Abstract
Circulating tumor cells (CTCs) are found in the blood of patients with cancer. Although these cells are rare, they can provide useful information for chemotherapy. However, isolation of these rare cells from blood is technically challenging because they are small in numbers. An integrated microfluidic chip, dubbed CTC chip, was designed and fabricated for conducting tumor cell isolation. As CTCs usually show multidrug resistance (MDR), the effect of MDR inhibitors on chemotherapeutic drug accumulation in the isolated single tumor cell is measured. As a model of CTC isolation, human prostate cancer cells were mixed with mouse blood cells and the label-free isolation of the tumor cells was conducted based on cell size difference. The major advantages of the CTC chip are the ability for fast cell isolation, followed by multiple rounds of single-cell measurements, suggesting a potential assay for detecting the drug responses based on the liquid biopsy of cancer patients.
INTRODUCTION
Circulating tumor cells (CTCs) were first described in 1869 by Ashworth,1 who observed small numbers of cells in patient blood that resembled cells of the primary tumor. These cells may constitute the seeds for subsequent metastasis in different organs.1–3 Although the nature of CTCs is not fully understood, these cells are reported to be drug resistant in some types of metastatic cancers (e.g., breast cancer, lung cancer, prostate cancer, and ovarian cancer),4–10 especially showing multidrug resistance (MDR) due to the expression of ATP-binding cassette (ABC) transporters.11,12 These transporters include P-glycoprotein (P-gp or ABCB1), multidrug resistant protein-1 (MRP1 or ABCC1), and breast cancer resistant protein (BCRP or ABCG2), which cause active transport of chemotherapeutic drugs (i.e., daunorubicin or paclitaxel) out of the cancer cell, termed as drug efflux, and this ultimately reduces the effectiveness of chemotherapy.13,14 Administration of MDR inhibitors that block drug efflux mediated by MDR transporters in combination with chemotherapeutic drugs that kill the tumor cells have been explored as a potential treatment strategy.15
Isolation of CTCs can be useful for personalized cancer chemotherapy because CTCs can be clinically important for providing predictive information for the adjustment of the therapeutic schemes.16 Our vision is that drug accumulation measured on CTCs can provide reliable information for patients undergoing chemotherapy. However, a key limitation in the capture of CTCs is their extreme rarity in blood (as low as ∼1–100 in 1 ml blood including 5 × 109 erythrocytes or red blood cells and 7 × 106 leukocytes or white blood cells (WBCs)).17–20
Currently, CellSearch™ is a CTC-based system to provide prognostic information for metastatic breast, prostate, and colon cancers.21–24 In this Food and Drug Administration (FDA)-approved system, CTCs are immunomagnetically captured from 7.5 ml of blood using magnetic labels conjugated to an antibody against the epithelial cell adhesion molecule (or EpCAM) on the cells and then fluorescently stained with labeled antibodies against epithelial cell-specific markers.25 While the system allows the CTCs to be remunerated for cancer prognosis, further cellular analysis cannot be applied because the CTCs are bound and fixed.26
Recently, worldwide efforts have been made to develop efficient and reliable CTC isolation techniques such as flow cytometry.27,28 Furthermore, a wide variety of microfluidic techniques used to isolate CTCs have been reported, and the isolation methods are based on immunoaffinity16,29–34 and immunomagnetic separation.35–37 The immuno-based methods depend on the use of an immunological label that recognizes the EpCAM biomarker in order to identify the presence of CTCs. Therefore, there are some limitations in this immuno-label method as some CTCs, particularly those of metastatic nature, might undergo epithelial-to-mesenchymal transition (EMT) thereby losing the EpCAM marker and potentially go undetected.26,38
However, there are some microfluidic methods that are label-free for CTC isolation,38 namely, dielectrophoresis (DEP)-based separation,39–42 density-based separation,37 deformability-based separation,43 and size-based separation.44 The last method is successful in isolating rare cells because most epithelial cells such as CTCs have sizes in order of 15–25 μm, which are larger than red blood cells (6–8 μm) and white blood cells (8–14 μm).45
In this paper, we designed and fabricated a microfluidic biochip (CTC chip) to isolate PCa among whole blood cells without the use of a label (i.e., EpCAM antibody), followed by multiple rounds of single-cell measurement. In our approach, the human prostate cancer cells (as a model of CTC) were mixed with mouse blood cells. After removal of red blood cells and plasma, the buffy coat (white blood cells) mixed with tumor cells was introduced into the CTC chip. Since the captured tumor cell had not been subjected to any immunoaffinity manipulations (i.e., antibody), the captured cell can be used for single-cell measurements such as the drug accumulation assay.46–48 This is an established assay used to measure the real-time effect of MDR inhibitors on accumulation of chemotherapeutic drugs (e.g., daunorubicin and labeled paclitaxel) in the same single prostate cancer cell.
MATERIALS AND METHODS
CTC chip design
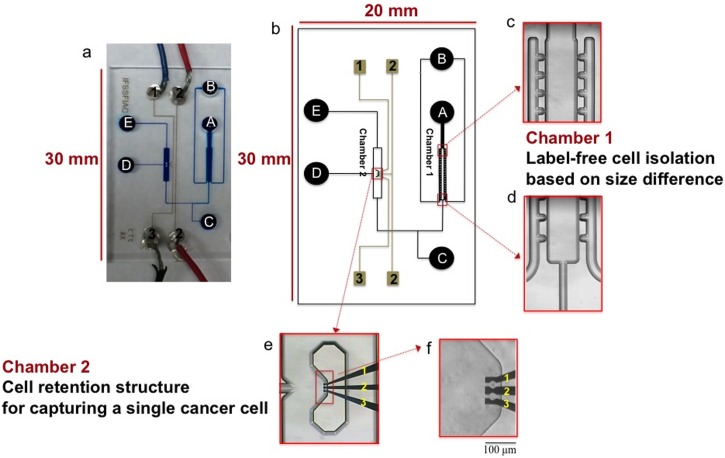
An integrated microfluidic chip, dubbed the CTC chip, was designed by the L-Edit software (Tanners). As shown in Fig. 1, the layout of the glass chip (20 mm × 30 mm) consisted of two chambers: Chamber 1 containing the sideward openings and chamber 2 containing the cell retention structure and DEP electrodes. Reservoirs A, B, and E served as the cell inlet, blood cell collector, and waste reservoirs, respectively; reservoir C was used to move cells from chamber 1 to chamber 2; reservoir D was used for drug delivery in chamber 2. The channels and chambers in the CTC chip were 40 μm deep, while the reservoirs were 0.6 mm deep and 2.5 mm in diameter. The CTC chip was fabricated by the standard micromachining processes at CMC Microsystems (Kingston, ON, Canada). The process includes standard chip cleaning, thin film deposition, photolithography, photoresist development, HF wet etching, reservoir forming, and chip bonding, as previously described.49
FIG. 1.
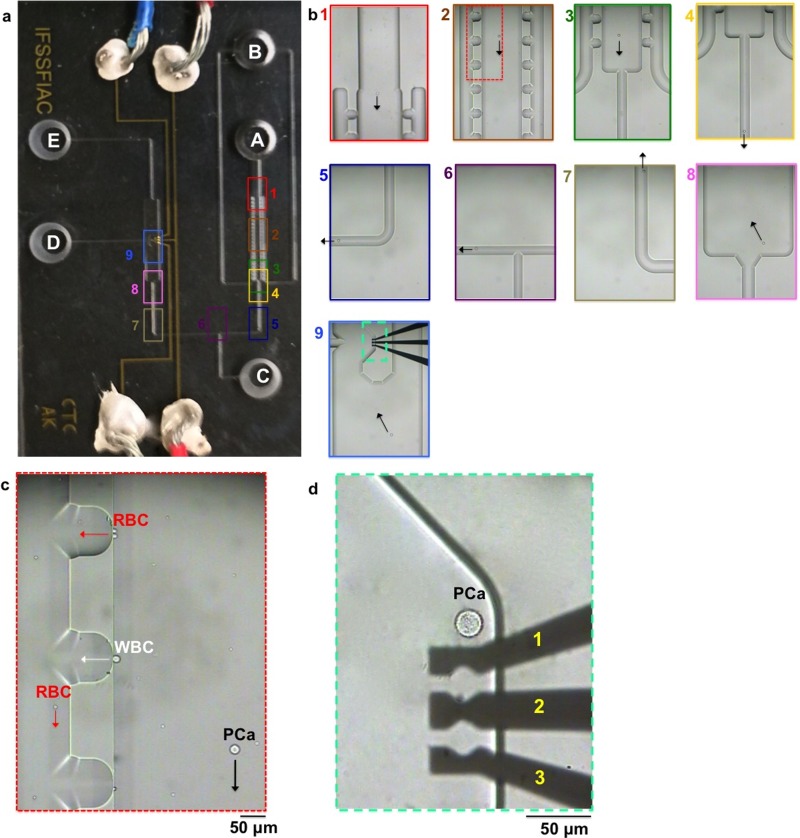
The CTC chip. (a) Image of the microchip consisting of channels filled with a blue food dye and of electrodes connected to electrical wires. (b) Layout of the microfluidic device showing reservoir D used for drug delivery, and reservoirs A and E served as the cell inlet and waste, respectively. Blood cells were collected in reservoir B. Reservoir C was used to move the selected cell from chamber 1 to chamber 2. (c and d) Close-up images of the cross-flow microfilters in chamber 1 in order to isolate prostate cancer cells among a large population of blood cells based on size difference. (e and f) Close-up images of the cell retention structure in chamber 2 and three DEP electrodes located inside the structure to capture the prostate cancer cell, followed by drug accumulation measurement. Scale bar: 100 μm.
As depicted in Fig. 1, chamber 1 in the CTC chip includes the cross-flow filter, with sideward openings perpendicular to the main flow, for separating cells based on size differences.50 Mouse blood cells, which are of similar size as human blood cells, were mixed with human prostate cancer cells, and the cell mixture was then introduced to reservoir A. Because of the small size (<15 μm) of blood cells, they can only pass through the sideward openings and be collected in reservoir B. Thereafter, as soon as an isolated prostate cancer cell could be found, it was captured in chamber 2. The DEP electrodes in chamber 2 (Pt 180 nm/Ta 20 nm) were used to capture the single-cell in a fixed location during drug accumulation measurement, when the drug was delivered from reservoir D.
Reagents
Daunorubicin (DNR), Oregon Green-labeled paclitaxel (OG-PTX), fumitremorgin C (FTC), and cyclosporine A (CsA) were purchased from Sigma-Aldrich (St Louis, MO, USA). RPMI 1460 medium, trypsin-ethylenediaminetetraacetic acid (Trypsin-EDTA) (0.025%), glutamine, penicillin/streptomycin (PEN/STR), and fetal bovine serum (FBS) were obtained from Life Technologies (Grand Island, NY). Hanks' balanced salt solution (HBSS) was from Invitrogen Corp (Grand Island, NY, USA, USA). DNR and OG-PTX were dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich) to make stock solutions of 350 μM and 300 μM, respectively. Similarly, stock solutions of CsA (500 μM) and FTC (1 mM) were made in DMSO. Alexa Fluor® 488-labeled anti-human P-gp monoclonal antibody was purchased from AbD Serotec (MorphoSys UK Ltd, Oxford, United Kingdom) and diluted in HBSS (1:20 ratio), and it was used to recognize human prostate cancer cell that expressed P-gp. The Ficoll-Paque PLUS solution from GE Healthcare (Pittsburgh, PA, USA) was kindly provided as a gift by Dr. Naveed Gulzar at the Department of Molecular Biology and Biochemistry (MBB), Simon Fraser University.
Cell samples
The prostate cancer (PCa) cell line, 22Rv1, obtained from ATCC, is an androgen-independent human cell line which naturally expresses ABCG2.51,52 This PCa cell line was grown in RPMI 1460 medium supplemented with 10% FBS, 1% PEN/STR, and 1% glutamine. For cell subculture, the cells were detached using trypsin-EDTA and re-seeded in fresh medium every 4 days. All cultures were maintained at 37 °C in a humidified 5% CO2 and 95% air incubator (NuAire, Plymouth, MN, USA). Prior to tumor cell isolation, the size of the cells was measured in order to determine the average sizes and the cells were counted using a hemocytometer. Mouse blood cells were obtained from the Animal Care Services at Simon Fraser University after protocol approval.
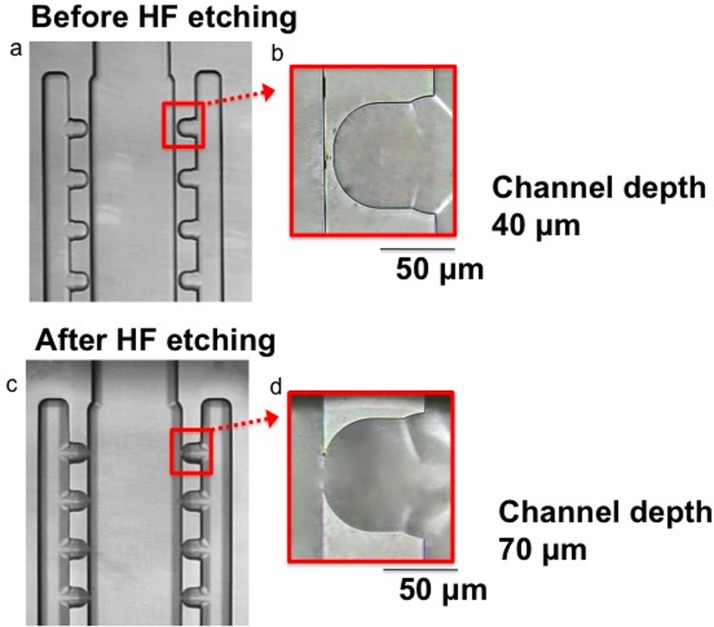
On-chip HF etching in chamber 1 to create the cross-flow microfilter
On-chip HF etching has been previously reported to enlarge a channel to create a weir structure to retain a single cardiomyocyte.49 We performed on-chip HF etching to create the cross-flow microfilter in chamber 1 in order to remove the small blood cells but not the big tumor cells. The spacing of the sideward openings was small enough to allow passing of the small blood cells (6–14 μm); whereas the large tumor cells (15–25 μm) did not even approach the sideward openings and leak through. Since the spacing smaller than 80 μm cannot be made by the initial glass etching process used to create the 40 μm-deep channels, HF etching was conducted after the glass chip was bonded. Briefly, 12% HF solution was put into reservoir A, which was close to chamber 1. In order to prevent HF from reaching the DEP electrodes located in chamber 2 and destroying them, water was introduced from reservoir C and the water flow allowed the HF solution to be localized in chamber 1. After 90 min, HF was removed from the chip to stop etching, and then the sideward openings were examined under the microscope (Fig. 2).
FIG. 2.
Creation of sideward openings in the cross-flow microfilter in chamber 1 inside a bonded glass chip. Images (top view) showing the microfilter: (a and b) before etching and (c and d) after etching. Scale bar: 50 μm.
Buffy coat preparation using a Ficoll gradient
A 2-ml sample of mouse blood was collected in a tube containing heparin as an anticoagulant. The white blood cells were isolated by centrifugation using a Ficoll gradient, according to the manufacturer's protocol (GE Healthcare, Pittsburgh, PA). Briefly, a diluted suspension of blood was layered over 3 ml of Ficoll-Paque solution in a 15 ml conical tube and centrifuged at 400g at 20 °C for 40 min. Afterwards, the top layer including plasma and platelets was removed, and the buffy coat that consisted of the white blood cells (see Fig. S1a in the electronic supplementary material)83 was then transferred to a new 15-ml conical tube and re-suspended with phosphate buffered saline (PBS). After the tube was centrifuged at 500g for 15 min, the cell pellet was collected. Since it was pink in color, it contained residual red blood cells (Fig. S1b).83 A micropipette tip was dipped into the cell pellet to gently remove the red blood cells. Thereafter, the white blood cells were re-suspended in PBS and centrifuged at 500g for 15 min. The cell pellet was washed one more time with PBS by spinning at 600g for 8 min (Fig. S1c).83 Cells were then re-suspended in RPMI-1460 medium (supplemented with 10% FBS and 1% PEN/STR and 1% glutamine). Thereafter, mouse blood cells, which are of similar size as human blood cells, were mixed with human prostate cancer cells.
Isolation of individual prostate cancer cells
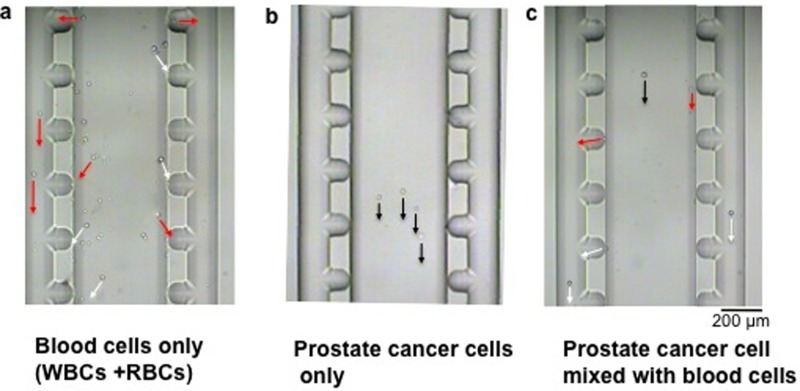
The prostate cancer cell (22Rv1) was isolated using the cross-flow microfilter in chamber 1. Prior to the CTC capture experiments, the channels and chambers were filled with culture medium (RPMI-1460 supplemented with 10% FBS). A cell sample containing a mixture of 22Rv1 cells and blood cells (in a ratio of 1:4000) was injected into the CTC chip from reservoir A. Fig. 3 describes the separation of the 22Rv1 cell among blood cells. Once the mixed cell sample (22Rv1 cells + blood cells) entered the wide chamber region (chamber 1), the larger 22Rv1 cells continued their straight path by the primary flow (Fig. 3(c)), while smaller and lighter cells followed the sideward flow (Fig. 3(c)). Comparison of Fig. 3(a) (blood cells only) and Fig. 3(b) (22Rv1 cells only) confirmed that the smaller cells moved toward the sideward openings, while the larger 22Rv1 cell continued the straight trajectory in the middle of chamber 1. This result suggested that the larger 22Rv1 cell did not reach the sideward openings regardless of the presence of blood cells (see Fig. S2 in the electronic supplementary material for the movement of cancer cells when mixed with blood cells at different ratios).83 The observation is counter-intuitive, but it is consistent with other studies that the contact of large cells (i.e., cell diameter > 10 μm) with the channel sidewall is very limited. It is because the opposed force generated by the hydrodynamic lift moves the cells away from the wall, and the force is stronger when the cell size is larger.40,44,53
FIG. 3.
Separation of the prostate cancer cell among other blood cells. (a) The blood cells (WBCs + RBCs) alone moved toward each of the sideward openings, splitting into two positions at right and left sides. (b) After injecting 22Rv1 cells alone, these big cells kept their straight path to reach chamber 2. (c) In a mixture of 22RV1 cell and blood cells, the WBCs + RBCs passed through the sideward exits and went to reservoir B; whereas the 22Rv1 cell moved to chamber 2. It took ∼2 s, 4 s, and 8 s for each RBC, WBC, and 22Rv1 cell to pass through chamber 1, respectively. The black, white, and red line arrows represent the movements of 22Rv1, WBC, and RBC, respectively. Scale bar: 200 μm.
For an actual CTC experiment, as soon as the 22Rv1 cells were observed in chamber 1, they were guided toward chamber 2 by manipulating the liquid flow using reservoirs A, C, and E. For instance, with the high liquid level at reservoir A and the low liquid level at reservoirs C and E, the 22Rv1 cells would leave chamber 1. As soon as the cells were near reservoir C, the liquid flow from it was increased to push the cell further toward the cell retention structure in chamber 2. The flow process has been controlled manually, but this process can be automated with the proper assistance of vision control and valving.
Dielectrophoresis electrodes to trap a single prostate cancer cell in chamber 2
The term dielectrophoresis was first introduced by Pohl54 in the 1950s to describe the behaviour of particles in a non-uniform electrical field. The dielectrophoretic (DEP) force can be created in such a field to move particles.11,55,56 The DEP force depends on factors such as cell membrane and cytoplasm electrical properties as well as cell size.57 When the DEP force and drag force that act on the cell reached equilibrium, the cell could be kept stationary. Based on this concept, we used DEP electrodes for single-cell trapping in fluorescent measurement. The proper frequency and magnitude of the alternating voltage have been optimized to retain the cell but not damage it by high voltage.58 Therefore, 11.5 V (3 MHz) was applied between electrodes 1 and 2 to keep the cell stationary for experiments. The DEP force was turned off at ∼5 min after trapping the single tumor cell.
On-chip drug measurement on isolated single prostate cancer cell
After the cell was kept stationary, the medium was introduced into reservoir D to induce a liquid flow and to make sure the cell is stationary before running the drug accumulation experiment. An optical detection system was employed for simultaneous fluorescence measurement and bright-field imaging.48,59 The procedure for drug accumulation measurement has previously been reported.46–48
Briefly, the anti-cancer drug (i.e., DNR or OG-PTX) was introduced via reservoir D and accumulation of the anti-cancer drug was measured in the single cell. In the next step, the MDR inhibitor (i.e., CsA and/or FTC) was introduced via reservoir D, and drug accumulation was measured in the same cell. Adding MDR inhibitors increased drug accumulation in the cell, and then the single-cell fluorescence intensity was enhanced. DNR was first used for drug accumulation measurement as it has inherent fluorescence (λex = 470 nm; λem = 585 nm). Thereafter, paclitaxel that was fluorescently labelled by Oregon Green was examined (λex = 488 nm; λem = 524 nm), since paclitaxel was the commonly used anti-cancer drug for prostate cancer treatment.
Statistical analysis
Data are presented as the mean ± SD (standard deviation). Statistical significance test was determined using the Student's t-test.
RESULTS AND DISCUSSION
Prostate cancer cell isolation among blood cell
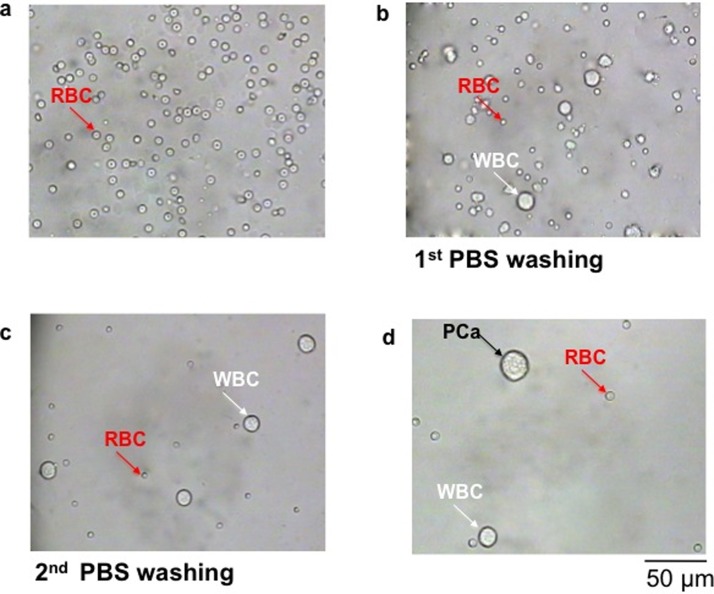
The morphology and size of prostate cancer cells and blood cells (WBCs + RBCs) were examined first. Fig. 4(a) shows the residual RBCs, and Fig. 4(b) and 4(c) illustrate the WBCs containing residual RBCs after one and two washes with PBS, respectively. Fig. 4(d) depicts the prostate cancer cell-WBCs mixture injected into reservoir A of the CTC chip. This is a model for the capture of CTCs in a condition of 1 CTC in 4000 WBCs, or 250 CTCs in 106 WBCs.20
FIG. 4.
Preparation of the mixture of tumor cells and blood cells. After isolating white blood cells through a Ficoll gradient, the cell pellet was resuspended in an appropriate medium solution. Images show the morphology and size of residual red blood cells only (a), of white and residual red blood cells after first PBS washing (b), of white blood cells after second PBS washing (c), and of prostate cancer (PCa) cell mixed with white blood cells (1:4000 ratio) (d). The first 3 images were taken on a slide, whereas the last image was taken in reservoir A of the CTC chip. Scale bar: 50 μm.
Fig. 5 shows the process for isolating the prostate cancer (PCa) cells among blood cells. Based on the small size of the blood cells, they were split into two directions and moved to the right and left side of the sideward openings in chamber 1 (Fig. 5(b-2,3)) and collected in waste reservoir B. The PCa cell moved in the middle of chamber 1 (Figs. 5(b-2,3) and 5(c)) without going near the sideward openings. Approximately 33 min after injecting the cell sample, the first PCa cell passed through the first channel, which connected reservoir A to chamber 1. An additional 4 min was taken to move the PCa cell from chamber 1 to chamber 2, followed by capturing the isolated single PCa cell by the DEP electrodes in the cell retention structure. This is a process for cell capturing without the use of an immunoaffinity label and for subsequent single-cell measurement, a process not currently feasible by using conventional methods.
FIG. 5.
Isolation of tumor cells from blood cells. (a) Images show different regions in the CTC chip (see also the schematic shown in Figure 1(b). (b1-9) Images to show isolation of the prostate cancer (PCa) cell among WBCs and RBCs, followed by capturing the cell inside the cell retention structure in chamber 2. (c) Close-up image from b2 to show the direction of the PCa cell movement. (d) Close-up image from b9 to show the capture of the PCa cell near electrode 1 for drug accumulation experiment. Scale bar: 50 μm.
We should point out that the concept of dielectric differences between cancer and normal cells has been used for separating cancer cells by DEP.60 In general, different cell types having various surface areas and size characteristics exhibit different DEP frequency responses. Mammalian cell dielectric properties may be described theoretically by using a shell model, in which the cell is represented as a homogenous core (i.e., cytoplasm) surrounded by a thin homogenous shell. This model describes the perfectly smooth idealized cell. However, in reality, cells that have surface morphological features on the lipid bilayer membrane increase the cell surface area as compared to the smooth idealized cell. These morphological features can be taken into account by introducing a fold factor (φ) to represent the ratio of actual membrane area to that of the idealized smooth shell. It has been reported that cancer cells have larger fold factors and larger radii (r) than both blood cells and normal cells of comparable origin.61,62 The DEP response of cancer and normal blood cells is expressed in terms of the reciprocal cell dielectric phenotype 1/(r·φ). As reported by Gascoyne and Shim,60 the parameter for prostate cancer cells and blood cells are 1.3–2.3 and 4.3–7.3, respectively. Therefore, the cell with a lower 1/(r·φ) value (e.g., tumor cell) will be attracted toward the electrode edge, while cells of higher 1/(r·φ) value (e.g., normal blood cell) will be repelled. This phenomenon suggests that the cell dielectric phenotype should be a widely applicable physical attribute of cancer cells. Interestingly, it has also been reported that even if the cancer cells undergo size reduction after maintenance in suspension, as compared to their original size when normally grown in contact with other cancer cells, the fold factor still remains high. Therefore, although the cancer cells exhibit a similar size to blood cells, it is still possible to use DEP to isolate the cancer cells.63 Based on this concept, even if any white blood cell with similar size as the PCa cell passed through chamber 2 in our CTC microchip, the blood cell could not be attracted toward the electrode edge and it would be washed away from the cell retention structure. In our hands, when only blood cells were injected into the CTC microchip, the white blood cells that entered chamber 2 were always repelled away from electrode 1.
The DEP process is selective for WBCs but not for RBCs because of their cell surface charges.64–66 Therefore, when the number of residual RBCs is high, a pre-screening of RBCs by a high flow is needed. Moreover, DEP-based separation has the best performance at a slow flow rate, otherwise the weaker DEP force will have a less chance of trapping targeted cancer cells due to a stronger flow-induced force. However, with a decrease in the flow rate, the throughput decreases, and so a higher flow is required for pre-screening whenever it is possible. In our microchip, a single cancer cell took ∼30 s to move along chamber 1, but this time was longer by a reduced liquid flow when the cell moved from the entrance of chamber 2 to the cell retention structure (∼60 s). Therefore, pre-screening of RBCs at a higher flow rate in chamber 1 is required, and then a slower flow rate is used to remove residual WBCs (∼5%) not trapped by DEP force in chamber 2.
Drug accumulation study on single 22Rv1 cell
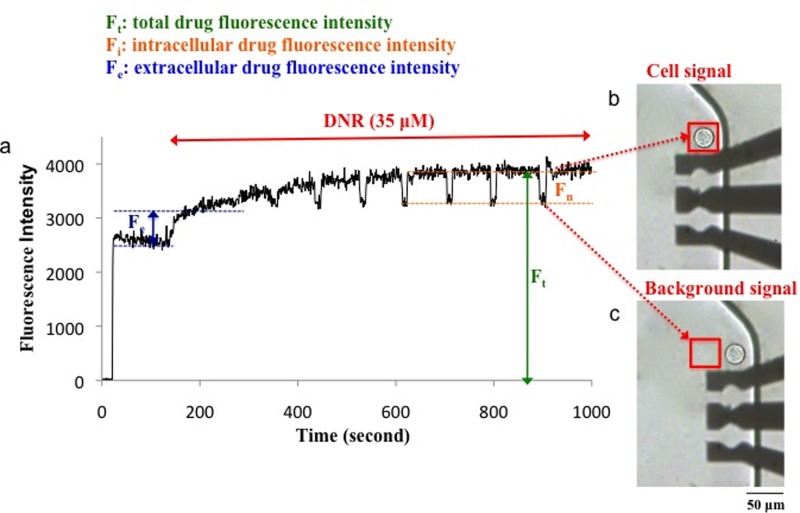
Drug accumulation measurement was conducted in chamber 2. In the first step, accumulation of the anticancer drug (i.e., DNR or OG-PTX) in the single 22Rv1 cell was measured in the absence of a MDR inhibitor. The 22Rv1 cells naturally express the ABCG2 transporter.51 As a well-known ABCG2 substrate,67 DNR was readily pumped out of the cell and the initial accumulation of DNR was low. As shown in Figs. 6(b) and 6(c), during data collection, the chip was shuttled left and right to allow for measurement of the cell and the background, respectively.
FIG. 6.
Drug accumulation measured in a single PCa cell. (a) The cellular fluorescence intensity due to accumulation of 35 μM DNR was measured in real time. The chip was shuttled to move the cell (b) into and (c) out of the detection window (red box) to measure the cellular signal and background, respectively. An AC electric field of 11.5 V at 3 MHz was applied to capture the cell close to the top DEP electrode before running the experiment. The electric field was turned off after 5 min as the cell was stationary.
The experiment was conducted using DNR at different concentrations (3.5, 7, 14, 35, 70, 350 μM) in order to determine the reasonable initial signal of drug accumulation in the cell. This signal should be around 10 times higher than the noise and not too high to allow for the room for fold-increase in the signal due to drug accumulation enhancement.
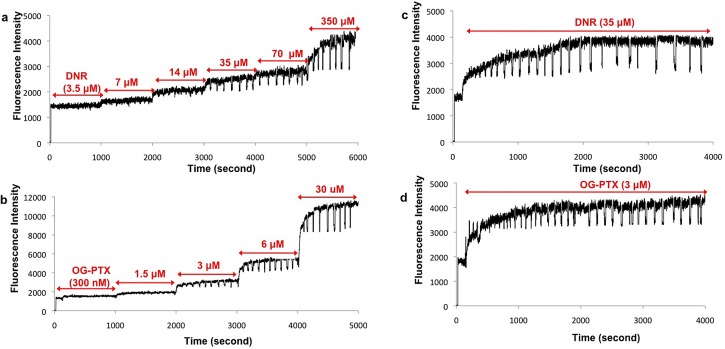
As shown in Fig. 7(a), the initial signal of DNR at the above 6 concentrations was 100 ± 37, 290 ± 39, 260 ± 41, 530 ± 42, 550 ± 47, and 1280 ± 50 counts per second (cps), respectively. The initial accumulation signal obtained from DNR at 35 μM provided the optimal signal. Similarly, experiments were conducted on a single 22Rv1 cell by treating it with OG-PTX at different concentrations (0.3, 1.5, 3, 6, 30 μM). As shown in Fig. 7(b), the initial signals of the OG-PTX were 60 ± 48, 100 ± 52, 500 ± 55, 1300 ± 56, 2900 ± 72 cps, respectively. Therefore, the optimal fluorescence signal was obtained after treating the single cell with 3 μM of OG-PTX. Subsequent experiments were carried out with 35 μM of DNR or 3 μM of OG-PTX. Fig. 7(c) shows a single 22Rv1 cell treated with only 35 μM of DNR for a long period of time, showing the cell has reached a saturated fluorescence level. A similar experiment was conducted on another single 22Rv1 cell by adding only 3 μM of OG-PTX, indicating saturation in the fluorescence intensity (Fig. 7(d)). We should point out that the alternating voltage used for DEP capture did not alter the cell membrane and reduce the MDR pump activity as no further increase in drug accumulation was observed after treating these single cells in multiple times by the anticancer drug alone (see Figs. 7(c) and 7(d)).
FIG. 7.
Optimization of DNR and OG-PTX concentrations for drug accumulation measurements. (a and b) The initial accumulation signal obtained from 35 μM of DNR and 3 μM of OG-PTX in 22Rv1 prostate cancer cells appeared to be reasonable. (c and d) Signal remained at the saturation level in the single 22Rv1 cells treated in multiple times with 35 μM of DNR alone (c) and 3 μM of OG-PTX only (d).
Effect of FTC on DNR accumulation
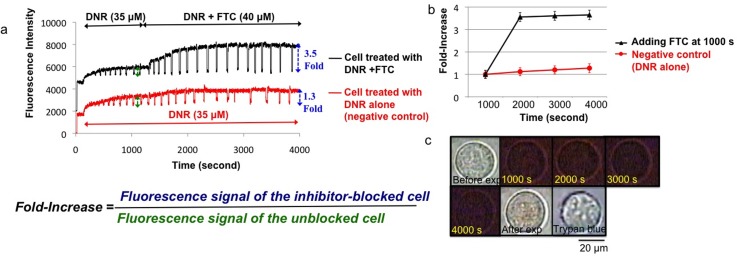
Since 22Rv1 cells highly express the ABCG2 transporter which leads to a low accumulation of DNR,51 adding FTC (as a specific ABCG2 inhibitor)68,69 should increase DNR accumulation, and single-cell fluorescence should be enhanced. As shown in Fig. 8(a), a steady state or plateau of the drug accumulation signal, due to a balance of the drug uptake and efflux processes, was first obtained. With the addition of FTC, the steady state was disturbed, and the DNR accumulation increased instantly. The effectiveness of the MDR inhibitor was indicated by the fold-increase in fluorescence, which was defined as the ratio between the fluorescence signals of the inhibitor-blocked cell and that of the unblocked cell. As shown in Fig. 8(a), adding FTC as a MDR inhibitor enhanced drug accumulation by 3.5 ± 0.2 fold (p < 0.0001), as compared with the 1.3 ± 0.2 fold (p > 0.1) obtained in the negative control. The fold-increases obtained at different time points before and after adding FTC were also plotted in Fig. 8(b). The value of 3.5 ± 0.2 is comparable, if not better than, the literature value of 2.0-fold obtained in the accumulation of D-luciferin (another well-known substrate of ABCG2) in 22Rv1 cells treated by FTC (25 μM), based on a time-consuming bioluminescence imaging experiment.70
FIG. 8.
Drug accumulation in a single 22Rv1cell in the presence of FTC as a MDR inhibitor. (a) Fluorescence intensity of the cell treated with 35 μM DNR measured in real time. The drug accumulation enhanced after adding 40 μM of FTC (black curve). The bottom curve in red shows the signal remained at a saturation level for the single cell treated with DNR only (negative control). (b) The fold-increase determined at different time points showing enhancement of drug accumulation after adding FTC (black line), as compared to no enhancement in the negative control (red line). (c) The images show the cell before, during, and after experiment, followed by trypan blue treatment. Scale bar: 20 μm.
Fig. 8(c) shows the images of this MDR cell before experiment (bright-field observation), during DNR treatment (simultaneous red-light bright-field observation during fluorescence measurement), after adding DNR in the presence of FTC, and after trypan blue treatment. Since the cell was not stained by trypan blue, it was viable and this occurred even after drug accumulation and DEP force were applied to capture the cell. Therefore, the cell was not killed even though 35 μM of DNR was used to treat the cell, and the xenon arc lamp was used to excite the drug molecules in the cell.
To illustrate how fast the reagents reach the cell trapped in the cell retention structure, Fig. S3 shows the flow of trypan blue at different time points, demonstrating how the dye quickly flows in chamber 2 to reach the cell (∼4 s) (see electronic supplementary material).83 This fast flow was consistent with the results of a recent simulation study about the liquid flow behavior in the cell retention structure.71
Our real-time fluorescence drug accumulation experiments also allow us to obtain the kinetics of the MDR inhibitor effects, using a previously reported mono-exponential drug uptake model.49 The curve fitting analysis was performed on the normalized drug accumulation data, using the SAS software (see Fig. S4 in the electronic supplementary material).83 Based on the curve fitting results (Tables S1 and S2),83 the fold-increases for DNR and OG-PTX accumulations were determined to be 3.4 ± 0.2 and 2.4 ± 0.4, which were believed to be caused by the action of FTC on the ABCG2-mediated drug efflux in the 22Rv1 cell.
Effective concentrations of MDR inhibitors (FTC and CsA)
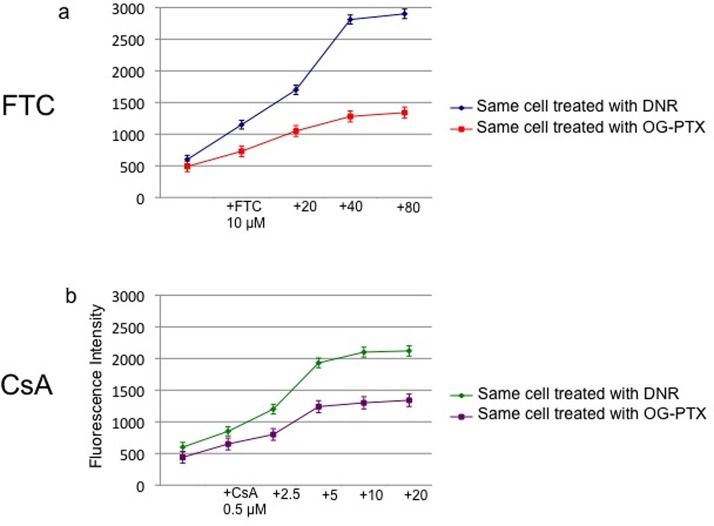
Although P-gp is weakly expressed in the normal prostate cells,72 its expression increases in the tumor epithelium,55 especially in androgen-independent prostate cancer.11 For instance, P-gp (ABCB1) was detected in 35% of cell samples collected from non-treated prostate cancer patients (Homma et al.56). However, the ABCG2 transporter has been found in androgen-independent prostate carcinoma cells such as 22Rv1 cells,51 and it has reported that this MDR transporter might mediate drug resistance in the prostate cancer stem cells resistant to androgen therapy.73 Our drug accumulation experiments on 22Rv1 cells were evaluated using both FTC (as a ABCG2 inhibitor) and CsA (as a ABCB1 inhibitor). As shown in Fig. 9(a) (blue line), the fluorescence signal of 35 μM DNR in a 22Rv1 cell was 630 ± 77 cps. After adding different concentrations of FTC (10, 20, 40, 80 μM), the signals were enhanced (1.9 ± 0.2, 2.8 ± 0.2, 4.7 ± 0.2, and 4.8 ± 0.2 fold-increase, respectively). Similar experiments were performed by adding different concentrations of OG-PTX (1.4 ± 0.2, 2.1 ± 0.2, 2.6 ± 0.2, and 2.7 ± 0.2 fold-increase, respectively) (Fig. 9(a): red line). Therefore, the optimal fluorescence signals were obtained after treating the single cells with both anticancer drugs in the presence of 40 μM of FTC. In a similar manner, experiments were performed on single cells to optimize the CsA concentration. As shown in Fig. 9(b) (green line), after adding DNR (35 μM) in the presence of different concentrations of CsA (0.5, 2.5, 5, 10, 20 μM), the signals were enhanced by 1.4 ± 0.2, 2.1 ± 0.2, 3.2 ± 0.2, 3.4 ± 0.2, 3.6 ± 0.2 fold, respectively. Similar CsA experiment was conducted by single-cell accumulation of 3 μM OG-PTX, and the fold-increases were 1.5 ± 0.2, 1.8 ± 0.2, 2.7 ± 0.2, 3.0 ± 0.2, 3.1 ± 0.2, respectively (Fig. 9(b): violet line). Therefore, the optimal fluorescence signals were found to be 5 μM of CsA after treating the single cells with both anticancer drug in the presence of this P-gp inhibitor.
FIG. 9.
Effective concentrations of MDR inhibitors for experiments on the 22Rv1 cells. (a) Use of FTC (10, 20, 40, 80 μM) to enhance DNR accumulation in the single-cell (blue line). Similar FTC experiments were conducted by the single-cell accumulation of OG-PTX (red line). The optimal fluorescence signal was obtained after treating the cell in the presence of 40 μM FTC. (b) Use of CsA (0.5, 2.5, 5, 10, 20 μM) to enhance DNR accumulation in the single cell (green line). Similar CsA experiments were conducted by the single-cell accumulation of OG-PTX (violet line). The optimal fluorescence signal was obtained after treating the cell in the presence of 5 μM CsA. 100 data points were obtained for each individual single cells.
We should point out that the half-saturation concentration for the effect of a MDR inhibitor on the steady-state accumulation level, termed as the half-maximal inhibitory concentration (IC50), can be determined by
| (1) |
where Ki is the intrinsic affinity of the inhibitor for P-gp at its inner leaflet on the cell membrane;74 FI represents fold-increase, which is the ratio of unblocked to blocked accumulation of drug. Litman et al.74 reported 0.50 μM as the mean Ki for the affinity of CsA for P-gp to DNR treatment, regardless of the cell lines. Based on this equation and the Ki value (0.5 μM), the half-maximal inhibitory concentration of pumping by P-gp (or IC50) of CsA was calculated based on the highest fold-increase of 3.57, resulting in a value of 1.8 μM. This IC50 was comparable with 1 μM, as reported for CsA on prostate cancer cell line 22Rv1.75 We should point out that IC50 depends on the number of pumps present in the plasma membrane; therefore, IC50 is greater in more highly multidrug resistant cells.
Effect of multiple MDR inhibitors on single prostate cancer cells
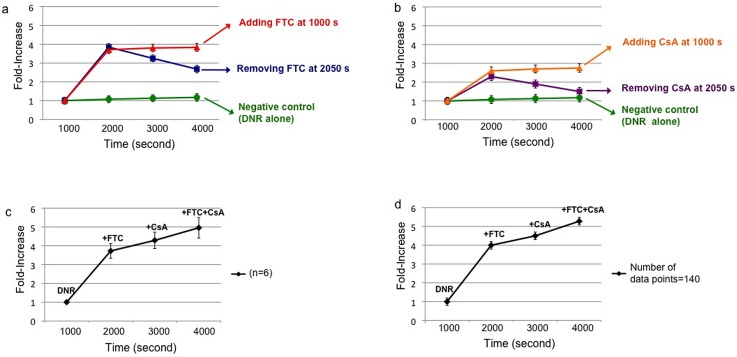
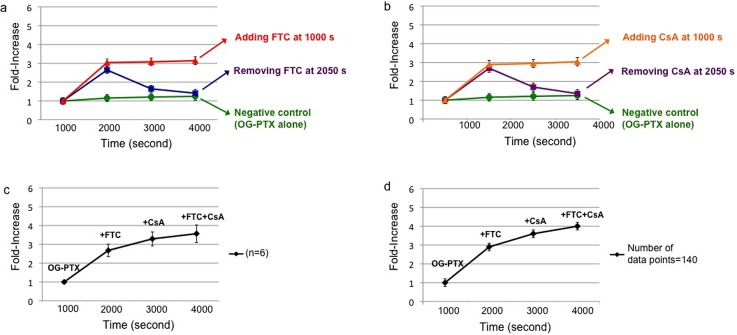
One of the requirements for the effectiveness of chemotherapy is the sufficient accumulation of chemotherapeutic drug in cancer cells.76–78 Since MDR is due to the function of ABC transporters that reduce intracellular drug accumulation in the cells, we examined whether FTC and CsA could alter BCRP and P-gp activity and improve the retention of DNR and OG-PTX in single prostate cancer cells. In single 22Rv1 cells treated with DNR in the presence of FTC (Fig. 10(a): red line), the fold-increase remained steady over 3000 s for a value of 3.8 ± 0.2. When CsA was used, an approximate 2.7 ± 0.2 fold-increase was observed. Similar steady values were obtained after applying OG-PTX in the presence of same MDR inhibitors (Fig. 11(a): red line for FTC and Fig. 11(b): orange line for CsA) (3.1 ± 0.2 and 3.0 ± 0.2 fold-increase, respectively). However, when the inhibitor was removed at 2050 s, there was drop in drug accumulation, showing a lower fold-increase. For instance, in the single cell treated with OG-PTX, the fluorescence signal decreased right after the MDR inhibitors were removed (fold-increase dropped from 2.7 ± 0.2 to 1.4 ± 0.2 for FTC and from 2.7 ± 0.2 to 1.3 ± 0.2 for CsA) (Fig. 11(a): blue line and Fig. 11(b): violet line). However, after accumulation of DNR in the presence of MDR inhibitors, DNR was more effectively retained in the cells right after removal of MDR inhibitors (Fig. 10(a): blue line and Fig. 10(b): violet line), particularly for FTC (fold-increase dropped from 3.8 ± 0.2 to 2.8 ± 0.2 for FTC and from 2.3 ± 0.2 to 1.5 ± 0.2 for CsA).
FIG. 10.
Enhancement of DNR accumulation in single 22Rv1 cells due to ABCG2 and ABCB1 inhibitors. (a) Use of FTC (40 μM) to enhance accumulation of DNR (35 μM) in a single-cell (red line). Blue line shows signal decrease after removing FTC at 2050 s. No significant enhancement was observed (p > 0.1) in single cells (green line) treated for 4000 s with 35 μM of DNR alone (negative control). (b) Use of CsA (5 μM) to enhance cellular drug accumulation (orange line). Violet line indicates signal decrease after removing CsA at 2050 s. (c) Averaged results (n = 6) of fold-increase in DNR accumulation were observed after adding FTC, followed by CsA and combination of FTC + CsA. (d) The fold-increases after treating a single cell with DNR in the presence of FTC, CsA, and FTC + CsA (number of data points = 140).
FIG. 11.
Enhancement of OG-PTX accumulation in single 22Rv1 cells due to ABCG2 and ABCB1 inhibitors. (a) Use of FTC (40 μM) to enhance accumulation of OG-PTX (3 μM) in a single cell (red line). Blue line indicates signal decrease after removing FTC at 2050 s. No significant enhancement was observed (p > 0.1) in single cells (green line) treated for 4000 s with 3 μM of OG-PTX alone (negative control). (b) Use of CsA (5 μM) to enhance cellular drug accumulation (orange line). Violet line shows signal decrease after removing CsA at 2050 s. (c) Averaged results (n = 6) of fold-increase in OG-PTX accumulation were observed after adding FTC, followed by CsA and combination of FTC + CsA. (d) The fold-increases after treating a single cell with OG-PTX in the presence of FTC, CsA, and FTC + CsA (number of data points = 140).
The more effective retention of DNR can be explained by its intercalation between the bases of DNA and impairments of its synthesis in the cell nucleus.79 This means DNR may get aggregated in the nucleus to form clusters that are too big to pass out through the nuclear pores in a short time (∼1 h).76–78 Unlike DNR, OG-PTX's site of action is in the cytoplasm, by stabilizing tubulin polymerization in it, ultimately interfering the cell division.80 Our results corroborate the notion that once DNR enters the cell, it may not be as easy as OG-PTX to be extruded from the cells once the MDR function is restored by removing the inhibitors.
More experiments were performed on DNR accumulation in single MDR cells (n = 6) using different inhibitors. Applying FTC, CsA and FTC + CsA produced fold-increases of 3.7 ± 0.4, 4.2 ± 0.6, 4.9 ± 0.7, respectively (Fig. 10(c)). Additional experiments on OG-PTX accumulation in six individual single cells with the above three MDR inhibitor treatments indicated similar results (2.7 ± 0.4, 3.4 ± 0.5, 3.7 ± 0.6, respectively) (Fig. 11(c)). It should be pointed out that ABCB1 appears to be the main transporter for PTX, while DNR is the substrate of both ABCB1 and ABCG2 transporters.81 Although there was an immediate decrease in drug accumulation after removing the MDR inhibitors, this signal decrease due to inhibitor loss could be compensated by adding CsA. We also found the treatment of FTC + CsA after treating with CsA enhanced further drug accumulation in the single cells. Although the inhibition mechanism of the combination of MDR inhibitors is not entirely clear, the simultaneous administration of MDR inhibitors has been reported. For instance, verapamil and CsA (as P-gp inhibitors) and MK571 (as a MRP1 inhibitor) have been reported to increase the intracellular levels of [3H]-labeled drug in leukemia cells only after co-administered simultaneously.82
Although further enhancement in drug accumulation might occur, the averaged data did not show a significant difference. It was interesting to find that the further enhancement of drug accumulation due to treatment of FTC + CsA was observed in the single-cell experiments, but not in the averaged results when the number of experiments was insufficient. For instance, the averaged results of several single-cell experiments did not result in the enhancement in a statistically significant manner (p > 0.05). However, the p-values were less than 0.0001 for the enhancement of DNR accumulation after treating the same single 22Rv1 cell with FTC + CsA, right after CsA treatment (Fig. 10(d)). In a similar manner, significant enhancement (p < 0.0001) of OG-PTX accumulation was observed when real-time measurement was conducted (number of data points = 140) on the same single cell after CsA treatment (Fig. 11(d)). We believe that more repeated experiments are demanded to obtain a significant enhancement in the averaged results. However, the same single cell experiment has the power to reveal the change in a significant manner when conducted real-time on the same single cell.
Comparison of drug accumulation in captured single prostate cancer cells and in normal white blood cells
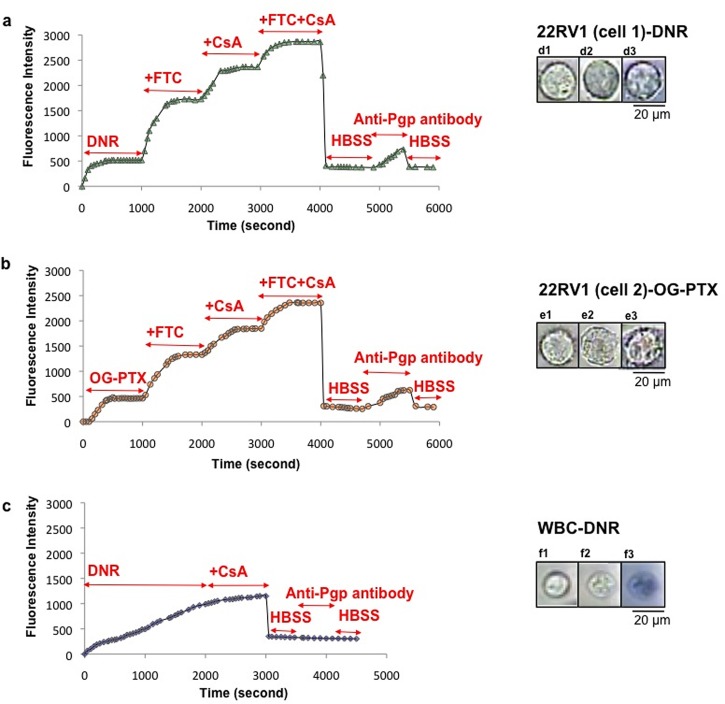
Multiple rounds of drug accumulation experiments were conducted on the single PCa cell isolated from blood cells. To ensure that the captured single cell was indeed cancerous, anti-human monoclonal P-gp antibody (anti-CD243) was introduced to detect P-gp on the 22Rv1 cell surface subsequent to drug accumulation experiments. Fig. 12(a) shows obvious enhancement in fluorescence intensity (at 585 nm) due to DNR accumulation in a 22Rv1 cell after undergoing various MDR inhibitors treatment (FTC, CsA, and CsA + FTC) (i.e., 3.3 ± 0.2, 4.5 ± 0.2, 5.4 ± 0.2 fold-increase, respectively (p < 0.0001). After washing the cell with HBSS (2×), anti-P-gp was applied and the fluorescence signal (524 nm) was found to increase. This result confirmed the cell was 22Rv1, but not a blood cell, since the cell was stained by the anti-P-gp antibody.
FIG. 12.
Anti-P-gp antibody binds to the 22Rv1 cells but not to white blood cell (WBC). (a) MDR inhibitors enhanced DNR accumulation on the captured single PCa cell. The fluorescence signal increased after adding P-gp antibody. (b) Similar results observed after treating another single PCa cell with OG-PTX in the presence of multiple MRR inhibitors (40 μM FTC, 5 μM CsA, and 40 μM FTC + 5 μM CsA), followed by adding anti-P-gp antibody. (c) Drug accumulation continued to rise in the WBC after treatment with DNR alone. No drug enhancement was observed after adding DNR in the presence of CsA. The WBC was not stained by anti-P-gp antibody (anti-CD243). (d, e, f) The cell images were depicted before and after experiment, followed by adding trypan blue in PCa cells treated with DNR (d), OG-PTX (e), and WBC (f). DNR concentration was 35 μM. Scale bar: 20 μm.
However, in a similar experiment conducted on a WBC, the cell was not stained by the anti-P-gp antibody, indicating it was not a 22Rv1 cell (Fig. 12(c)). As for the single-cell DNR accumulation measurement, the WBC demonstrated the behaviour of a non-cancerous cell, i.e., the drug accumulation reached only at a much longer time and there was no enhancement by adding CsA. The accumulation of DNR in the WBC also led to its staining by trypan blue (Fig. 12(f-3)). We should point out that this cell was examined when only blood cells but no cancer cells were used; therefore, the captured cell must be a WBC.
Fig. 12(b) depicts the accumulation of OG-PTX in a single 22Rv1 cell, showing an obvious enhancement in fluorescence intensity (at 524 nm) due to various MDR inhibitor treatments (FTC, CsA and FTC + CsA) (i.e., 2.8 ± 0.2, 4.4 ± 0.2, 4.9 ± 0.2 fold-increase, respectively (p < 0.0001). After washing the cell, there was a drop in fluorescence intensity, indicating the removal of OG-PTX from the cell. Thereafter, anti-P-gp antibody was applied, and the fluorescence signal (524 nm) was enhanced, confirming that the cell was 22Rv1.
CONCLUSIONS
In this study, we demonstrated the applicability of an integrated microfluidic chip for the label-free isolation of prostate cancer (PCa) cells. The microfilter in chamber 1 and DEP electrodes in chamber 2 have the capability to achieve the isolation of single cancer cells from blood cells, as a model of CTC capture, as well as to preserve cell viability for subsequent drug accumulation measurement. Multiple experiments can then be conducted on the viable single cell to investigate the effect of MDR inhibitors on anticancer drug accumulation. FTC (as a well-known ABCG2 inhibitor) and CsA (as a P-gp inhibitor) have been found to be effective in the enhancement of drug accumulation in the captured single PCa cells. The advantages of this integrated chip are the ability of fast isolation of PCa (< 1 h), of measuring drug accumulation (∼1 h) and of confirming the identity of the P-gp expressing cancerous cell. This new biochip requires a small number of cells to confirm the response of the cells to MDR inhibitors as compared to conventional methods, providing a potential for CTC research and for investigating multidrug resistance in CTCs.
ACKNOWLEDGMENTS
Financial support from Natural Science and Engineering Research Council (NSERC), the Australian Government Department of Health and the Australian National Health and Medical Research Council (APP:1031221 and APP:1031228) is acknowledged. The authors are thankful to Mary Dearden and Kim Buettner from Animal Care Services at Simon Fraser University for providing mouse blood samples. The authors also thank Chao Du for his assistance in curve fitting analysis.
Glossary
NOMENCLATURE
- ABCB1
ATP-binding cassette sub-family B member 1
- ABCG2
ATP-binding cassette sub-family G member 2
- BCRP
Breast cancer resistant protein
- CTC
Circulating tumor cell
- CsA
Cyclosporine A
- DEP
Dielectrophoretic
- DMSO
Dimethyl sulfoxide
- DNR
Daunorubicin
- EMT
epithelial-to-mesenchymal transition
- EpCAM
Epithelial cell adhesion molecule
- FBS
Fetal bovine serum
- FTC
Fumitremorgin C
- HBSS
Hanks' balanced salt solution
- MDR
Multidrug resistance
- OG-PTX
Oregon Green-labeled paclitaxel
- PBS
Phosphate buffered saline
- PEN/STR
Penicillin/streptomycin
- PCa
Prostate cancer
- P-gp
Permeability glycoprotein
- RPMI
Roswell Park Memorial Institute
References
- 1. Ashworth T. R. A., “ A case of cancer in which cells similar to those in the tumors was seen in the blood after death,” Med. J. Aust. 14, 146–149 (1869). [Google Scholar]
- 2. Caixeiro N. J., Kienzle N., Lim S. H., Spring K. J., Tognela A., Scott K. F., de Souza P., and Becker T. M., “ Circulating tumor cells-A bona fide cause of metastatic cancer,” Cancer Metastasis Rev. 33(2–3), 747–757 (2014). 10.1007/s10555-014-9502-8 [DOI] [PubMed] [Google Scholar]
- 3. Mi-Young K. M. Y., Oskarsson T., Acharyya S., Nguyen V. X., Zhang X. E., Norton L., and Massague J., “ Tumor self-seeding by circulating cancer cells,” Cell. 139, 1315–1326 (2009). 10.1016/j.cell.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maheswaran S. and Haber D., “ Circulating tumor cells: A window into cancer biology and metastasis,” Curr. Opin. Genet. Dev. 20(1), 96–99 (2010). 10.1016/j.gde.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alix-Panabières C., Schwarzenbach H., and Pantel K., “ Circulating tumor cells and circulating tumor DNA,” Annu. Rev. Med. 63, 199–215 (2012). 10.1146/annurev-med-062310-094219 [DOI] [PubMed] [Google Scholar]
- 6. Gazzaniga P., Naso G., Gradilone A., Cortesi E., Gandini O., Gianni W., Fabbri M., Agnese V., Vincenzi B., di Silverio F., and Frati L., “ Chemosensitivity profile assay of circulating cancer cells: Prognostic and predictive value in epithelial tumors,” J. Cancer 126(10), 2437–2447 (2010). 10.1002/ijc.24953 [DOI] [PubMed] [Google Scholar]
- 7. Hughes A. D., Marshall J. R., Keller E., Powderly J. D., Greene B. T., and King M. R., “ Differential drug responses of circulating tumor cells within patient blood,” Cancer Lett. 352, 28–35 (2014). 10.1016/j.canlet.2013.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gradilone A., Naso G., Raimondi C., Cortesi E., Gandini O., Vincenzi B., Saltarelli R., Chiapparino E., Spremberg F., Cristofanilli M., Frati L., Agliano A. M., and Gazzaniga P., “ Circulating tumor cells (CTCs) in metastatic breast cancer (MBC): Prognosis, drug resistance and phenotypic characterization,” Ann Oncol. 22(1), 86–92 (2011). 10.1093/annonc/mdq323 [DOI] [PubMed] [Google Scholar]
- 9. Sissung T. M., Baum C. E., Deeken J., Price D. K., Aragon-Ching J., Steinberg S. M., Dahut W., Sparreboom A., and Figg W. D., “ ABCB1 genetic variation influences the toxicity and clinical outcome of patients with androgen-independent prostate cancer treated with docetaxel,” Clin. Cancer Res. 14(14), 4543–4549 (2008). 10.1158/1078-0432.CCR-07-4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Seruga B., Ocana A., and Tannock I. F., “ Drug resistance in metastatic castration-resistant prostate cancer,” Nat. Rev. Clin. Oncol. 8, 12–23 (2011). 10.1038/nrclinonc.2010.136 [DOI] [PubMed] [Google Scholar]
- 11. Zhang H., Wang J., Cai K., Jiang L., Zhou D., Yang C., Chen J., Chen D., and Dou J., “ Down regulation of gene MDR1 by shRNA to reverse multidrug-resistance of ovarian cancer A2780 cells,” J. Cancer Res. Ther. 8(2), 226–231 (2012). 10.4103/0973-1482.98975 [DOI] [PubMed] [Google Scholar]
- 12. Sharom F. J., “ ABC multidrug transporters: structure, function, and role in chemoresistance,” Pharmacogenomics 9(1), 105–127 (2008). 10.2217/14622416.9.1.105 [DOI] [PubMed] [Google Scholar]
- 13. Marquez B. and Van Bambek F., “ ABC multidrug transporters: target for modulation of drug pharmacokinetics and drug–drug interactions,” Drug Targets 12, 600–620 (2011). 10.2174/138945011795378504 [DOI] [PubMed] [Google Scholar]
- 14. Massay P. R., Fojo T., and Bates S. E., “ ABC transporters: involvement in multidrug resistance and drug disposition,” Handbook of Anticancer Pharmacokinetics and Pharmacodynamics Cancer Drug Discovery and Development ( Springer, 2014), pp. 373–400. [Google Scholar]
- 15. Wu C. P., Hsieh C. H., and Wu Y. S., “ The emergence of drug transporter-mediated multidrug resistance to cancer chemotherapy,” Pharmaceutics 8(6), 1996–2011 (2011). 10.1021/mp200261n [DOI] [PubMed] [Google Scholar]
- 16. Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Haber D. A., and Toner M., “ Isolation of rare circulating tumor cells in cancer patients by microchip technology,” Nature 450, 1235–1239 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu M., Stott S., Toner M., Maheswaran S., and Haber D. A., “ Circulating tumor cells: Approaches to isolation and characterization,” J. Cell Biol. 192, 373–382 (2011). 10.1083/jcb.201010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller M. C., Doyle G. V., and Terstappen L. W. M., “ Significance of circulating tumor cells detected by CellSearch system in patients with metastatic breast colorectal and prostate cancer,” J. Oncol. 26, 17421 (2010). 10.1155/2010/617421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Allard J. W., Matera Jeri J., Miller C. M., Repollet M., Connelly M. C., Rao C., Tibbe A. G. J., Uhr J. W., and Terstappen L. W., “ Tumor cells circulates in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant disease,” Clin. Cancer Res. 10, 6897–6904 (2004). 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- 20. Hou H. M., Ebrahimi M., Khoo B. L., Li Z. R., Soo R. A., Daniel S. T., Lim W. T., Han J., Bhagat A. A., and Lim C. T., “ Isolation and retrieval of circulating tumor cells using centrifugal forces,” Sci. Rep. 3, 1259–1267 (2013). 10.1038/srep01259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Matsusaka S., Suenaga M., Mishima Y., Kuniyoshi R., Takagi K., Terui Y., Mizunuma N., and Hatake K., Cancer Sci. 102, 1188–1192 (2011). 10.1111/j.1349-7006.2011.01926.x [DOI] [PubMed] [Google Scholar]
- 22. Hayes D. F., Cristofanilli M., Budd T. G., Ellis M. J., Stopeck A., Miller C. M., Matera J., Allard J. W., Doyle G. V., and Terstappen L. W. M., “ Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival,” Clin. Cancer Res. 12, 4218–4224 (2006). 10.1158/1078-0432.CCR-05-2821 [DOI] [PubMed] [Google Scholar]
- 23. Danila D. C., Heller G., Gignac G. A., Gonzalez-Espinoza R., Anand A., Tanaka E., Lilja H., Schwartz L., Larson S., Fleisher M., and Scher H. I., “ Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer,” Clin. Cancer Res. 13, 7053–7058 (2007). 10.1158/1078-0432.CCR-07-1506 [DOI] [PubMed] [Google Scholar]
- 24. Cohen S. J., Punt C. J. A., Iannotti N., Saidman B. H., Sabbath K. D., Gabrail N. Y., Picus J., Morse M., Mitchell E., Miller C. M., Doyle G. V., Tissing H., Terstappen L. W. M., and Meropol N. J., “ Relationship of circulating tumor cells to tumor response, progression- free survival, and overall survival in patients with metastatic colorectal cancer,” J. Clin. Oncol. 26, 3213–3221 (2008). 10.1200/JCO.2007.15.8923 [DOI] [PubMed] [Google Scholar]
- 25. Shaffer D. R., Leversha M. A., Danila D. C., Lin O., Gonzalez-Espinoza R., Gu B., Anand A., Smith K., Maslak P., Doyle G. V., Terstappen L. W. M., Lilja H., Heller G., Fleisher M., and Scher H. I., “ Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer,” Clin. Cancer Res. 13, 2023–2029 (2007). 10.1158/1078-0432.CCR-06-2701 [DOI] [PubMed] [Google Scholar]
- 26. Willipinski-Stapelfeldt B., Riethdorf S., Assmann V., Woelfle U., Rau T., Sauter G., Heukeshoven J., and Pantel K., “ Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer,” Clin. Cancer Res. 11, 8006–8014 (2005). 10.1158/1078-0432.CCR-05-0632 [DOI] [PubMed] [Google Scholar]
- 27. Cruz I., Ciudad J., Cruz J. J., Ramos M., Gómez-Alonso A., Adansa J. C., Rodríguez C., and Orfao A., “ Evaluation of multiparameter flow cytometry for the detection of breast cancer tumor cells in blood samples,” Am. J. Clin. Pathol. 123(1), 66–74 (2005). 10.1309/WP3QWKVJFYDHHXQD [DOI] [PubMed] [Google Scholar]
- 28. Zabaglo L., Ormerod M. G., Parton M., Ring A., Smith I. E., and Dowsett M., “ Cell filtration laser scanning cytometry for the characterisation of circulating breast cancer cells,” Cytometry, Part A 55A(2), 102–108 (2003). 10.1002/cyto.a.10071 [DOI] [PubMed] [Google Scholar]
- 29. Esmaeilsabzali H., Beischlag T. V., Cox M. E., Parameswaran A. M., and Park E. J., “ Detection and isolation of circulating tumor cells: Principles and methods,” Biotechnol. Adv. 31(7), 1063–1084 (2013). 10.1016/j.biotechadv.2013.08.016 [DOI] [PubMed] [Google Scholar]
- 30. Chen J., Li J., and Sun Y., “ Microfluidic approaches for cancer cell detection, characterization, and separation,” Lab Chip 12(10), 1753–1767 (2012). 10.1039/c2lc21273k [DOI] [PubMed] [Google Scholar]
- 31. Pratt E. D., Huang C., Hawkins B. G., Gleghorn J. P., and Kirby B. J., “ Rare cell capture in microfluidic devices,” Chem. Eng. Sci. 66, 1508–1522 (2011). 10.1016/j.ces.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Didar T. F. and Tabrizian M., “ Adhesion based detection, sorting and enrichment of cells in microfluidic Lab-on-Chip devices,” Lab Chip 10(22), 3043–3053 (2010). 10.1039/c0lc00130a [DOI] [PubMed] [Google Scholar]
- 33. Kamande J. W., Hupert M. L., Witek M. A., Wang H., Torphy R. J., Dharmasiri U., Njoroge S. K., Jackson J. M., Aufforth R. D., Snavely A., Yeh J. J., and Soper S. A., “ Modular microsystem for the isolation, enumeration, and phenotyping of circulating tumor cells in patients with pancreatic cancer,” Anal Chem. 85(19), 9092–9100 (2013). 10.1021/ac401720k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stott S. L., Hsu C. H., Tsukrov D. I., Yu M., Miyamoto D. T., Waltman B. A., Rothenberg S. M., Shah A. M., Smas M. E., Korir G. K., Floyd F. P., Gilman A. J., Lord J. B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L. V., Lee R. J., Isselbacher K. J., Maheswaran S., Haber D. A., and Toner M., “ Isolation of circulating tumor cells using a microvortex-generating herringbone-chip,” Proc. Natl. Acad. Sci. 107(43), 18392–18397 (2010). 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Earhart C. E., Hughes R. S., Gaster C. C., Ooi R. J., Wilson L. Y., Zhou E. W., Xu L., Wong D. J., and Willingham S. B., “ Isolation and mutational analysis of circulating tumor cells from lung cancer patients with magnetic sifters and biochips,” Lab Chip 14, 78–88 (2014). 10.1039/C3LC50580D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoshino K., Huang Y. Y., Lane N., Huebschman M., Uhr J. W., Frenkel E. P., and Zhang X., “ Microchip-based immunomagnetic detection of circulating tumor cells,” Lab Chip 11(20), 3449–3457 (2011). 10.1039/c1lc20270g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xia N., Hunt T. P., Mayers B. T., Alsberg E., Whitesides G. M., Westervelt R. M., and Ingber D. E., “ Combined microfluidic-micromagnetic separation of living cells in continuous flow,” Microdevices 8(4), 299–308 (2006). 10.1007/s10544-006-0033-0 [DOI] [PubMed] [Google Scholar]
- 38. Polyak K. and Weinberg R. A., “ Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits,” Rev. Cancer 9, 265–273 (2009). 10.1038/nrc2620 [DOI] [PubMed] [Google Scholar]
- 39. Huang S. B., Wu M. H., Lin Y. H., Hsieh C. H., Yang C. L., Lin H. C., Tseng C. P., and Lee G. B., “ High-purity and label-free isolation of circulating tumor cells (CTCs) in a microfluidic platform by using optically induced-dielectrophoretic (ODEP) force,” Lab Chip 13(7), 1371–1383 (2013). 10.1039/c3lc41256c [DOI] [PubMed] [Google Scholar]
- 40. Moon H. S., Kwon K., Kim S. I., Han H., Sohn J., Lee S., and Jung H. I., “ Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP),” Lab Chip 11(6), 1118–1125 (2011). 10.1039/c0lc00345j [DOI] [PubMed] [Google Scholar]
- 41. Vahey M. D. and Voldman J., “ High-throughput cell and particle characterization using isodielectric separation,” Anal Chem. 81(7), 2446–2455 (2009). 10.1021/ac8019575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. An J., Lee J., Lee S. H., Park J., and Kim B., “ Separation of malignant human breast cancer epithelial cells from healthy epithelial cells using an advanced dielectrophoresis-activated cell sorter (DACS),” Anal. Bioanal. Chem. 394(3), 801–809 (2009). 10.1007/s00216-009-2743-7 [DOI] [PubMed] [Google Scholar]
- 43. McFaul S. M., Lin B. K., and Ma H., “ Cell separation based on size and deformability using microfluidic funnel ratchets,” Lab Chip 12(13), 2369–2376 (2012). 10.1039/c2lc21045b [DOI] [PubMed] [Google Scholar]
- 44. Hyuna K. A., Kwona K., Hanb H., Kimb S., and Jung H., “ Microfluidic flow fractionation device for label-free isolation of circulating tumor cells (CTCs) from breast cancer patients,” Biosens. Bioelectron. 40(1), 206–212 (2013). 10.1016/j.bios.2012.07.021 [DOI] [PubMed] [Google Scholar]
- 45. Harouaka R. A., Nisic M., and Zheng S. Y., “ Circulating tumor cell enrichment based on physical properties,” J. Lab. Automat. 18(6), 455–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khamenehfar A., Wan C. P. L., Li P. C. H., Letchford K., and Burt H. M., “ Same-single-cell analysis using the microfluidic biochip to reveal drug accumulation enhancement by an amphiphilic diblock copolymer drug formulation,” Anal. Bioanal. Chem. 406, 7071–7083 (2014). 10.1007/s00216-014-8151-7 [DOI] [PubMed] [Google Scholar]
- 47. Li X., Chen Y., and Li P. C. H., “ A simple and fast microfluidic approach of same-single-cell analysis (SASCA) for the study of multidrug resistance modulation in cancer cells,” Lab Chip 11(7), 1378–1384 (2011). 10.1039/c0lc00626b [DOI] [PubMed] [Google Scholar]
- 48. Li X., Ling V., and Li P. C. H., “ Same-single-cell analysis for the study of drug efflux modulation of multidrug resistant cells using a microfluidic chip,” Anal. Chem. 80(11), 4095–4102 (2008). 10.1021/ac800231k [DOI] [PubMed] [Google Scholar]
- 49. Li X., Huang J., Tibbits G. F., and Li P. C. H., “ Real-time monitoring of intracellular calcium dynamic mobilization of a single cardiomyocyte in a microfluidic chip pertaining to drug discovery,” Electrophoresis 28(24), 4723–4733 (2007). 10.1002/elps.200700312 [DOI] [PubMed] [Google Scholar]
- 50. Ji J., Samper V., Chen Y., Heng C., Lim T., and Yobas L., “ Silicon-based microfilters for whole blood cell separation,” Biomed Microdevices. 10, 251–257 (2008). 10.1007/s10544-007-9131-x [DOI] [PubMed] [Google Scholar]
- 51. Zhang Y., Bressler J. P., Neal J., Lal B., Bhang H. C., Laterra J., and Pomper M. G., “ ABCG2/BCRP expression modulates D-Luciferin-based bioluminescence imaging,” Cancer Res. 67(19), 9389–9397 (2007). 10.1158/0008-5472.CAN-07-0944 [DOI] [PubMed] [Google Scholar]
- 52. Liu T., Xu F., Du X., Lai D., Zhao Y., Huang Q., Jiang L., Huang W., Cheng W., and Liu Z., “ Establishment and characterization of multi-drug resistant, prostate carcinoma-initiating stem-like cells from human prostate cancer cell lines 22RV1,” Mol. Cell. Biochem. 340, 265–273 (2010). 10.1007/s11010-010-0426-5 [DOI] [PubMed] [Google Scholar]
- 53. Lenshof A. and Laurell T., “ Continuous separation of cells and particles in microfluidic systems,” Chem. Soc. Rev. 39(3), 1203–1217 (2010). 10.1039/b915999c [DOI] [PubMed] [Google Scholar]
- 54. Pohl H. A., “ The motion and precipitation of suspensoids in divergent electric fields,” J. Appl. Phys. 22, 869 (1951). 10.1063/1.1700065 [DOI] [Google Scholar]
- 55. Bhangal G., Halford S., Wang J., Roylance R., Shah R., and Waxman J., “ Expression of the multidrug resistance gene in human prostate cancer,” Urol Oncol. 5(3), 118–121 (2000). 10.1016/S1078-1439(99)00055-1 [DOI] [PubMed] [Google Scholar]
- 56. Homma S., Komohara Y., Harada M., Saya H., Todo S., Itoh K., and Noguchi M., “ Differential levels of human leukocyte antigen-class I, multidrug-resistance 1 and androgen receptor expressions in untreated prostate cancer cells: The robustness of prostate cancer,” Oncol. Rep. 18(2), 343–346 (2007). 10.3892/or.18.2.343 [DOI] [PubMed] [Google Scholar]
- 57. Voldman J., “ Electrical forces for microscale cell manipulation,” Annu. Rev. Biomed. Eng. 8, 425–454 (2006). 10.1146/annurev.bioeng.8.061505.095739 [DOI] [PubMed] [Google Scholar]
- 58. Khamenehfar A., Chen Y., Hogge D. E., and Li P. C. H., “ AML patient cells using a microfluidic dielectrophoresis (DEP) chip,” MicroTAS 1, 380–382 (2013). [Google Scholar]
- 59. Peng X. Y. and Li P. C. H., “ A three-dimensional flow control concept for single-cell experiments on a microchip,” Anal. Chem. 76(18), 5273–5281 (2004). 10.1021/ac049384s [DOI] [PubMed] [Google Scholar]
- 60. Gascoyne P. R. C. and Shim A., “ Isolation of circulating tumor cells by dielectrophoresis,” Cancers 6, 545–579 (2014). 10.3390/cancers6010545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mulhall H. J., Labeed F. H., Kazmi B., Costea D. E., Hughes M. P., and Lewis M. P., “ Cancer, pre-cancer and normal oral cells distinguished by dielectrophoresis,” Anal. Bioanal. Chem. 401, 2455–2463 (2011). 10.1007/s00216-011-5337-0 [DOI] [PubMed] [Google Scholar]
- 62. Broche L. M., Bhadal N., Lewis M. P., Porter S., Hughes M. P., and Labeed F. H., “ Early detection of oral cancer: Is dielectrophoresis the answer?” Oral Oncol. 43, 199–203 (2007). 10.1016/j.oraloncology.2006.02.012 [DOI] [PubMed] [Google Scholar]
- 63. Shim S., Gascoyne P., Noshari J., and Hale K. S., “ Dynamic physical properties of dissociated tumor cells revealed by dielectrophoretic field-flow fractionation,” Integr. Biol. 3, 850–862 (2011). 10.1039/c1ib00032b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Braschler T., Demierre N., Nascimento E., Silva T., Oliva A. G., and Renaud P., “ Continuous separation of cells by balanced dielectrophoretic forces at multiple frequencies,” Lab Chip 8(2), 280–286 (2008). 10.1039/B710303D [DOI] [PubMed] [Google Scholar]
- 65. Wang M. W., “ A microfluidic device for separating erythrocytes polluted by lead (II) from a continuous bloodstream flow,” Electrophoresis 33(5), 780–787 (2012). 10.1002/elps.201100354 [DOI] [PubMed] [Google Scholar]
- 66. Jubery T. Z. and Dutta P., “ A new design for efficient dielectrophoretic separation of cells in a microdevice,” Electrophoresis 34(5), 643–650 (2013). 10.1002/elps.201200560 [DOI] [PubMed] [Google Scholar]
- 67. Liu J., Shi J., Jiang L., Yamamoto F. S., Takano M., Chang M., Zhang H., and Chen Y., “ Segmented magnetic nanofibers for single-cell manipulation,” Appl. Surf. Sci. 258(19), 7530–7535 (2012). 10.1016/j.apsusc.2012.04.077 [DOI] [Google Scholar]
- 68. Rabindran S., He H., Singh M., Brown E., Collins K., Annable T., and Green-berger L., “ Reversal of a novel multidrug resistance mechanism in human colon carcinoma cells by fumitremorgin C,” Cancer Res. 58, 5850–5858 (1998). [PubMed] [Google Scholar]
- 69. Rabindran S., Ross D., Doyle L., Yang W., and Greenberger L., “ Fumitremorgin C reverses multidrug resistance in cells transfected with the breast cancer resistance protein,” Cancer Res. 60, 47–50 (2000). [PubMed] [Google Scholar]
- 70. Zhang Y., Pullambhatla M., Laterra J., and Pomper M. G., “ Influence of bioluminescence imaging dynamics by D-Luciferin uptake and efflux mechanisms,” Mol. Imaging 11(6), 499–506 (2012). 10.2310/7290.2012.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shen F., Li X. J., and Li P. C. H., “ Study of flow behaviors on single-cell manipulation and shear stress reduction in microfluidic chips using CFD simulations,” Biomicrofluidics 8, 1–12 (2014). 10.1063/1.4866358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kawai K., Sakurai M., Sakai T., Misaki M., Kusano I., Shiraishi T., and Yatani R., “ Demonstration of MDR1 P-glycoprotein isoform expression in benign and malignant human prostate cells by isoform-specific monoclonal antibodies,” Cancer Lett. 150, 147–153 (2000). 10.1016/S0304-3835(99)00384-5 [DOI] [PubMed] [Google Scholar]
- 73. Huss W. J., Gray D. R., Greenberg N. M., Mohler J. L., and Smith J. L., “ Breast cancer resistance protein–mediated efflux of androgen in putative benign and malignant prostate stem cells,” Cancer Res. 65(15), 6640–6652 (2005). 10.1158/0008-5472.CAN-04-2548 [DOI] [PubMed] [Google Scholar]
- 74. Litman T., Skovsgaard T., and Stein W. D., “ Pumping of drugs by PGlycoprotein: A two-step process?” J. Pharmacol. Exp. Ther. 307(3), 846–853 (2003). 10.1124/jpet.103.056960 [DOI] [PubMed] [Google Scholar]
- 75. Theyer G., Schimbock M., Thalhmmer T., Sherwood E. R., Baumgartner G., and Hamilton G., “ Role of the MDR-1-encoded multiple drug resistance phenotype in prostate cancer cell lines,” J. Urol. 150, 1544–1547 (1993). [DOI] [PubMed] [Google Scholar]
- 76. Fülöpa Z., Grefb R., and Loftsson T., “ A permeation method for detection of self-aggregation of doxorubicin in aqueous environment,” Int. J. Pharmacol. 454, 559–561 (2013). 10.1016/j.ijpharm.2013.06.058 [DOI] [PubMed] [Google Scholar]
- 77. Millenbaugh N. J., Wientjes M. G., and Au J. L., “ A pharmacodynamic analysis method to determine the relative importance of drug concentration and treatment time on effect,” Cancer Chemother. Pharmacol. 45, 265–272 (2000). 10.1007/s002800050039 [DOI] [PubMed] [Google Scholar]
- 78. Eksborg S., “ Extraction of daunorubicin and doxorubicin and their hydroxyl metabolites: Self-association in aqueous solution,” J. Pharm. Sci. 67(6), 782–785 (1978). 10.1002/jps.2600670613 [DOI] [PubMed] [Google Scholar]
- 79. Stein W. D., Robey R., Cardarelli C., Gottesman M. M., and Bates S. E., “ Low and high concentrations of the Topo II inhibitor Daunorubicin in NIH3T3 cells,” Cell Cycle 2(2), 134–142 (2003). 10.4161/cc.2.2.294 [DOI] [PubMed] [Google Scholar]
- 80. Anderson P. O., Knoben J. E., and Troutman W. G., Handbook of Clinical Drug Data ( McGraw-Hill Medical Publishing Division, 2002), pp. 237–253. [Google Scholar]
- 81. Klepsch F., Jabeen I., Chiba P., and Ecker G. F., “ Pharmacoinformatic approaches to design natural product type ligands of ABC-transporters,” Curr. Pharmaceut. Des. 16(15), 1742–1752 (2010). 10.2174/138161210791163992 [DOI] [PubMed] [Google Scholar]
- 82. Lainey E., Wolfromm N., Enot D., Scoazec M., Bouteloup C., Leroy C., Micol J. B., Botton S., Galluzz L., Fenaux P., and Kroemer G., “ Azacytidine and erlotinib exert synergistic effects against acute myeloid leukemia,” Oncogene 32, 4331–4342 (2013). 10.1038/onc.2012.469 [DOI] [PubMed] [Google Scholar]
- 83.See supplementary material at http://dx.doi.org/10.1063/1.4934715E-BIOMGB-9-025505 for the isolation of white blood cells from blood, the movement of cancer cells mixed with blood cells at different ratios, the flow of the blue dye from reservoir D, and the comparison of single-cell drug accumulation kinetics.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4934715E-BIOMGB-9-025505 for the isolation of white blood cells from blood, the movement of cancer cells mixed with blood cells at different ratios, the flow of the blue dye from reservoir D, and the comparison of single-cell drug accumulation kinetics.