xg at 4°C and lyophilized. Metabolites were derivatized with Trimethylsilyl (TMS) at 42°C for 30 minutes for GC-MS analysis. 13C enrichment analysis was performed by GC-MS as previously described48. For LC-MS analysis, lyophilized samples were resuspended in either 0.03% formic acid in water or in 5 mM Ammonium Acetate in water depending on the method of analysis. For 13C enrichment analysis of lactate, serine, and glycine by LC-MS/MS, we used the LC procedure described above for LC-MS/MS metabolomics analysis with the following modifications: the LC gradient was 0–3 minute 100% mobile phase A; 3–15 min 100%–0% A; 15–17 min 0% A; 17–17.5 min 0%–100% A; 17.5–20 min 100% A. For analysis of 3-phosphoglycerate (3-PG), the LC conditions were: mobile phase A, 5 mM ammonium acetate in water and mobile phase B, 5 mM ammonium acetate in acetonitrile, and a Fusion-RP HPLC column (150 × 2 mm, 4 μm, 80 Å, Phenomenex). The LC gradient was: 0–3 min 100% mobile phase A; 3–9 minute 100%–0% A; 9–11 minute 0% A; 11–12 minute 0%–100% A; 12–15 minute 100% A. For metabolite detection a triple quadrupole mass spectrometer (AB Sciex QTRAP 5500) was used on multiple reaction monitoring (MRM) mode as previously described, with some modifications48. The following transitions were used: positive mode: serine 106.1/60 (M+1: 107.1/60 and 107.1/61, M+2: 108.1/61 and 108.1/62, M+3: 109.1/62), glycine 76/30 (M+1 77/30 and 77/31, M+2 78/31); negative mode: lactate 89/43 (M+1 90/43 and 90/44, M+2 91/44 and 91/45, M+3 92/45), 3-PG 185/79 (M+1 186/79, M+2 187/79, M+3 188/79) and 185/97 (M+1 186/97, M+2 187/97, M+3 188/97). Unlabeled tissue was used as a negative control to confirm isotopic labeling in specific transitions.
Flow cytometric analysis of mitochondrial mass, mitochondrial membrane potential and ROS
Melanomas were generally dissociated enzymatically as described above. Equal numbers of dissociated cells (500,000–2,000,000) from subcutaneous tumours, Ficoll-depleted blood, or metastatic nodules were loaded with dyes to assess mitochondrial mass, mitochondrial membrane potential, and ROS levels. The dyes that were used to assess these parameters were all obtained from Life Technologies. We stained the dissociated cells for 20–45 minutes at 37°C with 5 μM Mitotracker Green, Mitotracker DeepRed, CellROX Green, or CellROX DeepRed in HBSS-free (Ca2+ and Mg2+ free) to assess mitochondrial mass, mitochondrial membrane potential, mitochondrial and cytoplasmic ROS, respectively. For each indicator, staining intensity per cell was assessed by flow cytometry in live human melanoma cells (positive for human HLA and dsRed and negative for DAPI and mouse CD45/CD31/Ter119).
In vivo treatment of xenografts with drugs
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center (protocol 2011-0118). Unless otherwise stated, 100 freshly dissociated melanoma cells were injected subcutaneously into the right flanks of NSG mice. When tumours became palpable, in some experiments mice were injected subcutaneously with N-acetyl-cysteine (NAC) (Sigma, 200 mg/kg/day in 200 μl PBS, pH 7.4) or PBS as a control. Mice were injected with their last NAC dose 10 minutes before being sacrificed for end point analysis. In experiments where mice received NAC via the drinking water, NAC was dissolved in PBS at 1mg/ml and the water was changed every 2 days. In other experiments methotrexate (Tocris, 1.25 mg/kg/day in 100 μl PBS) was injected intraperitoneally 5 days per week. Mice that received methotrexate were simultaneously administered Thymidine (Sigma, 3 mg/mouse/day in 100 μl PBS) and Hypoxanthine (Sigma, 750 μg/mouse/day in 100 μl PBS) to prevent suppression of nucleotide biosynthesis. Tumour growth was monitored weekly with a caliper. Experiments were terminated when any tumour in the cohort reached 2.5 cm in size. At the end of experiments, blood was collected by cardiac puncture. Organs were analyzed for micrometastases and macrometastases by bioluminescence imaging and visual inspection.
NADPH/NADP+ measurement
Subcutaneous tumours or metastatic nodules were surgically excised as quickly as possible after euthanizing the mice then melanoma cells were mechanically dissociated and NADPH and NADP+ were measured using NADPH/NADP Glo-Assay (Promega) following the manufactures instructions. Luminescence was measured using a using a FLUOstar Omega plate reader (BMG Labtech). Values were normalized to protein concentration, measured using a bicinchoninic acid (BCA) protein assay (Thermo).
Western Blot Analysis
Tissue lysates were prepared in Kontes tubes with disposable pestles using RIPA Buffer (Cell Signaling Technology) supplemented with phenylmethylsulphonyl fluoride (Sigma), and protease and phosphatase inhibitor cocktails (Roche). The BCA protein assay (Thermo) was used to quantify protein concentrations. Equal amounts of protein (15–30 μg) were separated on 4–20% Tris Glycine SDS gels (BioRad) and transferred to polyvinylidene difluoride membranes (BioRad). Membranes were blocked for 30 minutes at room temperature with 5% milk in TBS supplemented with 0.1% Tween20 (TBST) then incubated with primary antibodies overnight at 4°C. After incubating with horseradish peroxidase conjugated secondary antibodies (Cell Signaling Technology), membranes were developed using SuperSignal West Pico or Femto chemiluminescence reagents (Thermo). Blots were stripped with 1% SDS, 25mM glycine (pH 2) before re-probing. The following primary antibodies were used for western blot analyses:ALDH1L2 (LifeSpan Bio; LS-C178510), DHFR (LifeSpan Bio; LS-C138829), MTHFR (LifeSpan Bio; LS-C157974), SHMT1 (Cell Signaling; 12612S), SHMT2 (Cell Signaling; 12762S), MTHFD1 (ProteinTech; 10794-1-AP), MTHFD2 (ProtenTech; 12270-1-AP), Actin (Abcam, ab8227).
Immunofluorescence staining of frozen tissue sections
Tissues were fixed in 4% paraformaldehyde for 12 hours at 4°C, then transferred to 30% sucrose for 24 hours for cryoprotection. Tissues were then frozen in OCT. 10μm sections were permeabilized in PBS with 0.2% Triton (PBT), 3 times for 5 min each, and blocked in 5% goat serum in PBT for 30 minutes at room temperature. Sections were then stained with primary antibodies overnight: ALDH1L2 (LS-C178510, LifeSpan Bio; 1:50) and S100 (Z0311, Dako, 1:500). The next day, sections were washed in PBS with 0.2% Triton and stained with secondary goat anti-rabbit antibody (Invitrogen) at 1:500 for 30 minutes in the dark at room temperature. Sections were washed with PBT with DAPI (1:1000) and mounted for imaging.
Statistical methods
The data in most figure panels reflect multiple independent experiments performed on different days using melanomas derived from multiple patients. Variation is always indicated using standard deviation. For analysis of statistical significance, we first tested whether there was homogeneity of variation across treatments (as required for ANOVA) using Levene’s test, or when only two conditions were compared, using the F-test. In cases where the variation significantly differed among treatments, the data were log2-transformed. If the data contained zero values, 1/2 of the smallest non-zero value was added to all measurements before log2 transformation. If the data contained negative values, all measurements were log-modulus transformed (L(x) = sign(x) * log(|x| + 1)). In the rare cases when the transformed data continued to exhibit variation that significantly differed among treatments, we used a non-parametric Kruskal-Wallis test or a non-parametric Mann-Whitney test to assess the significance of differences among populations and treatments. Usually, variation did not significantly differ among treatments. Under those circumstances, two-tailed Student’s t-tests were used to test the significance of differences between two treatments. When more than two treatments were compared, a one-way ANOVA followed by Dunnett’s multiple comparisons tests were performed. A two-way ANOVA followed by Dunnett’s multiple comparisons tests were used in cases where more than two groups were compared with repeated measures. Hierarchical clustering was performed using Euclidean distance in Metaboanalyst49.
Mouse cages were randomized between treatments in all in vivo experiments (mice within the same cage had to be part of the same treatment). No blinding was used in any experiment. In all xenograft assays we injected 4–8 week old NSG mice, 5 mice per treatment. Both male and female mice were used. For long-term assays, we injected 10 mice per treatment to account for non-melanoma related deaths (NSG mice are susceptible to death from opportunistic infections). When mice died before the end of experiments due to opportunistic infections the data from those mice were excluded. There were only two experiments in which this occurred. In Figures 1c and d, 0–4 mice per melanoma line were found dead due to an opportunistic bacterial infection prior to termination of the experiment and were excluded from the reported results. In Figure 2b, 0–3 mice per melanoma line were found dead due to opportunistic infections, before the first imaging time point after transplantation. These mice were excluded from the reported results.
Extended Data
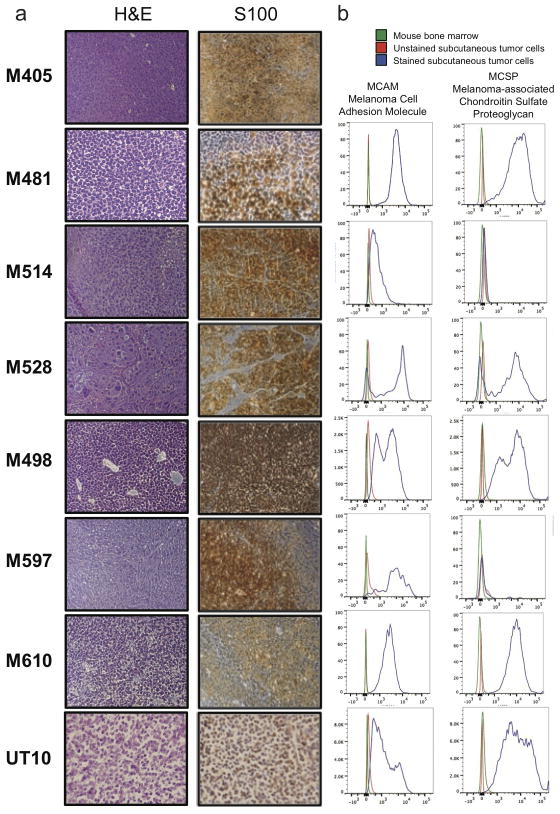
Extended data figure 1. Expression of melanoma markers by xenografted melanomas.
a) M405, M481, M514, M528, M498, M597, M610, and UT10 tumours were consistently positive for S100, a marker used clinically to diagnose melanoma. b) Flow cytometric analysis of xenografted tumour cells that were HLA-ABC+ and negative for mouse CD31/CD45/Ter119 showed that these cells were usually positive for Melanoma Cell Adhesion Molecule (MCAM) and Melanoma-associated Chondroitin Sulfate Proteoglycan (MCSP). Both of the tumors that lacked MCSP staining (M514 and M597) were heavily pigmented and expressed other melanoma markers (such as S100 and MCAM).
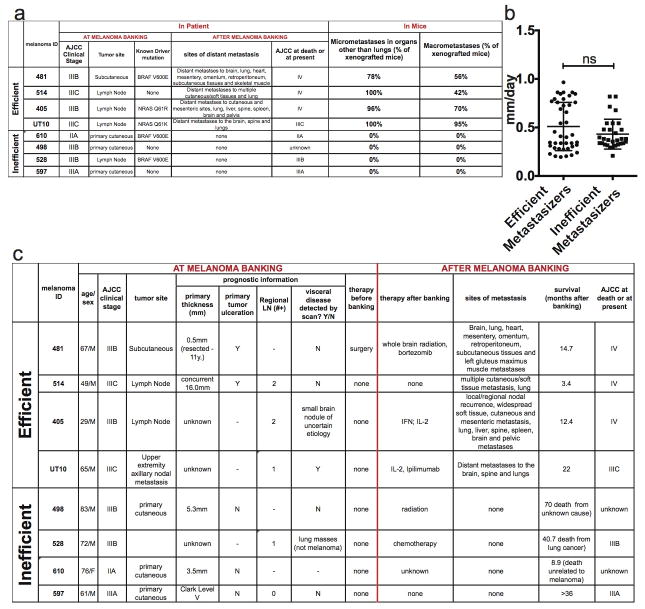
Extended data figure 2. Clinical data on the melanomas used in this study and summary of their metastatic behavior in NSG mice.
a) Summary of the clinical characteristics of the melanomas used in this study at the time of banking, as well as patient outcome after banking, and metastasis patterns upon transplantation of banked tumours into NSG mice. Melanomas were stratified into efficient and inefficient metastasizers. Efficient metastasizers formed distant metastases in patients and in NSG mice. Inefficient metastasizers did not form distant metastases in patients or distant macrometastases in NSG mice. They did form micrometastases in the lung, but not outside of the lung in the period of time it took for subcutaneous tumours to grow to 2 cm in diameter (when the mice had to be euthanized in these experiments27). Nonetheless, most of the inefficient metastasizers have the ability to form macrometastases if given enough time (data not shown). b) Growth rates of subcutaneous tumours in NSG mice after subcutaneous transplantation of 100 cells. Statistical significance was assessed using two-tailed Student’s t-test. c) Clinical characteristics of the patients from whom melanomas were obtained at the time of banking and upon subsequent clinical follow up. The tumours were confirmed to be melanomas by clinical dermatopathology. The tumours were independently confirmed to be melanomas after xenografting in mice by histological and flow cytometric analysis of melanoma markers (Extended data figure 1) as well as examination by a clinical dermatopathologist.
Extended data figure 3. Barriers to distant metastasis in vivo.
a) Live human melanoma cells were identified by flow cytometry based on the expression of DsRed (all melanomas in this study stably expressed DsRed) and human HLA and the lack of expression of mouse CD45, CD31 and Ter119 (to exclude mouse hematopoietic and endothelial cells). Human melanoma cells were observed in the blood of NSG mice bearing efficiently metastasizing melanomas. b) Mice xenografted with efficiently metastasizing melanomas (n=43 mice with tumours derived from 4 patients) had significantly higher frequencies of circulating melanoma cells (CMCs) in their blood than mice xenografted with inefficiently metastasizing melanomas (n=13 mice with tumours derived from 4 patients) or control mice that had not been xenografted (n=18 mice). Blood was collected by cardiac puncture. Statistical significance was assessed using one-way analysis of variance (ANOVA) followed by Tukey’s test for multiple comparisons (**, p< 0.005). c–f) Bioluminescence analysis of total photon flux (photons/second) from mouse organs after intravenous injection (c, d) or intrasplenic injection (e, f) of luciferase-tagged melanoma cells derived from efficiently metastasizing (c, e) or inefficiently metastasizing (d, f) melanomas. Each melanoma was derived from a different patient and was studied in an independent experiment. g) Schematic of the experiment shown in Table 2a. h) Schematic of the experiment shown in Table 2b. i) Summary of mean limiting dilution frequencies of tumour-forming cells after subcutaneous, intravenous, or intrasplenic transplantation into NSG mice.
Extended data figure 4. Unsupervised clustering suggests that melanoma cells undergo reversible metabolic changes during metastasis.
Hierarchical clustering from two independent experiments reflecting (a) subcutaneous (SQ) tumours and metastatic nodules from the liver, pancreas, lung, and kidneys of mice transplanted with melanomas M405, M481 and M514 (see Extended data table 1 for data on individual metabolites) and (b) subcutaneous (SQ) tumours and metastatic nodules from the liver, pancreas, and kidneys of mice transplanted with melanomas M405, M481 and UT10 (n=2–3 mice/melanoma in each experiment; see Extended data table 2 for data on individual metabolites). c) Hierarchical clustering of metabolites extracted from flow cytometrically sorted human melanoma cells isolated from subcutaneous tumours or metastatic nodules (UT10, M481, n=3 mice/melanoma in two independent experiments). d and e) Hierarchical clustering of metabolites extracted from subcutaneous tumours and metastatic nodules from mice transplanted subcutaneously with either subcutaneous, circulating or metastatic melanoma cells (n=4 mice for each melanoma in two independent experiments). f) GSH/GSSG ratios from each of the experiments that compared subcutaneous tumours and metastasizing cells. i) Metabolites were extracted in the presence of 0.1% formic acid to inhibit spontaneous oxidation42 in two independent experiments comparing subcutaneous and metastatic tumours from mice with three different melanomas in each experiment (M405, M481, and UT10). ii and iii) GSH/GSSG ratios from the experiments shown in panels a and b, respectively. iv) GSH/GSSG ratios in melanoma cells isolated by flow cytometry from subcutaneous tumours and the blood (CMCs) of mice bearing M405 and M481. v) Metabolites were extracted in the presence of 0.1% formic acid in two independent experiments in which melanoma cells were isolated by flow cytometry from subcutaneous and metastatic tumours (M405 and M481). While the GSH/GSSG ratio was always significantly higher in melanoma cells from subcutaneous tumours as compared to circulating cells or metastatic nodules the ratio varied among experiments as a result of technical differences in cell isolation and metabolite extraction as well as differences in MS sensitivity to GSH and GSSG. g) GSH/GSSG ratios in subcutaneous tumours that arose from the transplantation of melanoma cells obtained from subcutaneous tumours or metastatic nodules, as well as the metastatic nodules from the same mice (M405; n=2 to 3 mice per treatment in one experiment). These data suggest that the decline in GSH/GSSG ratio in metastasizing melanoma cells is reversible upon subcutaneous transplantation. h) Histogram showing mitochondrial mass in subcutaneous tumour cells that arose from the transplantation of subcutaneous cells (SQ from SQ), subcutaneous tumour cells that arose from the transplantation of metastatic cells (SQ from Mets), and metastatic cells (Metastases). These histograms reflect the data shown in Figure 1g. All error bars represent standard deviation. Statistical significance was assessed using, two-tailed Student’s t-tests (f and g) (*, p<0.05)
Extended data figure 5. Metastatic nodules exhibited increased enrichment of labeled serine and glycine as compared to subcutaneous tumours.
In vivo isotope tracing of uniformly 13C-labelled (a) lactate (M+3), (b) 3-phosphoglycerate (M+3), (c) serine (M+3), and (d) glycine (M+2) in subcutaneous tumours versus metastatic nodules from the same mice (UT10, n=3–4 mice per time point in 2 independent experiments). The fractional enrichment of labeled lactate, and 3-PG did not significantly differ among plasma, subcutaneous tumours, or metastatic tumours at any time point. In contrast, the fractional enrichment of labeled serine and glycine were significantly higher in metastatic as compared to subcutaneous tumors. This is consistent with increased de novo serine synthesis in metastatic tumors but could also reflect altered serine/glycine exchange with circulating serine/glycine pools in metastatic as compared to subcutaneous tumors. e) NADPH/NADP ratios in subcutaneous tumours and metastatic nodules from the same mice shown in Figure 2d and 2e. f and g) Western blot of ALDH1L2 (f) and MTHFD1 (g) protein after shRNA knockdown in melanoma cells. Uncropped western blots are shown in Supplementary Figure 1. h and i) Amount of GSH (h) and GSSG (i) per mg of subcutaneous or metastatic tumour as measured by LC-MS (M405, M481 and UT10, n=2–3 mice/melanoma in 2 independent experiments). All data represent mean±sd. Statistical significance was assessed using two-tailed Student’s t-tests (e, h, and i) and one-way analyses of variance (ANOVAs) followed by Dunnett’s tests for multiple comparisons (a – d; *, p<0.05; ***, p< 0.0005). (j) Schematic of the folate pathway including NADPH generating (green box) and NADPH consuming (red box) reactions.
Extended data figure 6. Immunofluorescence analysis of ALDH1L2 in subcutaneous melanoma as well as metastatic nodules in the liver, pancreas and lung.
Melanoma cells can be distinguished from host stromal cells based on staining for the melanoma marker, S100.
Extended data table 1.
Fold change and statistical significance corrected for multiple comparisons of metabolites detected by LC-MS/MS in the experiment in Extended data figure 2a.
| Metabolite | Fold Change of Metastasis/SQ Tumor | FDR |
|---|---|---|
| citraconic acid/ketohexanoic acid | 12.311 | 0.054768 |
| glucarate | 11.472 | 0.13347 |
| sedoheptulose 7-phosphate | 5.6121 | 0.084567 |
| GABA | 4.8296 | 0.44062 |
| betaine | 4.8132 | 6.12E-05 |
| AICAR | 4.5004 | 0.15975 |
| carnitine-C18 | 4.4942 | 0.13347 |
| spermidine | 4.4416 | 0.016469 |
| adenosine 5-diphosphoribose | 4.1336 | 0.041494 |
| xanthosine | 3.9645 | 0.11311 |
| glutamine | 3.9371 | 0.000419 |
| hexose-phosphate | 3.0656 | 0.086761 |
| GSSG | 2.8724 | 0.081173 |
| S-methyl-5-thioadenosine | 2.8234 | 0.21534 |
| gluconate | 2.7963 | 0.052588 |
| xanthine | 2.4873 | 0.084567 |
| pyridoxamine | 2.4531 | 0.24821 |
| spermine | 2.3796 | 0.19553 |
| aminoadipic acid | 2.3513 | 0.44062 |
| carnitine-C16 | 2.3105 | 0.27824 |
| uridine | 2.2733 | 0.13025 |
| citrate | 2.1797 | 0.081173 |
| methionine sulfoxide | 2.0674 | 0.22847 |
| ribose 5-phosphate | 2.0193 | 0.40173 |
| glycerol 3-phosphate | 2.0132 | 0.40173 |
| IMP | 1.987 | 0.018053 |
| S-adenosylmethionine | 1.8423 | 0.17083 |
| inosine | 1.8253 | 0.24736 |
| riboflavin | 1.7888 | 0.19553 |
| hypoxanthine | 1.6282 | 0.23352 |
| acetyllysine | 1.5539 | 0.5435 |
| NAD | 1.5512 | 0.47123 |
| taurine | 1.5405 | 0.21332 |
| thiamine | 1.4869 | 0.47274 |
| aspartate | 1.3202 | 0.68641 |
| putrescine | 1.3107 | 0.64286 |
| kynurenine | 1.2857 | 0.56968 |
| dAMP | 1.2535 | 0.61138 |
| thymidine | 1.2419 | 0.6972 |
| ornithine | 1.2218 | 0.70841 |
| phenylalanine | 1.2154 | 0.49828 |
| AMP | 1.2057 | 0.69525 |
| S-adenosylhomocysteine | 1.1992 | 0.77036 |
| acetylcarnitine | 1.1403 | 0.68641 |
| trimethyllysine | 1.1395 | 0.83403 |
| carnitine-C8 | 1.1074 | 0.78839 |
| carnitine-C3 | 1.0818 | 0.83403 |
| asparagine | 1.0678 | 0.72161 |
| choline | 1.0661 | 0.83403 |
| malate | 1.0497 | 0.78839 |
| glycerophosphorylcholine | 1.0439 | 0.9381 |
| 1-methylhistidine | 1.0248 | 0.9381 |
| nicotinamide | 1.0018 | 0.99511 |
| glyoxylate | 0.98478 | 0.9381 |
| dimethylglycine | 0.97783 | 0.91723 |
| glycine | 0.94707 | 0.91453 |
| succinate | 0.94473 | 0.83403 |
| alanine | 0.92949 | 0.78839 |
| carnitine-C6 | 0.92856 | 0.83403 |
| indole | 0.92843 | 0.83403 |
| tryptophan | 0.92275 | 0.80122 |
| urea | 0.90704 | 0.66141 |
| Guanidoacetic acid | 0.88659 | 0.70463 |
| histidine | 0.86073 | 0.67732 |
| valine | 0.8468 | 0.66141 |
| tyrosine | 0.82787 | 0.47274 |
| leucine | 0.82491 | 0.29642 |
| citrulline | 0.82483 | 0.44062 |
| S-lactoylglutathione | 0.81622 | 0.78839 |
| guanosine | 0.81517 | 0.55117 |
| pyroglutamic acid | 0.81294 | 0.61138 |
| GMP | 0.81159 | 0.64286 |
| 3-hydroxybutyrate | 0.80538 | 0.72186 |
| hydroxyphenylpyruvate | 0.798 | 0.26547 |
| glycerate | 0.77694 | 0.66141 |
| phenyllactic acid | 0.77196 | 0.02971 |
| homoserine/threonine | 0.7596 | 0.44062 |
| methionine | 0.74125 | 0.28847 |
| carnitine | 0.73503 | 0.38386 |
| cADP-ribose | 0.73299 | 0.12096 |
| O-acetylserine | 0.72801 | 0.19553 |
| homocysteine | 0.72727 | 0.16015 |
| methylglyoxal | 0.72483 | 0.08875 |
| creatine | 0.72291 | 0.24821 |
| fumarate | 0.7092 | 0.17141 |
| lactate | 0.70716 | 0.13819 |
| proline | 0.69532 | 0.27038 |
| alpha-ketoisovaleric acid | 0.6943 | 0.14034 |
| cystathionine | 0.6937 | 0.56124 |
| UMP | 0.67863 | 0.13819 |
| carbamoylphosphate | 0.67623 | 0.33231 |
| N-acetylglucosamine phosphate | 0.65886 | 0.05477 |
| carnitine-C4 | 0.63354 | 0.52908 |
| CMP | 0.60248 | 0.11112 |
| orotate | 0.60141 | 0.15634 |
| pipecolic acid | 0.6 | 0.05259 |
| fructose | 0.59758 | 0.09166 |
| adenine | 0.59213 | 0.03863 |
| hydroxyproline | 0.59052 | 0.00051 |
| GSH | 0.58564 | 0.12364 |
| pantothenic acid | 0.57614 | 0.11592 |
| cytidine | 0.54462 | 0.05231 |
| phospho-hydroxybenzoate | 0.53605 | 0.31067 |
| phosphocholine | 0.53561 | 0.06755 |
| propionylcholine | 0.53061 | 0.13303 |
| 1-methyladenosine | 0.52411 | 0.00529 |
| glutamate | 0.48917 | 0.01647 |
| N-acetylaspartate | 0.48911 | 0.15634 |
| hydroxyglutarate | 0.48629 | 0.01468 |
| acetylcholine | 0.48281 | 0.03065 |
| shikimic acid | 0.46778 | 0.05604 |
| phosphocreatine | 0.46674 | 0.13819 |
| carnitine-C5 | 0.45886 | 0.19875 |
| alpha-ketoglutarate | 0.4554 | 0.08972 |
| arginine | 0.44217 | 0.02915 |
| 2-deoxycytidine | 0.43995 | 0.23352 |
| uracil | 0.43712 | 0.01468 |
| 5-aminolevulinic acid | 0.436 | 0.00012 |
| nicotinate | 0.4316 | 0.19553 |
| lysine | 0.40191 | 6.12E-05 |
| 1-methylnicotinamide | 0.39599 | 0.33556 |
| N,N-dimethylarginine | 0.38777 | 0.00093 |
| carnitine-C14 | 0.3696 | 0.24821 |
| myo-inositol | 0.34547 | 0.13347 |
| creatinine | 0.29845 | 0.00047 |
| ascorbic acid | 0.25524 | 1.57E-06 |
| XMP | 0.15598 | 0.06755 |
| carnitine-C12 | 0.15514 | 0.09166 |
| ADP | 0.071143 | 2.54E-07 |
Extended data table 2.
Fold change and statistical significance corrected for multiple comparisons of metabolites detected by LC-MS/MS in the experiment shown in Extended data figure 2b.
| Metabolite | Fold Change Metastasis/SQ Tumor | FDR |
|---|---|---|
| citraconic acid/ketohexanoic acid | 34.483 | 0.45284 |
| carnitine-C18 | 10.963 | 0.02495 |
| NADP | 10.741 | 0.02463 |
| adenosine 5-diphosphoribose | 6.9242 | 0.01199 |
| carnitine-C16 | 4.962 | 0.03569 |
| hypoxanthine | 4.8303 | 0.01049 |
| pyridoxamine | 4.7958 | 0.02923 |
| betaine | 4.7812 | 0.03627 |
| S-adenosylhomocysteine | 4.3483 | 0.01917 |
| gluconate | 4.2564 | 0.03675 |
| xanthine | 4.1876 | 0.00192 |
| GSSG | 4.0565 | 0.00339 |
| AICAR | 3.8575 | 0.02826 |
| xanthosine | 3.7994 | 0.07897 |
| adenosine | 3.555 | 0.00983 |
| uridine | 3.497 | 9.33E-05 |
| glutamine | 3.318 | 0.01391 |
| aconitate | 3.2415 | 0.04627 |
| sedoheptulose 7-phosphate | 2.9576 | 0.19981 |
| NAD | 2.9139 | 0.00151 |
| riboflavin | 2.8081 | 0.0222 |
| adenosine | 2.7184 | 0.00181 |
| guanosine | 2.4328 | 0.01321 |
| ZMP | 2.4059 | 0.12097 |
| inosine | 2.3785 | 0.14043 |
| IMP | 2.3314 | 0.01049 |
| deoxyinosine | 2.2956 | 0.20059 |
| uridine | 2.2759 | 0.01549 |
| SAICA | 2.0448 | 0.13492 |
| malate | 1.9365 | 1.81E-06 |
| aminoadipic acid | 1.9042 | 0.01321 |
| carnitine-C8 | 1.8744 | 0.0222 |
| kynurenine | 1.869 | 0.00672 |
| dAMP | 1.802 | 0.00099 |
| fructose 6-phosphate | 1.7965 | 0.26537 |
| thymidine | 1.6716 | 0.27532 |
| cADP-ribose | 1.5868 | 0.0663 |
| cytidine | 1.5195 | 0.21432 |
| carnitine-C6 | 1.512 | 0.08539 |
| S-lactoylglutathione | 1.46 | 0.6708 |
| glycerol 3-phosphate | 1.4129 | 0.42166 |
| 7-methylguanosine | 1.4076 | 0.55385 |
| glyoxylate | 1.3983 | 0.00203 |
| S-adenosylmethionine | 1.3577 | 0.15136 |
| glycerophosphorylcholine | 1.335 | 0.16611 |
| tryptophan | 1.318 | 0.19633 |
| 1-methyladenosine | 1.2813 | 0.03412 |
| AMP | 1.2804 | 0.43896 |
| citrate | 1.2214 | 0.67146 |
| nicotinamide | 1.2063 | 0.62079 |
| succinate | 1.196 | 0.50829 |
| hydroxyphenylpyruvate | 1.1937 | 0.26901 |
| phenylalanine | 1.1838 | 0.1462 |
| carnitine-C3 | 1.1524 | 0.54363 |
| tyrosine | 1.1166 | 0.50877 |
| methionine | 1.0869 | 0.70454 |
| 2-aminooctanoic acid | 1.0803 | 0.71185 |
| adenine | 1.0676 | 0.74371 |
| asparagine | 1.0509 | 0.69494 |
| pipecolic acid | 1.0216 | 0.96295 |
| thiamine | 1.0179 | 0.97565 |
| ribose 5-phosphate | 1.0132 | 0.97843 |
| homocysteic acid | 1.0076 | 0.97843 |
| fumarate | 1.0022 | 0.9897 |
| indole | 1.0017 | 0.9897 |
| pantothenic acid | 0.97947 | 0.9674 |
| homocysteine | 0.97252 | 0.96295 |
| Guanidoacetic acid | 0.97007 | 0.79908 |
| valine | 0.94431 | 0.69728 |
| orotate | 0.93458 | 0.90099 |
| methionine sulfoxide | 0.90754 | 0.71378 |
| citrulline | 0.90545 | 0.69494 |
| leucine | 0.90323 | 0.39527 |
| GSH | 0.88817 | 0.69787 |
| S-methyl-5-thioadenosine | 0.87643 | 0.60788 |
| putrescine | 0.87293 | 0.3561 |
| cysteine | 0.86437 | 0.7895 |
| 3-hydroxybutyrate | 0.86329 | 0.5511 |
| N-acetylaspartate | 0.78325 | 0.48791 |
| serine | 0.78075 | 0.79908 |
| alanine | 0.76685 | 0.11668 |
| carnitine-C14 | 0.76026 | 0.70454 |
| GMP | 0.74002 | 0.18657 |
| histidine | 0.72959 | 0.0396 |
| O-acetylserine | 0.72727 | 0.01269 |
| glutamate | 0.72558 | 0.14273 |
| acetylcarnitine | 0.68306 | 0.00192 |
| 2-deoxycytidine | 0.67868 | 0.29585 |
| ascorbic acid | 0.67473 | 0.28456 |
| carnitine-C12 | 0.67254 | 0.5056 |
| lactate | 0.65954 | 0.00047 |
| choline | 0.65842 | 0.00389 |
| acetylcholine | 0.6573 | 0.13515 |
| creatinine | 0.6543 | 0.16525 |
| y-aminobutyric acid | 0.64139 | 0.2191 |
| glucose/fructose | 0.63849 | 0.00925 |
| homoserine/threonine | 0.62876 | 0.02462 |
| dimethylglycine | 0.62167 | 0.00068 |
| carnitine-C4 | 0.59432 | 0.16312 |
| N-acetylglucosamine phosphate | 0.59273 | 0.00095 |
| glucuronate | 0.57813 | 0.00338 |
| UMP | 0.57702 | 0.0163 |
| uridine 5-diphosphoglucuronic acid | 0.57518 | 0.02632 |
| phenyllactic acid | 0.57402 | 0.00024 |
| N,N-dimethylarginine | 0.5534 | 0.00047 |
| propionylcholine | 0.55055 | 0.00853 |
| allantoin | 0.54186 | 2.44E-05 |
| urea | 0.52852 | 7.29E-07 |
| 1 -methylnicotinamide | 0.51941 | 0.02008 |
| 5-aminolevulinic acid | 0.51688 | 8.12E-05 |
| deoxyribose phosphate | 0.50607 | 0.01321 |
| uracil | 0.50113 | 0.06104 |
| carnitine | 0.49567 | 1.66E-07 |
| taurine | 0.46303 | 2.17E-05 |
| cytosine | 0.45951 | 6.21E-07 |
| proline | 0.4591 | 4.23E-06 |
| carnitine-C5 | 0.44249 | 0.00483 |
| ornithine | 0.43836 | 0.00505 |
| 1-methylhistidine | 0.4023 | 2.31E-06 |
| uracil | 0.3904 | 0.01116 |
| creatine | 0.37961 | 1.26E-07 |
| trimethyllysine | 0.37893 | 7.58E-05 |
| arginine | 0.36929 | 0.00047 |
| CMP | 0.36814 | 3.24E-09 |
| phosphocholine | 0.36461 | 7.43E-09 |
| alpha-ketoglutarate | 0.35719 | 0.02197 |
| spermidine | 0.35496 | 0.03329 |
| acetyllysine | 0.35375 | 3.01E-05 |
| phosphocreatine | 0.34497 | 0.00048 |
| agmatine | 0.32422 | 8.26E-06 |
| hydroxyglutarate | 0.30783 | 3.24E-09 |
| lysine | 0.27436 | 3.91E-10 |
| phosphoenolpyruvic acid | 0.27364 | 1.99E-08 |
| XMP | 0.2693 | 4.33E-05 |
| cystathionine | 0.26823 | 0.02462 |
| glycine | 0.24469 | 3.32E-09 |
| carbamoylphosphate | 0.17283 | 8.74E-09 |
| myo-inositol | 0.14292 | 5.08E-07 |
| cadaverine | 0.06894 | 0.09355 |
| nicotinate | 0.066878 | 0.12347 |
Extended data table 3.
Treatment with the anti-oxidant N-acetyl-cysteine increased tumour formation by intravenously transplanted melanoma cells.Limiting dilution analysis of tumour formation by intravenously transplanted efficiently metastasizing (a, M405, M481, UT10) or inefficiently metastasizing (b, M528 and M597) melanoma cells. In half the mice, the cells were pre-treated with NAC prior to transplantation and the mice were maintained on water supplemented with NAC after transplantation (n=5 mice/treatment/melanoma for a total of 110 mice in 5 independent experiments). Statistical significance was assessed by a chi-square test using ELDA software46
| a | ||||||
|---|---|---|---|---|---|---|
| Efficient Metastasizers | ||||||
| Cell Dose | Mice Injected | Mice that formed tumours | Frequency of tumour-forming cells | |||
| Total | With NAC | 1000 | 15 | 13 | 1/443 | * |
| 100 | 15 | 4 | * | |||
| Without NAC | 1000 | 15 | 3 | 1/4983 | * | |
| 100 | 15 | 0 | * | |||
| b | |||||
|---|---|---|---|---|---|
| Inefficient Metastasizers | |||||
| Cell Dose | Mice Injected | Mice that formed tumours | Frequency of tumour-forming cells | ||
| Total | With NAC | 10000 | 5 | 2 | 1/16157 |
| 1000 | 10 | 1 | |||
| 100 | 10 | 0 | |||
| Without NAC | 10000 | 5 | 0 | - | |
| 1000 | 10 | 0 | |||
| 100 | 10 | 0 | |||
p<0.00005.
Extended data table 4.
Fold increase in metastatic nodules as compared to subcutaneous tumours of transcripts involved in antioxidant responses and NADPH generation.Data are based on RNAseq analysis of two subcutaneous tumours and two metastatic nodules for each of M405 and M481.
| Gene Name | Gene | Metastatic/Subcutaneous (Fold Change) | p-value* |
|---|---|---|---|
| GCLC | Glutamate-cysteine ligase (catalytic subunit) | 1.20 | 0.560 |
| GCLM | Glutamate-cysteine ligase (modifier subunit) | 0.86 | 0.300 |
| GSS | Glutathione synthase | 0.90 | 0.240 |
| SOD1 | Superoxide disumutase 1 | 1.09 | 0.355 |
| SOD2 | Superoxide dismutase 2 | 0.86 | 0.454 |
| SOD3 | Superoxide dismutase 3 | 0.64 | 0.189 |
| CAT | Catalase | 1.09 | 0.548 |
| GPX1 | Glutathione Peroxidase 1 | 1.30 | 0.117 |
| GPX2 | Glutathione Peroxidase 2 | 1.00 | 0.998 |
| GPX3 | Glutathione Peroxidase 3 | 0.40 | 0.048 |
| GPX4 | Glutathione Peroxidase 4 | 1.07 | 0.459 |
| GPX7 | Glutathione Peroxidase 7 | 0.83 | 0.183 |
| GPX8 | Glutathione Peroxidase 8 | 0.87 | 0.631 |
| GSR | Glutathione Reductase | 1.12 | 0.372 |
| HMOX1 | Heme Oxygenase 1 | 0.86 | 0.186 |
| HMOX2 | Heme Oxygenase 2 | 1.41 | 0.008 |
| TXN | Thioredoxin 1 | 1.16 | 0.297 |
| TXN2 | Thioredoxin 2 | 1.15 | 0.480 |
| TXNRD1 | Thioredoxin redutase 1 | 0.97 | 0.859 |
| TXNRD2 | Thioredoxin redutase 2 | 0.91 | 0.701 |
| TXNRD3 | Thioredoxin redutase 3 | 1.15 | 0.176 |
| PRDX1 | Peroxiredoxin 1 | 1.67 | 0.334 |
| PRDX2 | Peroxiredoxin 2 | 0.94 | 0.289 |
| PRDX3 | Peroxiredoxin 3 | 1.31 | 0.013 |
| PRDX4 | Peroxiredoxin 4 | 1.13 | 0.075 |
| PRDX5 | Peroxiredoxin 5 | 1.28 | 0.032 |
| PRDX6 | Peroxiredoxin 6 | 1.05 | 0.172 |
| NFE2L2 | Nuclear factor, erythroid 2-like 2 | 0.90 | 0.106 |
| G6PD | Glucose-6-phosphate Dehydrogenase | 2.20 | 0.156 |
| PGD | Phosphogluconate dehydrogenase | 0.98 | 0.758 |
| ME2 | Malic Enzyme 2 | 1.05 | 0.865 |
| CBS | Cystathione beta Synthase | 1.20 | 0.469 |
based on Bonferroni correction for multiple comparisons a p-value of 0.0016 would be considered statistically significant in this analysis
Supplementary Material
Acknowledgments
S.J.M. is a Howard Hughes Medical Institute (HHMI) Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, the director of the Hamon Laboratory for Stem Cells and Cancer, and a Cancer Prevention and Research Institute of Texas Scholar. We thank K. Correll and M. Gross for mouse colony management; N. Loof and the Moody Foundation Flow Cytometry Facility. We thank Nisha Meireles and the University of Michigan Melanoma Biobank, for Biobank database and melanoma clinical data management.
Footnotes
AUTHOR CONTRIBUTION STATEMENT
E.P. and S.J.M. conceived the project, designed, and interpreted experiments. M.A. and R.J.D. participated in the design and interpretation of experiments related to metabolic mechanisms. E.P. performed all the experiments with technical assistance from S.E.H., metabolomics assistance from M.A and imaging assistance from M.M.M. Z.H. and M.A developed metabolomics methods. Z.Z. assisted with statistical analyses. A.M.L. and T.M.J. provided melanoma specimens and associated clinical data. E.P., M.A., and S.J.M wrote the manuscript.
AUTHOR INFORMATION
The authors declare no competing financial interests.
References
- 1.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stott SL, Lee RJ, Nagrath S, Yu M, Miyamoto DT, Ulkus L, Inserra EJ, Ulman M, Springer S, Nakamura Z, Moore AL, Tsukrov DI, Kempner ME, Dahl DM, Wu CL, Iafrate AJ, Smith MR, Tompkins RG, Sequist LV, Toner M, Haber DA, Maheswaran S. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Science Translational Medicine. 2010;2:25ra23. doi: 10.1126/scitranslmed.3000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, Desai R, Zhu H, Comaills V, Zheng Z, Wittner BS, Stojanov P, Brachtel E, Sgroi D, Kapur R, Shioda T, Ting DT, Ramaswamy S, Getz G, Iafrate AJ, Benes C, Toner M, Maheswaran S, Haber DA. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24:410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, Groom AC. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. American Journal of Pathology. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron MD, Schmidt EE, Kerkvliet N, Nadkarni KV, Morris VL, Groom AC, Chambers AF, MacDonald IC. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Research. 2000;60:2541–2546. [PubMed] [Google Scholar]
- 8.Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nature Medicine. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 9.Engell HC. Cancer cells in the blood; a five to nine year follow up study. Annals of Surgery. 1959;149:457–461. doi: 10.1097/00000658-195904000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths JD, McKinna JA, Rowbotham HD, Tsolakidis P, Salsbury AJ. Carcinoma of the colon and rectum: circulating malignant cells and five-year survival. Cancer. 1973;31:226–236. doi: 10.1002/1097-0142(197301)31:1<226::aid-cncr2820310130>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 11.Salsbury AJ. The significance of the circulating cancer cell. Cancer Treatment Reviews. 1975;2:55–72. doi: 10.1016/s0305-7372(75)80015-6. [DOI] [PubMed] [Google Scholar]
- 12.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nature Reviews Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 13.Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 14.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris IS, Treloar AE, Inoue S, Sasaki M, Gorrini C, Lee KC, Yung KY, Brenner D, Knobbe-Thomsen CB, Cox MA, Elia A, Berger T, Cescon DW, Adeoye A, Brustle A, Molyneux SD, Mason JM, Li WY, Yamamoto K, Wakeham A, Berman HK, Khokha R, Done SJ, Kavanagh TJ, Lam CW, Mak TW. Glutathione and Thioredoxin Antioxidant Pathways Synergize to Drive Cancer Initiation and Progression. Cancer Cell. 2015 doi: 10.1016/j.ccell.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Sayin VI, Ibrahim MX, Larsson E, Nilsson JA, Lindahl P, Bergo MO. Antioxidants accelerate lung cancer progression in mice. Science Translational Medicine. 2014;6:221ra215. doi: 10.1126/scitranslmed.3007653. [DOI] [PubMed] [Google Scholar]
- 17.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs SY, Diehl JA, Koumenis C. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. Journal of Clinical Investigation. 2015;125:2592–2608. doi: 10.1172/JCI78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T, Lorkiewicz P, St Clair D, Hung MC, Evers BM, Zhou BP. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamarajugadda S, Cai Q, Chen H, Nayak S, Zhu J, He M, Jin Y, Zhang Y, Ai L, Martin SS, Tan M, Lu J. Manganese superoxide dismutase promotes anoikis resistance and tumor metastasis. Cell Death & Disease. 2013;4:e504. doi: 10.1038/cddis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qu Y, Wang J, Ray PS, Guo H, Huang J, Shin-Sim M, Bukoye BA, Liu B, Lee AV, Lin X, Huang P, Martens JW, Giuliano AE, Zhang N, Cheng NH, Cui X. Thioredoxin-like 2 regulates human cancer cell growth and metastasis via redox homeostasis and NF-kappaB signaling. Journal of Clinical Investigation. 2011;121:212–225. doi: 10.1172/JCI43144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, 3rd, Felding-Habermann B. Adaptation of energy metabolism in breast cancer brain metastases. Cancer research. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Bennet B, Mu E, Rabinowitz J, Kang Y. Metabolomic changes accompanying transformation and acquisition of metastatic potential in a syngeneic mouse mammary tumor model. Journal of Biological Chemistry. 2010;285:9317–9321. doi: 10.1074/jbc.C110.104448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, Feron O, Michiels C, Gallez B, Sonveaux P. A mitochondrial switch promotes tumor metastasis. Cell Reports. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 25.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, Kalluri R. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nature Cell Biology. 2014;16:992–1003. 1001–1015. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 27.Quintana E, Piskounova E, Shackleton M, Weinberg D, Eskiocak U, Fullen DR, Johnson TM, Morrison SJ. Human melanoma metastasis in NSG mice correlates with clinical outcome in patients. Science Translational Medicine. 2012;4:159ra149. doi: 10.1126/scitranslmed.3004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nature Reviews Drug Discovery. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 29.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular Cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, Qing G, Liu Z, Simon MC, Rabinowitz JD, Thompson CB. Serine Catabolism Regulates Mitochondrial Redox Control during Hypoxia. Cancer Discovery. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature Genetics. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chandel NS, Tuveson DA. The promise and perils of antioxidants for cancer patients. The New England Journal of Medicine. 2014;371:177–178. doi: 10.1056/NEJMcibr1405701. [DOI] [PubMed] [Google Scholar]
- 36.Gao P, Zhang H, Dinavahi R, Li F, Xiang Y, Raman V, Bhujwalla ZM, Felsher DW, Cheng L, Pevsner J, Lee LA, Semenza GL, Dang CV. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teoh-Fitzgerald ML, Fitzgerald MP, Zhong W, Askeland RW, Domann FE. Epigenetic reprogramming governs EcSOD expression during human mammary epithelial cell differentiation, tumorigenesis and metastasis. Oncogene. 2014;33:358–368. doi: 10.1038/onc.2012.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glasauer A, Chandel NS. Targeting antioxidants for cancer therapy. Biochemical Pharmacology. 2014;92:90–101. doi: 10.1016/j.bcp.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 39.Glasauer A, Sena LA, Diebold LP, Mazar AP, Chandel NS. Targeting SOD1 reduces experimental non-small-cell lung cancer. Journal of Clinical Investigation. 2014;124:117–128. doi: 10.1172/JCI71714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fortmann SP, Burda BU, Senger CA, Lin JS, Beil TL, O’Connor E, Whitlock EP. Vitamin, Mineral, and Multivitamin Supplements for the Primary Prevention of Cardiovascular Disease and Cancer: A Systematic Evidence Review for the U.S. Preventive Services Task Force U.S. Preventive Services Task Force Evidence Syntheses, formerly Systematic Evidence Reviews. 2013. [PubMed] [Google Scholar]
- 41.The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. The Alpha-Tocopherol, Beta Carotene Cancer Prevention Study Group. The New England Journal of Medicine. 1994;330:1029–1035. doi: 10.1056/NEJM199404143301501. [DOI] [PubMed] [Google Scholar]
- 42.Klein EA, Thompson IM, Jr, Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, Minasian LM, Ford LG, Parnes HL, Gaziano JM, Karp DD, Lieber MM, Walther PJ, Klotz L, Parsons JK, Chin JL, Darke AK, Lippman SM, Goodman GE, Meyskens FL, Jr, Baker LH. Vitamin E and the risk of prostate cancer: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2011;306:1549–1556. doi: 10.1001/jama.2011.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodman GE, Thornquist MD, Balmes J, Cullen MR, Meyskens FL, Jr, Omenn GS, Valanis B, Williams JH., Jr The Beta-Carotene and Retinol Efficacy Trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. Journal of the National Cancer Institute. 2004;96:1743–1750. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- 44.Deghan Manshadi S, Ishiguro L, Sohn KJ, Medline A, Renlund R, Croxford R, Kim YI. Folic acid supplementation promotes mammary tumor progression in a rat model. PloS one. 2014;9:e84635. doi: 10.1371/journal.pone.0084635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebbing M, Bonaa KH, Nygard O, Arnesen E, Ueland PM, Nordrehaug JE, Rasmussen K, Njolstad I, Refsum H, Nilsen DW, Tverdal A, Meyer K, Vollset SE. Cancer incidence and mortality after treatment with folic acid and vitamin B12. JAMA. 2009;302:2119–2126. doi: 10.1001/jama.2009.1622. [DOI] [PubMed] [Google Scholar]
- 46.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of Immunological Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 47.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, Chandel NS, DeBerardinis RJ. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell reports. 2014;7:1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic acids research. 2012;40:W127–133. doi: 10.1093/nar/gks374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.