Abstract
Microbial communities are complex heterogeneous systems that are influenced by physical and chemical interactions with their environment, host, and community members. Techniques that facilitate the quantitative evaluation of how microscale organization influences the morphogenesis of multispecies communities could provide valuable insights into the dynamic behavior and organization of natural communities, the design of synthetic environments for multispecies culture, and the engineering of artificial consortia. In this work, we demonstrate a method for patterning microbes into simple arrangements that allow the quantitative measurement of growth dynamics as a function of their proximity to one another. The method combines parylene-based liftoff techniques with microfluidic delivery to simultaneously pattern multiple bacterial species with high viability using low-cost, customizable methods. Quantitative measurements of bacterial growth for two competing isolates demonstrate that spatial coordination can play a critical role in multispecies growth and structure.
I. INTRODUCTION
In natural systems, bacterial growth occurs in complex heterogeneous communities. The spatial organization of community members combined with the local physical and chemical architecture of the environment play an important role in community development, robustness, and function.1–3 Spatial structure contributes to bacterial community function in plant and animal microbiomes4,5 and in dental plaques.6,7 The physical structure has been studied in multiple model systems2,8–10 and examined using computational tools.1,3 Interactions between bacterial community members will depend on the spatial orientation in the community.
The traditional method for studying bacterial interactions is to screen growth on agar plates by hand spotting bacterial cells in close proximity to one another. This simple and rapid approach remains a first pass screen for qualitatively identifying interactions between bacterial strains. Bacterial interactions can also be investigated using pin replicator systems that provide higher throughput for pairwise patterning.11 An alternate approach to bacterial patterning is to use surface chemical modification to immobilize bacteria. This has been shown for E. coli systems12,13 and non-model organisms,14 but can significantly alter cell viability and development. Another approach to patterning is direct printing (ink jet), which can produce spot patterns with a resolution of approximately 200 μm.15,16 Other variations for studying interactions include contact printing,17–19 direct write patterning,20 fluidic immobilization,21,22 and mixing,23 as well as electrical control of patterning.24 These and other techniques have been reviewed in the literature.25 Stencil approaches have used fluidic patterning with dry liftoff or surface chemical patterning to form multi-component, bio-molecular patterns at micron-scale resolution.26,27
In this work, we adapt parylene dry lift-off techniques,28,29 fluidic patterning, and transfer printing to generate robust bacterial patterns that exhibit high viability and growth commensurate with conventional culture methods. By aligning a microfluidic network to a stencil etched in parylene, we demonstrate the ability to “ink” agar pucks loaded into silicon micro-wells with different species of bacteria. The bacteria are then transferred onto agar surfaces, imaged during growth, and quantitatively evaluated to assess interactions. The approach demonstrated here is flexible for patterning of diverse cultivable bacteria.
II. RESULTS AND DISCUSSION
A. Delivery of multiple bacterial species in spatially defined patterns
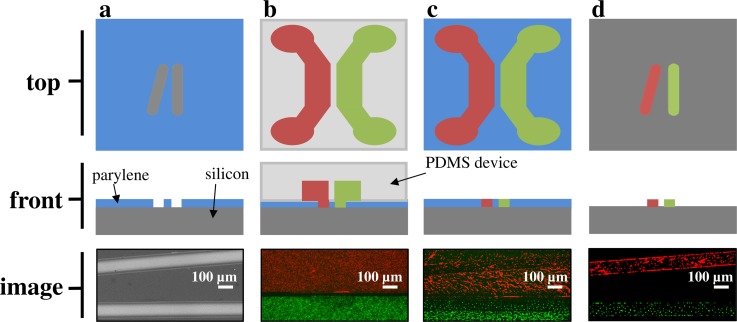
A two-layer patterning method was designed to deliver bacterial strains to specific locations on a surface. PDMS microfluidic networks, designed to align to predefined parylene coated silicon microwell substrates, were created using conventional soft lithography techniques.30 Figure 1 shows the schematic of the patterning process with representative images from patterning experiments. First, the parylene stencil and PDMS device are aligned manually under a dissecting scope, and then channels are filled with targeted bacteria in growth media. After a 30 min incubation period, fluid is removed from the channels and the PDMS device is removed. Then, the parylene is peeled to leave the stencil pattern. Pattern fidelity is reproducible along the length of the pattern (∼5 mm), and the two strains were patterned as close at 120 μm as indicated in Figure 1(d). For this process, the proximity of strains is limited by the width of the boundary between adjacent channels in the PDMS delivery device.
FIG. 1.
Patterning workflow. (a) The stencil pattern is etched through the parylene layer (blue) to the silicon surface (gray). SEM image shows the parylene coated silicon surface with target pattern. (b) Bacteria are delivered to the pattern with microfluidic channels aligned to the stencil pattern. Fluorescent image shows bacteria in adjacent channels. (c) The microfluidic device is removed, leaving imprinted bacteria. Fluorescent image shows pattern delivered to surface. (d) The parylene is peeled, leaving the bacteria patterned with the stencil. Fluorescent image shows final pattern delivery.
B. Pattern intensity and fidelity depends on seeding density
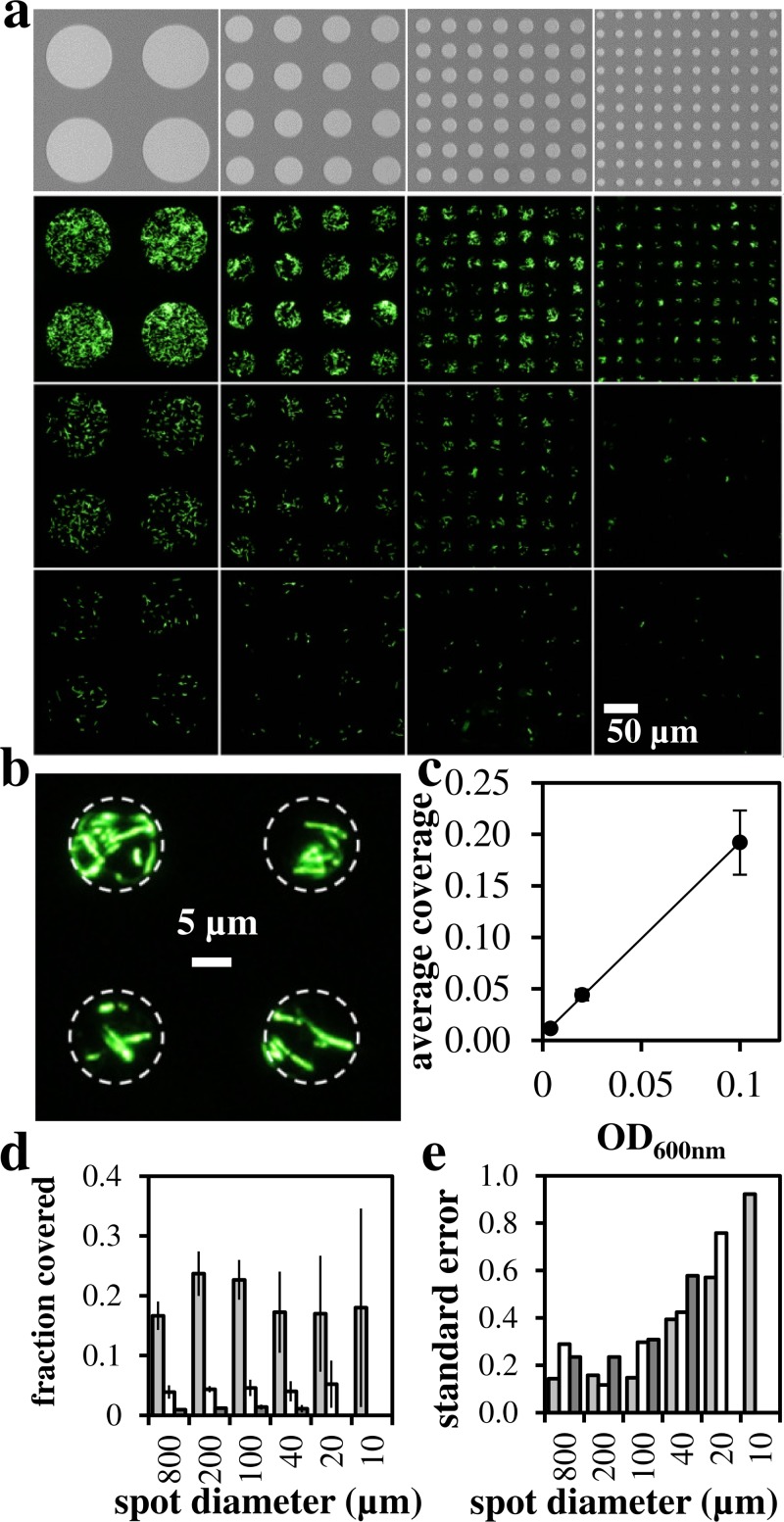
To determine effect of culture density on pattern intensity and fidelity, bacteria were patterned on spots of 800–10 μm diameter onto silicon surfaces (Figure 2). At OD600 nm = 0.1, coverage of spots was observed down to spot diameters approaching the size of single cells (Figure 2(b)). Pattern coverage decreased with decreasing bacterial load (Figures 2(c) and 2(d)). At OD600 nm = 0.004, we observe greater variability in spot coverage as quantified by standard deviation divided by the mean shown in Figure 2(e). These results indicate that higher inoculum optical density produces higher fidelity patterns. With the control of optical density and spot size, this patterning approach could be adapted to deliver single bacterial cells to defined locations using parylene stencils. In this study, we used a parylene layer with thickness of ∼1 μm to ensure that the parylene would have the mechanical strength required for removal without tearing. At the densities and incubation times used in this study, we observed an approximate monolayer of cells deposited to the surface.
FIG. 2.
Microfluidic delivery of bacteria to surface. (a) SEM images of spot array target (top) and fluorescent microscopy images of patterned GFP-labeled E. coli. Culture density was decreased from top to bottom using OD600 nm = 0.1, 0.02, 0.004. Spot diameter in microns (left–right): 100, 40, 20, 10. (b) High resolution image of patterned GFP-E. coli cells (OD600 nm = 0.1, 10 μm diameter). (c) Quantification of delivery vs. inoculum optical density. (d) Quantification of delivery as measured by fraction of spot covered. Gray bars: OD600 nm = 0.1, white bars: OD600 nm = 0.02, black bars: OD600 nm = 0.004. (e) Variability in spot coverage as standard error = standard deviation/mean.
C. Pattern transfer to agar surfaces
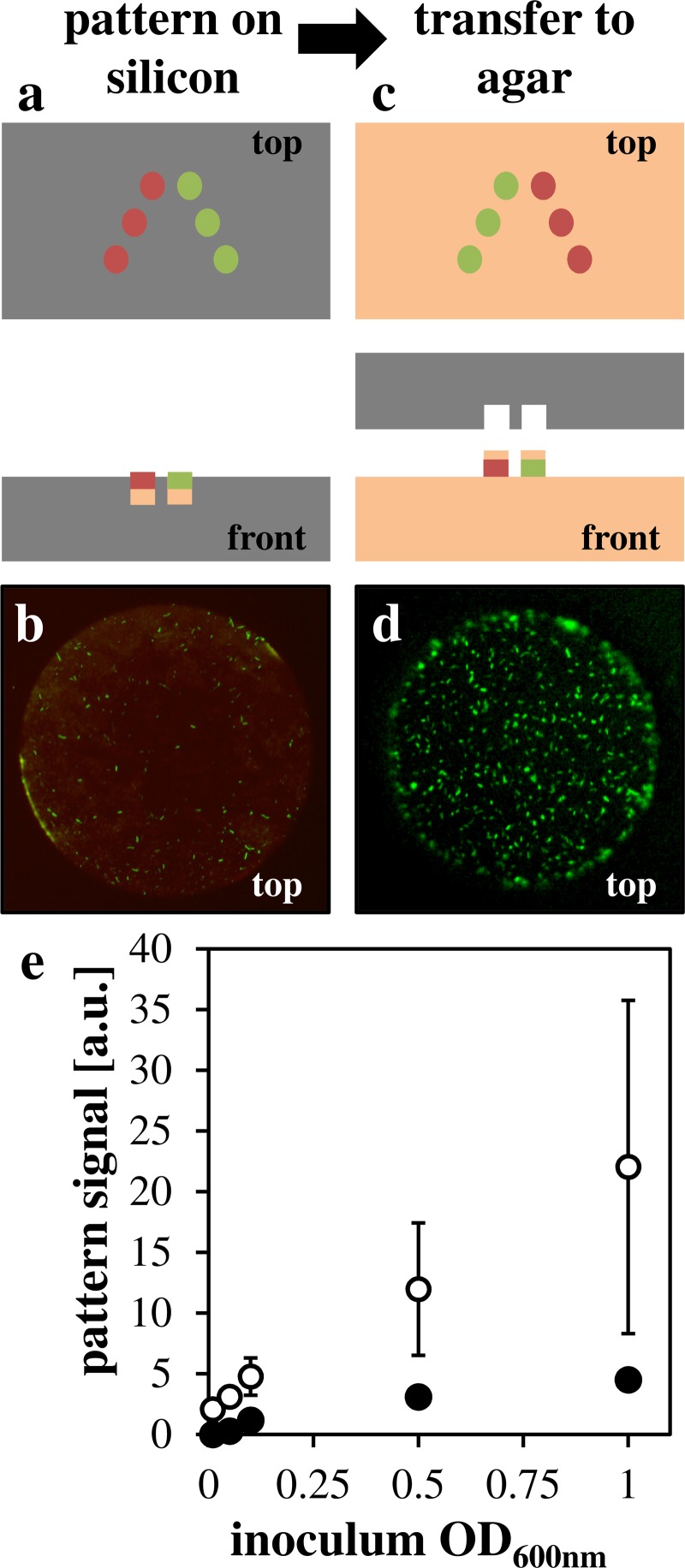
While patterning on silicon results in well-controlled intensity and fidelity, we observed that cells would not grow after stamping to growth medium. Analysis of cell viability showed that the majority of cells were not viable after the drying process (Figure S1).35 To enhance cell viability during transfer to growth medium, patterns were further etched into the silicon layer using the Bosch process then filled with agar before aligning the microfluidic delivery network. This process generates a cylindrical agar pad as a cell carrier which must be efficiently transferred to growth medium. Figure 3 shows the schematic of the process and the images of bacterial cells on agar pads before (Figures 3(a) and 3(b)) and after (Figures 3(c) and 3(d)) transfer to semi-solid growth medium. As observed with pattern transfer to silicon, transfer to growth medium was dependent on optical density (Figure 3(e)). At higher cell concentration, a clear edge effect is observed where cells are biased towards the perimeter of the agar pad. Such edge effects have been observed in similar patterning strategies14 and may be attributed to surface roughness generated as a byproduct of the Bosch etch process. Transfer of agar pads to growth medium was dependent on aspect ratio of wells (Figure S2).35 While well depth increases agar volume and thus the ability to keep cells hydrated, shallow wells are required for consistent transfer. Development of automated transfer processes would likely improve both the pad transfer fidelity at higher aspect ratios and the inter-substrate reproducibility. Further, the ability to shape the agar surface potentially allows for testing the effect of surface topology on microbial growth.
FIG. 3.
Agar pad incorporation for cell transfer. (a) Schematic showing agar pads in silicon microwells (b) representative image of GFP E. coli on RFP-labeled agar pad in silicon well (c) schematic showing agar pad transferred to agar surface (d) representative image of GFP E. coli on agar growth medium with agar pad. (e) Quantification of green pixels on target spot (white points) or off-target background (black points) plotted against OD600 nm.
D. Demonstration of multispecies patterning
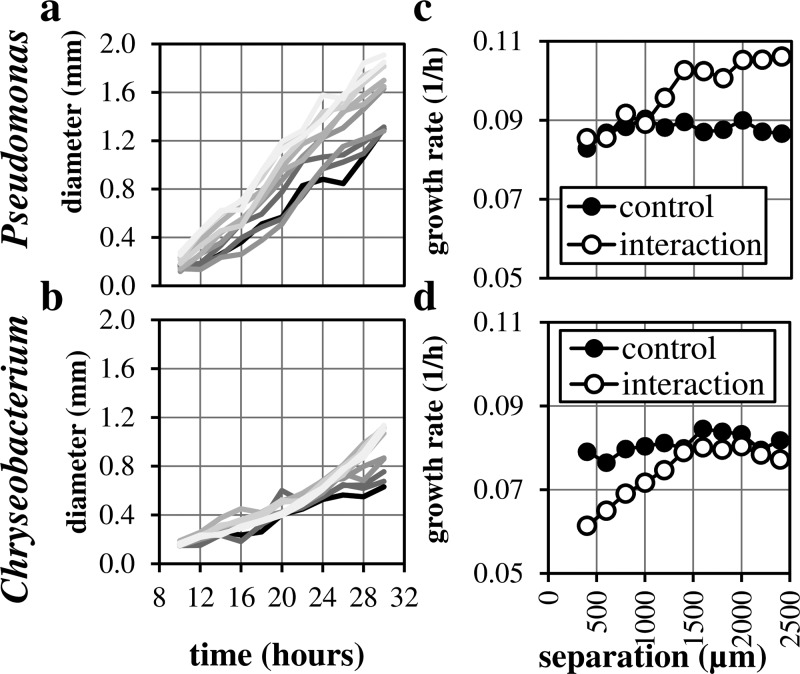
To demonstrate multispecies patterning, growth, and interaction, we chose two bacterial strains isolated from poplar roots.31 Strains were patterned from cultures at OD600 = 1.0 in a V-pattern (Figure S3)35 then imaged over a 30-h period (supplementary Video 1).35 Colony diameters were measured, and colony growth rates were extracted by fitting an exponential growth model (Figure 4). The high density was chosen to minimize the possibility of stochastic effects on interactions. Despite the fusing of colonies during growth, colonies of the different organisms that were closer together grew more slowly than when they were spaced farther apart, suggesting competitive interactions that are distance dependent. This trend was qualitatively conserved in replicate experiments (not shown). Quantification of colony growth rates by measuring diameter reveals that colonies of both species are affected by proximity to the competitor strain when compared to control experiments where both sides were patterned with the same bacteria. In control experiments, we did not observe a dependence of colony growth rate on separation distance. For the Pseudomonas isolate, the early colony growth rate at separation distances between 400 and 1000 μm is slow relative to that observed at greater separation distances. At greater separation distances, the growth rate is higher, presumably where no interaction is occurring. At large separation distances, growth is increased relative to the no-interaction control growth, potentially explained by swarming behavior as a response to a competitor. This behavior is also observed in traditional hand-spotting interaction measurements. Further work is required to distinguish the cause of this bimodal growth behavior. For the Chryseobacterium isolate, we observe a steady decrease in colony growth rate with proximity to the competitor. This interaction demonstrates the utility of this assay and ability to extract and quantify changes in growth due to proximity to a competitor.
FIG. 4.
Colony growth curves in interaction experiment between Pseudomonas and Chryseobacterium isolates. (a) Quantification of Pseudomonas colony diameter over time. (b) Quantification of Chryseobacterium colony diameter over time. Darker line colors represent colonies closer to the competitor. (c) Growth rate vs. separation distance for Pseudomonas. (d) Growth rate vs. separation distance for Chryseobacterium. Control experiments were performed using the same pattern without competitors.
III. CONCLUSIONS
Microfluidic inking of patterned parylene/silicon microwells combined with agar transfer was used to demonstrate a method for selectively patterning multiple bacterial species for the study of microbe-microbe interactions. This method can provide more information than simple spotting techniques and provides data that reveal distance- and time-dependencies of interactions. This information-dense assay can be used to infer mechanisms of interaction between bacterial strains. The method was demonstrated by studying contact-independent interactions between Pseudomonas and Chryseobacterium strains from our isolate collection. There was a clear, distance-dependent interaction between the two strains that could be attributed to secretion of inhibitory molecules or competition for resources.
The minimum resolution for multi-species patterns depends on the achievable separation between the fluidic channels used to ink the stencil. We showed patterning as close as 120 μm, but this could be decreased using optimized patterns and microfluidics, automated alignment of the fluidic device to the stencil layer (manual alignment was used here), or chemical surface modification. We did observe edge effects as has been reported previously,32 and presumably this could be harnessed to improve pattern resolution. The resolution presented here is better than contemporary methods of pin-deposition or ink-jet printing.15,16
In addition to the simple pattern used in this study, more complex patterns of multiple bacterial species could be configured to generate spatially coordinated microbial communities. Such communities could be used in studies of metabolite transfer3,10 or to mimic spatially defined multi-member biofilms found in natural systems.6
The method developed here is generally applicable to cultivable bacteria. The simple approach involves cells grown in liquid media and delivered to a solid media surface. The agar pads keep the cells hydrated through the pattern transfer process. The robustness of the patterning method and fidelity of the pattern eliminates the need for fluorescent labels to distinguish bacterial strains, allowing for rapid screening of quantitative interactions. The method described here offers a simple and general method for patterning bacteria and observing spatial behavior and interactions.
IV. METHODS
A. Parylene deposition
Parylene C polymer is deposited using ∼0.8 g of parylene C monomer (Specialty Coating Systems, Indianapolis, IN, USA) in the Labcoater PDS 210 (Specialty Coating Systems, Indianapolis, IN, USA) according to manufacturer instructions. Parylene coated wafers are stored under vacuum before and after processing until use for experiments. The thickness of the parylene layer used in this study was 1.1–1.4 μm.
B. Plasma etching and photolithography
Stencil patterns were developed with microfluidic inking patterns using CAD software. Masks were written using a Heidelberg DWL 66 (Heidelberg Instruments, Heidelberg, Germany). Parylene coated ⟨100⟩ silicon wafers were pre-coated with Shin-Etsu MicroSi MicroPrime P20 at 3000 rpm for 45 s followed by positive photoresist at 3000 rpm for 45 s. Wafers were then exposed for 15 s with a Quintel contact mask aligner (Neutronix Quintel, CA, USA) and developed in CD-26 for 4 min, rinsed with DI H2O, and dried with N2. Wafers were then treated with RIE O2 plasma (Oxford Instruments). After etching through the parylene, the silicon wells were etched using the Bosch process (Oxford Instruments) to the desired depth. Photoresist was removed using acetone (no agitation), and the substrates were thoroughly rinsed in water and dried with nitrogen prior to use. All wafer processing steps were carried out in the Center for Nanophase Materials clean room at Oak Ridge National Laboratory.
C. Microfluidic fabrication
Clean silicon wafers and standard photolithography techniques were used to transfer the pattern used for the fluidic master. Poly-dimethyl-siloxane was mixed with curing agent at a 10:1 ratio and frozen for future use. On the day of device production, the PDMS mixture was poured onto the silicon master and degassed. The device was baked at 80 °C for 45 min, allowed to cool, then peeled and punched using 1 mm or 1.5 mm Harris punches for fluidic access.
D. Pattern inking and transfer
Microfluidic delivery devices were designed for specific parylene stencil patterns. Following fabrication of the stencil and microfluidic device, the patterned substrates are filled with a semi-solid agar that provides a hydrated and buffered environment to enhance bacterial viability. The microfluidic network is aligned to the stencil, assembled, and sealed using a vacuum pump to apply negative pressure in a post region around features, and the different network channels are filled with the bacteria of interest using 1 ml syringes (BD). Bacteria were incubated for 30 min in the channels and attached to the filled microwells and surrounding parylene stencil. The channels are emptied, and the microfluidic overlay is removed. Parylene was peeled using clear adhesive tape, and the pattern was immediately covered with liquefied agar (1.5% agar in R2A or LB media at 42 °C). After cooling, the pattern was inverted, allowed to dry for 30 min, then covered and imaged.
E. Cell culture
E. coli K12 strains expressing GFP or RFP from plasmid pBBR1-MCS533 were grown in LB media with 10 μg/ml Gentamycin at 37 °C with shaking. Pseudomonas sp. GM41 and Chryseobacterium sp. CF314 were isolated from the roots of Populus trees31 and were grown in R2A media at 29 °C with shaking. Optical density (600 nm) was measured using a Nanodrop (Thermo Scientific, USA). Cells were pelleted and resuspended in appropriate volumes of fresh R2A media for patterning. Bacterial viability was measured using the BacLight Bacterial Viability Kit (Invitrogen, USA) according to the manufacturer's instructions.
F. Imaging
Still images of bacterial patterns were collected using a Zeiss Axioskop 2 (Carl Zeiss Microscopy, NY, USA) with brightfield, phase contrast, and fluorescent capabilities. Images were collected using constant illumination with white light and optimized exposure times based on plate-to-plate variation. Time-lapse of colony growth was imaged using the Zeiss Stereoscope SteREO Lumar V12 (Carl Zeiss Microscopy, NY, USA) every 30 min for 30 h after patterning. Images collected in 2 h intervals between 10 and 30 h were used for quantification. Electron microscopy images were collected using a Hitachi TM3000 using standard settings.
G. Image analysis
Pixel densities were collected in ImageJ using standard analysis scripts.34 Colony growth rates were collected by measuring colony diameter as vertical diameter in images and movies using ImageJ. Growth rate was extracted by fitting an exponential growth curve in MSExcel.
H. Colony growth rate analysis
For each colony, diameter data were fit to the exponential growth function with D0 set to 100 μm (the agar pad diameter in the pattern used for experiments). The sum-of-squares error between measured diameter and fitted diameter D was minimized using the Solver add-in in MSExcel, allowing only growth rate r to change.
ACKNOWLEDGMENTS
The authors would like to acknowledge Amber Bible for creating the fluorescent E. coli K12 strains used in this study. This research was funded by the U.S. DOE Office of Biological and Environmental Research, Genomic Science Program and the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory. A portion of this research was conducted at the Center for Nanophase Materials Sciences, which is sponsored at Oak Ridge National Laboratory by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under Contract No. DEAC05-00OR22725. This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains, and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
References
- 1. Xavier J. B. and Foster K. R., Proc. Natl. Acad. Sci. U.S.A. 104, 876 (2007). 10.1073/pnas.0607651104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Denef V. J., Mueller R. S., and Banfield J. F., ISME J. 4, 599 (2010). 10.1038/ismej.2009.158 [DOI] [PubMed] [Google Scholar]
- 3. Harcombe W. R., Riehl W. J., Dukovski I., Granger B. R., Betts A., Lang A. H., Bonilla G., Kar A., Leiby N., Mehta P., Marx C. J., and Segrè D., Cell Rep. 7, 1104 (2014). 10.1016/j.celrep.2014.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., and Schulze-Lefert P., Annu. Rev. Plant Biol. 64, 807 (2013). 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- 5. Gillilland M. G., Erb-Downward J. R., Bassis C. M., Shen M. C., Toews G. B., Young V. B., and Huffnagle G. B., Appl. Environ. Microbiol. 78, 2359 (2012). 10.1128/AEM.05239-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kolenbrander P. E., Palmer R. J., Periasamy S., and Jakubovics N. S., Nat. Rev. Microbiol. 8, 471 (2010). 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- 7. Kuramitsu H. K., He X., Lux R., Anderson M. H., and Shi W., Microbiol. Mol. Biol. Rev. 71, 653 (2007). 10.1128/MMBR.00024-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hansen S. K., Rainey P. B., Haagensen J. A. J., and Molin S., Nature 445, 533 (2007). 10.1038/nature05514 [DOI] [PubMed] [Google Scholar]
- 9. Hassell M. P., Comins H. N., and May R. M., Nature 370, 290 (1994). 10.1038/370290a0 [DOI] [Google Scholar]
- 10. Kim H. J., Boedicker J. Q., Choi J. W., and Ismagilov R. F., Proc. Natl. Acad. Sci. U.S.A. 105, 18188 (2008). 10.1073/pnas.0807935105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sánchez-Hidalgo M., Pascual J., de la Cruz M., Martín J., Kath G. S., Sigmund J. M., Masurekar P., Vicente F., Genilloud O., and Bills G. F., Antonie Van Leeuwenhoek 102, 361 (2012). 10.1007/s10482-012-9746-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Costello C. M., Kreft J.-U., Thomas C. M., Hammes D. M., Bao P., Evans S. D., and Mendes P. M., Soft Matter 8, 9147 (2012). 10.1039/c2sm26111a [DOI] [Google Scholar]
- 13. Dwidar M., Leung B. M., Yaguchi T., Takayama S., and Mitchell R. J., PLoS One 8, e67165 (2013). 10.1371/journal.pone.0067165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hansen R. R., Hinestrosa J. P., Shubert K. R., Morrell-Falvey J. L., Pelletier D. A., Messman J. M., Kilbey S. M., Lokitz B. S., and Retterer S. T., Biomacromolecules 14, 3742 (2013). 10.1021/bm4011358 [DOI] [PubMed] [Google Scholar]
- 15. Xu T., Petridou S., Lee E. H., Roth E. A., Vyavahare N. R., Hickman J. J., and Boland T., Biotechnol. Bioeng. 85, 29 (2004). 10.1002/bit.10768 [DOI] [PubMed] [Google Scholar]
- 16. Stubblefield B. A., Howery K. E., Islam B. N., Santiago A. J., Cardenas W. E., and Gilbert E. S., Appl. Microbiol. Biotechnol. 86, 1941 (2010). 10.1007/s00253-010-2473-y [DOI] [PubMed] [Google Scholar]
- 17. Eichinger C. D., Hsiao T. W., and Hlady V., Langmuir 28, 2238 (2012). 10.1021/la2039202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H., Shepherd N. H., and Nuzzo R. G., Soft Matter 6, 2238 (2010). 10.1039/c000385a [DOI] [Google Scholar]
- 19. Weibel D. B., Lee A., Mayer M., Brady S. F., Bruzewicz D., Yang J., Diluzio W. R., Clardy J., and Whitesides G. M., Langmuir 21, 6436 (2005). 10.1021/la047173c [DOI] [PubMed] [Google Scholar]
- 20. Kim J., Shin Y.-H., Yun S.-H., Choi D.-S., Nam J.-H., Kim S. R., Moon S.-K., Chung B. H., Lee J.-H., Kim J.-H., Kim K.-Y., Kim K.-M., and Lim J.-H., J. Am. Chem. Soc. 134, 16500 (2012). 10.1021/ja3073808 [DOI] [PubMed] [Google Scholar]
- 21. Li B., Qiu Y., Glidle A., McIlvenna D., Luo Q., Cooper J., Shi H.-C., and Yin H., Anal. Chem. 86, 3131 (2014). 10.1021/ac5001306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moffitt J. R., Lee J. B., and Cluzel P., Lab Chip 12, 1487 (2012). 10.1039/c2lc00009a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma L., Datta S. S., Karymov M. A., Pan Q., Begolo S., and Ismagilov R. F., Integr. Biol. 6, 796 (2014). 10.1039/C4IB00109E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pranzetti A., Mieszkin S., Iqbal P., Rawson F. J., Callow M. E., Callow J. A., Koelsch P., Preece J. A., and Mendes P. M., Adv. Mater. 25, 2181 (2013). 10.1002/adma.201204880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Velve-Casquillas G., Le Berre M., Piel M., and Tran P. T., Nano Today 5, 28 (2010). 10.1016/j.nantod.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ghosh M., Alves C., Tong Z., Tettey K., Konstantopoulos K., and Stebe K. J., Langmuir 24, 8134 (2008). 10.1021/la8006525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Atsuta K., Suzuki H., and Takeuhi S., J. Micromech. Microeng. 17, 496 (2007). 10.1088/0960-1317/17/3/011 [DOI] [Google Scholar]
- 28. Tan C. P., Cipriany B. R., Lin D. M., and Craighead H. G., Nano Lett. 10, 719 (2010). 10.1021/nl903968s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tan C. P., Seo B. R., Brooks D. J., Chandler E. M., Craighead H. G., and Fischbach C., Integr. Biol. 1, 587 (2009). 10.1039/b908036h [DOI] [PubMed] [Google Scholar]
- 30. Khademhosseini A., Langer R., Borenstein J., and Vacanti J. P., Proc. Natl. Acad. Sci. U.S.A. 103, 2480 (2006). 10.1073/pnas.0507681102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown S. D., Utturkar S. M., Klingeman D. M., Johnson C. M., Martin S. L., Land M. L., Lu T. S., Schadt C. W., Doktycz M. J., and Pelletier D. A., J. Bacteriol. 194, 5991 (2012). 10.1128/JB.01243-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hansen R. R., Shubert K. R., Morrell-Falvey J. L., Lokitz B. S., Doktycz M. J., and Retterer S. T., Biosensors 4, 63 (2014). 10.3390/bios4010063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kovach M. E., Elzer P. H., Steven Hill D., Robertson G. T., Farris M. A., Roop R. M. II, and Peterson K. M., Gene 166, 175 (1995). 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- 34. Schneider C. A., Rasband W. S., and Eliceiri K. W., Nat. Methods 9, 671 (2012). 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.See supplementary material at http://dx.doi.org/10.1063/1.4935938E-BIOMGB-9-007506 for supplementry figures and time lapse video.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4935938E-BIOMGB-9-007506 for supplementry figures and time lapse video.