Abstract
We report a microfluidic blood-brain barrier model that enables both physiological shear stress and optical transparency throughout the device. Brain endothelial cells grown in an optically transparent membrane-integrated microfluidic device were able to withstand physiological fluid shear stress using a hydrophilized polytetrafluoroethylene nanoporous membrane instead of the more commonly used polyester membrane. A functional three-dimensional microfluidic co-culture model of the neurovascular unit is presented that incorporates astrocytes in a 3D hydrogel and enables physiological shear stress on the membrane-supported endothelial cell layer.
INTRODUCTION
The blood-brain barrier (BBB) is composed of endothelial cells separating the systemic blood circulation from the brain and characterized by a high electrical resistivity, low permeability, and regulated transport of biomolecules. The BBB plays a critical role in brain drug delivery and pathogenesis of neurological conditions such as Alzheimer's disease and stroke. Despite increasing knowledge regarding regulation of the BBB,1,2 delivery of therapeutics into the brain remains a persistent challenge in drug development for the central nervous system (CNS). BBB function depends not only on the phenotype of the endothelial cells but also on their dynamic interactions with other cells and the extracellular matrix within the “neurovascular unit.”2,3 Several groups, including ours, have reported efforts to develop organotypic microphysiological systems representative of the neurovascular unit.4 We previously demonstrated that culturing astrocytes (the chief supporting cell of the BBB) in a three-dimensional (3D) collagen matrix differentially affects their ability to modulate brain endothelial barrier function.5 Moreover, it is well documented that fluid shear stress stimulates formation of higher-resistance endothelial cell layers generally6 and supports development of barrier properties in brain-derived endothelial cells in particular.7
Several membrane-integrated microfluidic devices have recently been reported for modeling the BBB.7–12 To date, the membrane materials used in microfluidic devices have represented a tradeoff between robust cell adhesion enabling shear stress studies (polycarbonate, PC) and optical transparency (polyester, PE). PC membranes are optically translucent, resulting in an inability to visualize cells by phase contrast microscopy, requiring fluorescence staining, and thus precluding observation over time. Transendothelial electrical resistance (TEER) can be monitored over the course of a study with electrodes integrated into the microfluidic device, but at the cost of added complexity in fabrication and handling. PE membranes are optically transparent, but data supporting their use for culture of brain endothelial cells and other endothelial cells (human umbilical vein endothelial cells (HUVEC), dermal) under physiologically relevant shear stress are lacking4 or only indicate tolerance of shear stress for a limited period of time.13,14
Previously, we reported a novel bonding procedure to integrate, in polydimethylsiloxane (PDMS) microfluidic devices, a commercial nanoporous Teflon membrane (polytetrafluoroethylene, PTFE). This PTFE membrane, used in well inserts (Millicells®, EMD Millipore), has a proprietary coating to make it hydrophilic. PTFE membrane-integrated microfluidic devices demonstrated the ability to support primary cell culture.15
Here, we report brain endothelial cell growth on PTFE for the first time, to our knowledge, and demonstrate this optically transparent membrane supports bEnd.3 (murine brain endothelial) cells grown under physiological fluid shear stress with functional protein expression. Using this membrane, we demonstrate viability and differentiation of a microfluidic version of our previously described static 3D model of the neurovascular unit, including astrocytes in a hydrogel,5 and demonstrate the ability of this 3D microfluidic co-culture to accommodate physiological shear stress in the endothelial channel in a format convenient for cellular assays.
METHODS
Two-compartment microfluidic devices were obtained by sandwiching a nanoporous membrane between two PDMS micromolded channels. Membrane-integrated devices had a cell culture area of 10 mm (l) × 1 mm (w) with an apical channel height of 150 μm and basolateral compartment height of 150–300 μm (Fig. 1(a)). Commercial PE and PTFE nanoporous membranes (pore size 0.4 μm, TClear 3450, Corning, and BGCM00010, Millipore) were used. Fabrication details of membrane-integrated devices are described elsewhere.15,16 In order to achieve a physiologically relevant fluid shear stress of 5 dyn/cm2 (Refs. 11 and 17) on a 150 μm tall flow channel, a fluid flow of 120 μl/min was used. The average shear stress τ was calculated using the equation , where η is the viscosity of water, Q is the flow rate, h is the height, and w is the width of the channel. The apical compartment was connected to a programmable syringe pump (Harvard Apparatus, PHD Ultra) operating in aspiration and a 10 ml medium reservoir for low-flow studies. For high fluid shear studies (5 dyn/cm2), a medium recirculation system was adopted with the device connected in aspiration to a peristaltic pump (Masterflex L/S) connected to a 100-ml reservoir. Both PE and PTFE devices withstood this fluid flow in the incubator for up 4 days, with no leaks, indicating effective bonding of the membrane to PDMS.
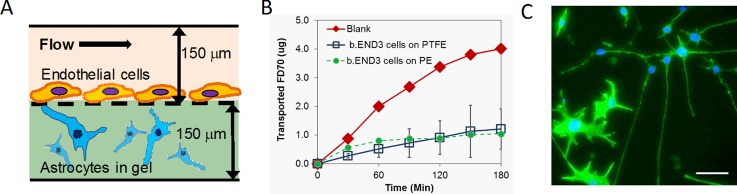
FIG. 1.
(a) Schematic of the microfluidic co-culture. (b) FITC-dextran transport across bEnd.3 cells on PTFE (n = 3, error bars are S.D.) and PE (n = 2) membrane-integrated microfluidic devices. (c) Phalloidin (green)-stained C8D1A astrocytes in collagen hydrogel in co-culture with bEnd.3 at day 14. Blue: Hoechst 33 342 counterstain. Scale bar: 50 μm.
Astrocytes (C8D1A, ATTC) were encapsulated at 1 × 106 cells/ml in 2 mg/ml collagen hydrogels obtained by neutralizing and diluting rat tail collagen (10.08 mg/ml, Corning 354249) on ice. Hydrogel-encapsulated cells were injected with a chilled 50-μl Hamilton syringe into the basolateral compartment and polymerized in situ for 30 min before seeding of endothelial cells in the apical compartment.
bEnd.3 cells (ATTC) were seeded on PE membranes coated with 1.8 μg/cm2 collagen IV–fibronectin. The optimal coating for PTFE was found to be 1.6 μg/cm2 collagen I. After phase contrast microscopy indicated complete coverage of the membrane channel, barrier properties of bEnd.3 cells were measured by apical to basolateral transport of 70 kDa fluorescein isothiocyanate (FITC)-dextran (2 mg/ml in phenol red-free Dulbecco's Modified Eagle Medium (DMEM)) after flushing out the collagen-embedded astrocytes. Media from the basolateral compartment were collected every 30 min for 180 min, and fluorescence was measured using a plate reader (Flipr Tetra, Molecular Devices). The apparent permeability coefficient was calculated using the equation , where ΔQ/ΔT is the slope of the linear portion of the compound transported vs. time curve, A is the area of the membrane, and C0 is the initial concentration.
Phalloidin staining for actin and immunofluorescence staining for claudin-5 were performed as previously described.5,15
RESULTS AND DISCUSSION
bEnd.3 cells cultured on PTFE membranes demonstrated barrier properties with 70-kDa FITC dextran transport clearly reduced (Papp = 5.9 ± 2.3 × 10−7 cm/s, n = 3) from a blank membrane (Papp = 2.1 × 10−6 cm/s) (Fig. 1(b)), under both high and low shear stress. The barrier was superior to the one measured for bEnd.3 on PE (Papp = 1.2 × 10−6 cm/s, n = 2), which could only be obtained at low shear stress. Dextran permeabilities on both membranes in devices were as good or superior to those measured in our static 3D BBB co-culture model with the same cell types (Papp = 1–7 × 10−6 cm/s).5 Permeability in the PTFE-integrated device is comparable with the PC membrane-based microfluidic bEnd.3/C8D1A co-culture described by Booth,12 (Papp ∼ 9 × 10−7 cm/s), although in that case the C8D1A are grown on the membrane and not in a 3D matrix.
The flow in the apical compartment was adequate to ensure viability of C8D1A in the basolateral compartment, with viability over 90% at day 4 (n = 3) as measured by live/dead staining (L-3224, Life Technologies). As observed by phase microscopy, bEnd.3 in co-culture with C8D1A reached confluence within a period of 1.6 ± 0.9 days, faster than in monoculture for both PE and PTFE devices (5.8 ± 1.0 and 4.5 ± 0.5 days, respectively). The co-cultures could be maintained for as long as 14 days (Fig. 1(c)), but were typically used within one week.
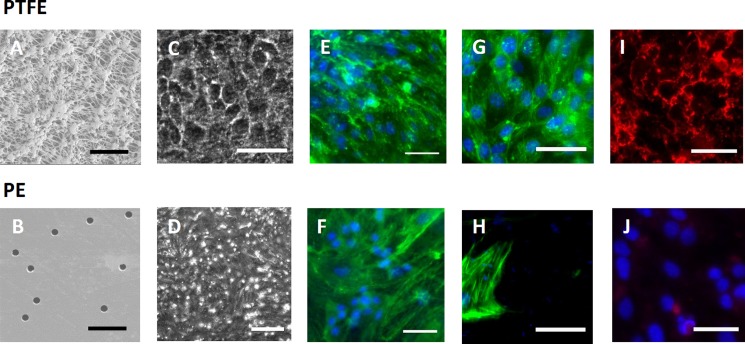
Both PE and PTFE membranes enable phase contract imaging of cells throughout their cultures (to conveniently monitor growth and confluence, Figs. 2(c) and 2(d)). bEnd.3 formed confluent monolayers with similar cell morphology on both membranes as illustrated by phalloidin (Molecular Probes) staining (Figs. 2(e) and 2(f)). Monolayers remained intact at 5 dyn/cm2 shear stress for 24 h on PTFE (Fig. 2(g)) and exhibited barrier properties (Fig. 1(b)), while on PE the cells were peeled off (Fig. 2(h); supplementary material).18
FIG. 2.
(a) and (b) SEM images of PTFE and PE membranes, respectively, scale bar: 5 μm. (c)–(j) Optical microscope images of bEnd.3 microfluidic cultures, scale bar: 50 μm unless otherwise noted. (c) and (d) Phase contrast (200-μm scale bar). (e)–(h) Phalloidin (green) and Hoechst (blue) stains of bEnd.3 cells with minimal flow ((e) and (f)) and at 5 dyn/cm2 ((g) and (h)). (i) and (j) Claudin-5 staining (red).
Protein expression of claudin-5, an integral membrane protein highly expressed in brain endothelial cells and a major structural component of tight junctions responsible for the size-selectivity of the BBB,19 was observed on PTFE membranes (Fig. 2(i)), while it was consistently negative on PE membrane devices (n = 10 tests, Fig. 2(j)).
In summary, we demonstrate for the first time a functional neurovascular unit model in a microfluidic platform incorporating a 3D matrix for supporting cells and an optically transparent membrane for endothelial cells that supports cell attachment at fluid shear stress comparable to that in the cerebral microvasculature in vivo. An in vitro platform recapitulating these key features of the neurovascular unit will facilitate better evaluation of brain pharmacokinetics in vitro, improving the pace and quality of preclinical CNS drug development.
ACKNOWLEDGMENTS
This research was supported by RTI International internal R&D funds. We thank Steven Hall, RTI, for technical assistance with device microfabrication.
References
- 1. Engelhardt B. and Liebner S., Cell Tissue Res. 355, 687 (2014). 10.1007/s00441-014-1811-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Obermeier B., Daneman R., and Ransohoff R. M., Nat. Med. 19, 1584 (2013). 10.1038/nm.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hawkins B. T. and Davis T. P., Pharmacol. Rev. 57, 173 (2005). 10.1124/pr.57.2.4 [DOI] [PubMed] [Google Scholar]
- 4. Alcendor D. J., Block F. E., Cliffel D. E., Daniels J. S., Ellacott K. L. J., Goodwin C. R., Hofmeister L. H., Li D. Y., Markov D. A., May J. C., McCawley L. J., McLaughlin B., McLean J. A., Niswender K. D., Pensabene V., Seale K. T., Sherrod S. D., Sung H. J., Tabb D. L., Webb D. J., and Wikswo J. P., Stem Cell Res. Ther. 4, S18 (2013). 10.1186/scrt379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hawkins B. T., Grego S., and Sellgren K. L., Brain Res. 1608, 167 (2015). 10.1016/j.brainres.2015.02.025 [DOI] [PubMed] [Google Scholar]
- 6. Gulino-Debrac D., Tissue Barriers 1, e24180 (2013). 10.4161/tisb.24180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cucullo L., Hossain M., Puvenna V., Marchi N., and Janigro D., BMC Neurosci. 12, 40 (2011). 10.1186/1471-2202-12-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Achyuta A. K. H., Conway A. J., Crouse R. B., Bannister E. C., Lee R. N., Katnik C. P., Behensky A. A., Cuevas J., and Sundaram S. S., Lab Chip 13, 542 (2013). 10.1039/C2LC41033H [DOI] [PubMed] [Google Scholar]
- 9. Booth R., Noh S., and Kim H., Lab Chip 14, 1880 (2014). 10.1039/c3lc51304a [DOI] [PubMed] [Google Scholar]
- 10. Douville N. J., Tung Y. C., Li R., Wang J. D., El-Sayed M. E. H., and Takayama S., Anal. Chem. 82, 2505 (2010). 10.1021/ac9029345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griep L. M., Wolbers F., de Wagenaar B., ter Braak P. M., Weksler B. B., Romero I. A., Couraud P. O., Vermes I., van der Meer A. D., and van den Berg A., Biomed. Microdevices 15, 145 (2013). 10.1007/s10544-012-9699-7 [DOI] [PubMed] [Google Scholar]
- 12. Booth R. and Kim H., Lab Chip 12, 1784 (2012). 10.1039/c2lc40094d [DOI] [PubMed] [Google Scholar]
- 13. Song J. W., Cavnar S. P., Walker A. C., Luker K. E., Gupta M., Tung Y.-C., Luker G. D., and Takayama S., PLoS One 4, e5756 (2009). 10.1371/journal.pone.0005756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shao J. B., Wu L., Wu J. Z., Zheng Y. H., Zhao H., Jin Q. H., and Zhao J. L., Lab Chip 9, 3118 (2009). 10.1039/b909312e [DOI] [PubMed] [Google Scholar]
- 15. Sellgren K. L., Butala E. J., Gilmour B. P., Randell S. H., and Grego S., Lab Chip 14, 3349 (2014). 10.1039/C4LC00552J [DOI] [PubMed] [Google Scholar]
- 16. Gregory C. W., Sellgren K. L., Gilchrist K. H., and Grego S., Biomicrofluidics 7, 56503 (2013). 10.1063/1.4827600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mairey E., Genovesio A., Donnadieu E., Bernard C., Jaubert F., Pinard E., Seylaz J., Olivo-Marin J. C., Nassif X., and Dumenil G., J. Exp. Med. 203, 1939 (2006). 10.1084/jem.20060482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.See supplementary material at http://dx.doi.org/10.1063/1.4935594E-BIOMGB-9-004506 for images showing endothelial cell adhesion on PE membranes at different flows.
- 19. Nitta T., Hata M., Gotoh S., Seo Y., Sasaki H., Hashimoto N., Furuse M., and Tsukita S., J. Cell Biol. 161, 653 (2003). 10.1083/jcb.200302070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4935594E-BIOMGB-9-004506 for images showing endothelial cell adhesion on PE membranes at different flows.