Abstract
Long non-coding RNAs (lncRNAs), transcripts as longer than 200 nt in length with a great number of varieties in human genomics, play important roles in the regulation of genetics and epigenetics including gene transcription and post-transcription. Increasing evidence have demonstrated the upregulation of lncRNAs in tumorigenesis and metastasis of esophageal cancer (EC), a type of malignant tumors particularly in Asia. In this review, we briefly discuss the profiles and functions of lncRNAs involved in the progression of EC, which may provide a new approach to improve EC diagnosis and treatment.
Keywords: long non-coding RNAs, esophageal cancer, biomarker, review
Introduction
Esophageal cancer (EC) is the sixth common cause of cancer death worldwide and has become a major health concern, especially in Asia.1 There are two primary forms of EC, eg, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). ESCC is the most prevalent type in eastern countries while EAC often occurs in the west. Despite wide application of radical esophagectomy and systemic chemo-radiotherapy, the overall 5-year survival rate of patients with ESCC remains extremely low. The morbidity and mortality of EC ranks fourth in the People’s Republic of China, by following after lung cancer, gastric cancer, and liver cancer.2 One of the main reasons for the low overall survival is the lack of appropriate molecular biomarkers for the early detection of EC.
Over the past decades, a large number of studies focused on aberrant expression of protein-coding RNAs in cancers, which provided many promising approaches for cancer diagnosis and treatment. In recent years, however, along with the advance of high-resolution microarray and genome-wide sequencing technology, increasing evidence obtained from the studies of genomics and transcriptomics suggest that non-coding RNAs can also be applied as new biomarkers for cancer detection and molecular targets for cancer therapy.3 Based on the transcript lengths, the non-coding RNAs contain small non-coding RNAs, long non-coding RNAs (lncRNAs), and very long non-coding RNA.4,5 Among non-coding RNAs, lncRNAs have recently caught more attentions by identifying numerous of lncRNAs during the progression of multiple types of cancer.6,7
We collected potentially eligible studies through searching electronic databases PubMed and Web of Science. We used lncRNAs, long non-coding RNA, or –esophageal cancer as the keywords for the search. The latest search was updated on March 15, 2015. This review aims to outline the biogenesis of lncRNA by highlighting a correlation of several lncRNAs to EC progression for discovery of new biomarkers and molecular targets for improving EC diagnosis and treatment.
LncRNAs
Definition
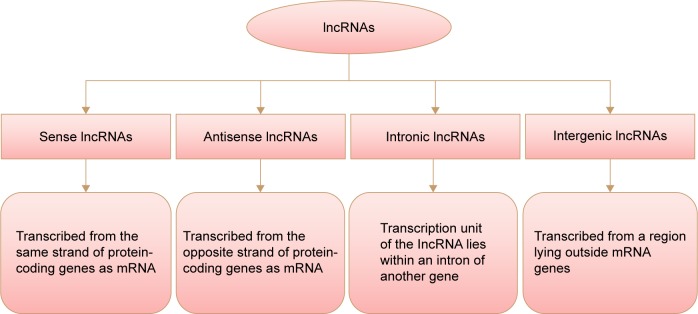
LncRNAs are commonly defined as a non-protein-coding molecule longer than 200 nucleotides in length.8,9 They can be classified depending on their location with respect to the mRNA gene, as shown in Figure 1.10
Figure 1.
Schematic representation of transcripted lncRNAs.
Abbreviation: lncRNAs, long non-coding RNAs.
Biological functions of lncRNAs
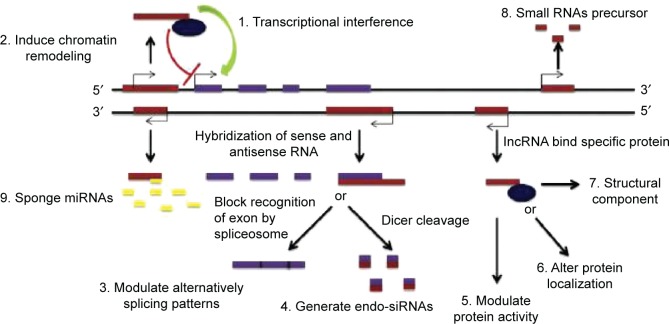
In past decades, the common knowledge suggested that long non-coding transcripts were simply transcriptional noises or cloning artifacts.11,12 Nonetheless, tight evolutionary conservation and strict gene regulation of lncRNAs predict that these transcripts are important in both normal physiological processes and abnormal pathological developments.13–15 Wang and Chang distilled the myriad functions of lncRNAs into four archetypes of molecular mechanisms (Figure 2).16
Figure 2.
Molecular mechanisms of lncRNAs.
Abbreviation: lncRNAs, long non-coding RNAs.
LncRNAs and cancers
Correlation of lncRNAs to cancers
A number of studies demonstrated the deregulation of lncR-NAs in cancers, suggesting that the aberrant expression of lncRNAs is associated with tumorigenesis, metastasis, and prognosis of cancer. For instance, H19, the first non-coding RNA was discovered as a tumor promoter in liver and bladder cancers; however, it appears to be a tumor suppressor shown in teratoma and nephroblastoma.17,18 Hirata et al reported that metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is highly expressed in human renal cell carcinoma tissues and associated with reduced patient survival. MALAT1 silencing decreased renal cell carcinoma cell proliferation and invasion and increased apoptosis.19 In gliomas, the transcripts of the gene of Colorectal Neoplasia Differentially Expressed (CRNDE) are upregulated in both primary specimens from glioma patients and in vitro with cell lines.20 Shi et al reported that the SPRY4 intronic transcript 1 (SPRY4-IT1) expression was significantly upregulated in 48 breast cancer tumor tissues as compared with the corresponding normal tissues. Additionally, the increased SPRY4-IT1 expression was found to be associated with a larger tumor size and advanced pathological stages in breast cancer patients.21 Altogether, these results suggest that lncRNAs may play important roles in the development of cancer by their effects on either tumor promotion or tumor suppression.
Gene regulation by lncRNAs
In order to determine whether aberrant expression of lncR-NAs is a cause for cancer or only a consequence of cancerous phenotype, numerous studies have investigated the biological functions of lncRNAs during the progression of cancer as well as mechanistic associations involved. To date, it has been reported that lncRNAs participate in genetic and epigenetic regulation including transcription, splicing, RNA decay, and translation.22
Correlation of lncRNAs to EC
Compared with the protein markers, lncRNAs are likely to be more useful biomarkers for cancer detection. Li et al screened lncRNA expression profiles in paired tumor and normal tissues from 119 patients with ESCC and predicted that three long-chain lncRNA signatures can be applied as independent prognostic factors for patients with different stages of ESCC.23 These results suggest that lncRNA expression profiles may serve as new biomarkers for the prognosis of ESCC. In this review, we conclude the expression profiles of lncRNAs in EC and discuss several lncRNAs as the potential biomarkers for EC in Table 1.
Table 1.
Aberrant expression of lncRNAs in EC
| Name | Genome position | Pathological types | Expression level | Relevant to prognosis |
|---|---|---|---|---|
| HOTAIR | 12q13.13 | ESCC | ↑ | Negative correlation |
| POU3F3 | 2q12.1 | ESCC | ↑ | Unclear |
| TUG1 | 22q12.2 | ESCC | ↑ | Unclear |
| FOXCUT | 6p25.3 | ESCC | ↑ | Negative correlation |
| PlncRNA-1 | 8q24.21 | ESCC | ↑ | Negative correlation |
| MALAT1 | 11q13.1 | ESCC | ↑ | Negative correlation |
| SPRY4-IT | 15q31.3 | ESCC | ↑ | Unclear |
| H19 | 11p15.5 | ESCC | ↑ | Negative correlation |
| UCA1 | 19p13.12 | ESCC | ↑ | Negative correlation |
| PEG10 | 7q21 | ESCC | ↑ | Unclear |
| LOC285194 | 3q13.31 | ESCC | ↓ | Positive correlation |
| AFAP1-AS1 | 4p16.1 | EAC | ↑ | Unclear |
| HNF1A-AS1 | 12q24.31 | EAC | ↑ | Unclear |
Note: The upward arrow signifies upregulation.
Abbreviations: EAC, esophageal adenocarcinoma; EC, esophageal cancer; ESCC, esophageal squamous cell carcinoma; lncRNAs, long non-coding RNAs.
HOTAIR
HOTAIR is transcribed from HOXC locus on chromosome 12, it binds to polycomb and targets it to HOXD genes on chromosome 2. It has been shown to be highly induced in metastatic breast cancer and highly expressed in primary breast tumors.24 The aberrant expression of overexpressed HOTAIR in liver, gastrointestinal stromal, pancreatic, and colorectal cancer has been found as well.25–27 Additionally, we recently detected that HOTAIR was also overexpressed in ESCC tissues. In 78 cases of ESCC tissues vs adjacent noncancerous tissues, we found that elevated levels of HOTAIR expression were likely to be clinicopathologically associated with increased invasiveness, tumor metastasis, and advanced TNM (2009) stage classification. Furthermore, HOTAIR expression in ESCC tissues was determined to be closely associated with poor prognosis, indicating that HOTAIR may also be an indispensable prognostic indicator for patients in clinical settings. Elevated HOTAIR expression was correlated with increased occurrence of lymph node metastasis and lower overall survival rates. In vitro study, inhibition of HOTAIR expression was shown to decrease the invasiveness and migration of ESCC cells and significantly increase the response of ESCC cells to apoptosis induction.28 Thus, elucidation of HOTAIR function relevant to ESCC may provide a new approach for ESCC diagnosis and treatment. The development of HOTAIR-based therapeutic strategies for the downregulation of such oncogenic lncRNAs may provide a novel and promising alternative therapeutic approach for future cancer treatment.
POU3F3
Lnc-POU3F3, a 747 nt transcript containing four exons, is located in the chromosome 2q12.1 on the reverse strand. It was reported that functional lnc-POU3F3 was overexpressed in high-grade glioma tissues and promoted cell viability and proliferation in glioma cells.29 Li et al found that the level of POU3F3 was significantly higher in ESCC as compared with neighboring non-cancerous tissues. Overexpression of lnc-POU3F3 resulted in reduced expression of POU3F3 mRNA but increased cell tumorigenicity, whereas knockdown of lnc-POU3F3 increased the levels of POU3F3 mRNA. CpG islands in POU3F3 were densely methylated when lnc-POU3F3 was overexpressed in the cells. Consistently, the methylation at these sites was reduced by silencing lnc-POU3F3. Pharmacologic inhibition of EZH2 led to increased levels of POU3F3 mRNA by reducing binding of DNA methyltransferases to POU3F3, such as DNMT1, DNMT3A, and DNMT3B.30 Subsequently, our previous studies demonstrated that plasma levels of POU3F3 are significantly higher in ESCC patients as compared with normal controls.31 Therefore, lnc-POU3F3 may provide a useful tool for the early detection of ESCC.
TUG1
Taurine upregulated gene 1 (TUG1), a 7.1 kb lncRNA, was originally detected in a genomic screen for genes upregulated in response to taurine treatment of developing mouse retinal cells.32 Xu et al collected 62 pairs of primary ESCC tissues and adjacent normal tissues from patients who had undergone surgery and discovered that TUG1 was significantly over-expressed in ESCC, and upregulation of TUG1 was associated with family history and upper segmental EC. A higher TUG1 expression of upper EC might be a predictive factor of the poorer clinical outcome. Detecting the free TUG1 abundance might be more convenient as a risk or prognostic factor in the future. Further, in vitro silencing TUG1 via siRNA inhibited the proliferation and migration of ESCC cells and blocked the progression of cell cycle.33 Therefore, this study indicates that as a potential oncogene, TUG1 promotes proliferation and migration of ESCC cells.
FOXCUT
Mechanistic studies reported that a number of lncRNAs can functionally contact with their adjacent mRNAs to form lncRNA mRNA pair complexes in the gene regulatory networks.34
Using bioinformatics analysis, Pan et al identified a pair of lncRNA–mRNA, FOXCUT, and FOXC1, which highly interacted in ESCC tissues compared with adjacent noncancerous tissues. FOXC1 and FOXCUT expression levels were significantly elevated in tumor samples from patients with the presence of metastasis and poor differentiation. Upregulation of both FOXC1 and FOXCUT was correlated with aggressive clinicopathological characteristics, such as poor differentiation, advanced lymph node classification, and metastasis. And, patients with high expression of FOXCUT or FOXC1 had a significantly poor prognosis. These data suggest that high expression of FOXCUT or FOXC1 might play an important role in the tumorigenesis, development, progression of ESCC, and both of them may serve as prognostic biomarkers for ESCC patients. In general, patients with high levels of FOXCUT or FOXC1 experienced poor prognoses.35
PlncRNA-1
Prostate cancer long non-coding RNA 1 (PlncRNA-1), located on the human chromosome 8q24.21, was firstly discovered to be highly related to prostate cancer susceptibility.36 Its silence can significantly inhibit the growth of prostate cancer cells by inactivation of androgen receptor. In addition, our study team used quantitative real-time polymerase chain reaction (qRT-PCR) to detect the expression level of PlncRNA-1 in 73 pairs of ESCC and their matched normal tissues and showed that PlncRNA-1 is also highly expressed in ESCC compared with adjacent non-cancerous tissues. The high level of PlncRNA-1 expression was significantly correlated with advanced clinical stage and lymph node metastasis. Furthermore, knockdown of PlncRNA-1 reduced cell proliferation and increased the apoptosis in vitro.37 In conclusion, PlncRNA-1 plays an important role in ESCC cell proliferation. Overexpression of PlncRNA-1 is correlated with advanced tumor stage and lymph node metastasis, and may serve as a potential prognostic marker and therapeutic target for ESCC.
MALAT1
MALAT1 has been shown to express at a high level in multiple types of tumor such as non-small cell lung cancer and hepatocellular carcinoma.38,39 Recent studies demonstrated that overexpression of MALAT1 was found in 46.3% of ESCC tissues, mostly in advanced tumor tissues with larger tumor size and lymph nodes metastasis, indicating that MALAT1 may primarily participate in ESCC advancing but not initialization, which may be a promising biomarker for late-stage ESCC with metastasis. Inhibition of MALAT1 suppressed tumor growth in vitro and in vivo, as well as cell migratory and invasive capacity, confirming its oncogenic roles in ESCC.40 Wang et al provided the first evidence that a post-transcriptional regulatory mechanism by which miR-101 and miR-217 negatively regulate MALAT1 in ESCC cells. Silencing of MALAT significantly suppressed the proliferation of ESCC cells by arresting cell cycle at G2/M, which may be due to MALAT1-mediated upregulation of p21 and p27 expression and/or the inhibition of B-MYB expression. Consistently, the capacities of invasion and migration of ESCC cells were inhibited after overexpression of miR-101, miR-217, or MALAT1 siRNA, which may be attributed to the deregulation of downstream target genes of MALAT1, such as MIA2, HNF4G, ROBO1, CCT4, and CTHRC1.41 However, much more work is still required to determine the detailed mechanisms it functions in ESCC and the potentiality of MALAT1 as a therapeutic target for ESCC.
SPRY4-IT1
LncRNA SPRY4-IT1, a transcript from the second intron of the SPRY4 gene, was originally identified in adipose tissue. Our study showed that SPRY4-IT1 was also highly expressed in ESCC patients. The relative expression level of SPRY4-IT1 was associated with T stage, lymph node metastasis, and advanced pathologic stage of ESCC patients. Moreover, SPRY4-IT1 expression was not related to patients’ age, sex, smoking status, alcohol consumption, or tumor location. In addition, SPRY4-IT1 high expression was correlated with lower overall survival rates and could be an independent prognostic factor in patients with ESCC. In vitro experiments showed that siRNA-mediated knockdown of SPRY4-IT1 significantly decreased proliferation and metastasis capability of KYSE30 and Eca109 cells. Knockdown of SPRY4-IT1 also significantly reduced tumor weight and volume in nude mice compared with control, suggesting that knockdown of SPRY4-IT1 can suppress the development of ESCC in vivo.42 These findings indicate that SPRY4-IT1 may be a prognostic biomarker and a therapeutic target of ESCC in future.
H19
Bartolomei et al firstly discovered lncRNA H19 in 1991, which was associated with multiple human carcinomas.43–46 Gao et al detected loss of imprinting (LOI) of IGF2 in 276 samples and analyzed methylation status of H19 DMR. Patients with LOI of IGF2 in their tumor had deeper degree of lymph node involvement and were more likely to occur metastasis than those with MOI of IGF2.
In addition, they found that H19 CBS6 hypermethylation is related to LOI of IGF2 in human ESCC and IGF2 LOI usually leads to an overexpression of IGF2, which was correlated with lymph node involvement, neoplastic grade, and tumor metastasis.47 Therefore, H19 CBS6 methylation potentially represents a novel clinically relevant epigenetic marker to identify individuals at increased risk for the occurrence, progression, and prognosis of ESCC.
UCA1
It was discovered that the relative levels of UCA1 were significantly higher in ESCC tissues than the adjacent non-cancerous tissues. The expression level of UCA1 was associated with tumor differentiation, clinical stage, and lymph node metastasis of ESCC patients. However, lncRNA UCA1 expression was not correlated with patient’s age, sex, and tumor location. Consistently, remarkably higher expression of UCA1 was also observed in EC cell lines compared with the immortalized esophageal epithelial cell line NE1 60. SiRNA-mediated knockdown of UCA1 significantly decreased proliferation, migration, and invasion capability of EC cells compared with the control group, which indicated that downregulation of UCA1 can suppress the development of EC.48 These findings demonstrated that UCA1 is a potential diagnostic and therapeutic target in patients with ESCC in the future.
PEG10
PEG10 is a retrotransposon-derived gene adapted through mammalian evolution located on the human chromosome 7q21, which codes for at least two proteins, PEG10-RF1 and PEG10RF1/2. Zang et al investigated the expression of lncRNA PEG10 in a cohort of EC to assess its impact on EC cell proliferation, apoptosis, and invasion. qRT-PCR was used to quantify the expression levels of lncRNA PEG10 in 43 paired EC tissues and adjacent non-neoplastic tissues. Cell growth, apoptosis, and transwell invasion assays were used to evaluate the effects of lncRNA PEG10 in EC cells. The result showed that lncRNA PEG10 was expressed at higher levels in EC tissues than in adjacent non-neoplastic tissues. This relatively high expression was significantly associated with the occurrence of lymph node metastases. Downregulation of lncRNA PEG10 decreased the expression of PEG10. These results indicated that PEG10 is upregulated in EC tissues, and its downregulation inhibits proliferation and invasion, and promotes apoptosis in EC cells.49
LncRNA PEG10 may serve as a therapeutic agent in EC.
AFAP1-AS1
Alterations in methylation of protein-coding genes are associated with Barrett’s esophagus and EAC. Wu et al have shown that AFAP1-AS1 expression is substantially increased in EAC vs NE tissues as well as in EAC cell lines. This elevated expression of AFAP1-AS1, its role in cell proliferation and apoptosis, and its effect on cell migration and invasion suggest that dysregulated expression of AFAP1-AS1 is involved in development or progression of EAC. Proliferation assays showed that inhibition of AFAP1-AS1 by siRNA diminished cell proliferation. Furthermore, treatment with siRNA inhibited colony formation and reduced migration and invasion.50 Taken together, these findings suggest that AFAP1-AS1 is a functional lncRNA in human EAC cells and the potential utility of AFAP1-AS1 as a biomarker of EAC.
HNF1A-AS1
LncRNA HNF1A-AS1 is located on the human chromosome 12q24.31. Yang et al reported that HNF1A-AS1 is remarkably upregulated in human primary EACs compared with their corresponding normal esophageal tissues. Knockdown of HNF1A-AS1 in EAC cells significantly inhibited proliferation and anchorage-independent growth by inhibiting S-phase entry, invasion, and migration. Further mechanistic studies revealed that HNF1A-AS1 knockdown preferentially affect gene expression due to assembly of chromatin and the nucleosome, which is essential for cell-cycle progression. As discussed earlier, H19, as a well-known cancer-related lncRNA, is inhibited by HNF1A-AS1 knockdown, indicating a functional correlation between HNF1A-AS1 and H19 in primary EACs.51
LOC285194
The lncRNA LOC285194, also called LSAMP antisense RNA 3, consisting of four exons with 2105 bases in length is located on the human chromosome 3q13.31. It was first reported to be a tumor suppressor unit in osteosarcoma and depletion of this lncRNA promoted proliferation of normal osteoblasts through regulation of apoptotic and cell-cycle transcripts as well as VEGF receptor 1.52 Decreased LOC285194 expression has been reported in several cancers, and was strongly associated with malignant potential and poor patient prognosis.53,54 Our previous studies demonstrated that LOC285194 expression was significantly downregulated in ESCC tumor tissues when compared with the adjacent normal tissues. Low expression of LOC285194 was associated with larger tumor size, advanced TNM stage, more lymph node metastases, and distant metastases. Univariate analysis revealed that low expression of LOC285194 was significantly correlated with chemoradiotherapy response. Furthermore, decreased expression of LOC285194 could potentially serve as a molecular marker to predict the clinical outcomes of ESCC patients after surgery, and select patients who will benefit from the preoperative chemoradiotherapy.55
Conclusion
Although increasing evidence showed that multiple lncRNAs are involved in the progression of EC, the biological function of lncRNA in EC retains elusive, in particular lncRNAs do not have strong genomic footprints either inherently or heterogeneously and lack the underlying mechanism involved in EC metastasis as well. The relationship between lncRNAs and EC is currently under investigation. Particularly, high technologies such as gene chip, high-throughput profiling, and bioinformatics will be greatly beneficial for increasing our understanding of the expression profile and function of lncRNA in cancer. Overall, integration of biogenesis of lncRNA in the progression of EC is considered to provide a promising approach for EC diagnosis and treatment.
Acknowledgments
This work was supported by the Foundation of Nanjing City Committee of Science and Technology (grant no 201108027), the Innovation Team of Science and Education Health Project of Jiangsu in 2011 (grant no 15), and the National Natural Science Foundation of China (H1617/81201881) to Dr Xiu-Feng Cao, and the National Natural Science Foundation of China (81372199) to Dr Yong Xu.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27(1):2–12. doi: 10.3978/j.issn.1000-9604.2015.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 4.Vikram R, Ramachandran R, Shameem K, Abdul M. Functional significance of long noncoding RNAs in breast cancer. Breast Cancer. 2014;21(5):515–521. doi: 10.1007/s12282-014-0554-y. [DOI] [PubMed] [Google Scholar]
- 5.Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361(1):13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 9.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman M, Donaghey J, Carey BW, et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 15.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matouk IJ, DeGroot N, Mezan S, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2(9):e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matouk IJ, Mezan S, Mizrahi A, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta Mol Cell Res. 2010;1803(4):443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75(7):1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31(1):358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 21.Shi Y, Li J, Liu Y, et al. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. doi: 10.1186/s12943-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Chen Z, He J, et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–1710. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 26.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 27.Niinuma T, Suzuki H, Nojima M, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72(5):1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 28.Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52(11):908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 29.Guo H, Wu L, Yang Q, Ye M, Zhu X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene. 2015;554(1):114–119. doi: 10.1016/j.gene.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Zheng J, Deng J, et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146(7):1714–1726. doi: 10.1053/j.gastro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Tong YS, Wang XW, Zhou XL, et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 33.Xu Y, Wang J, Qiu M, et al. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumor Biol. 2014;36(3):1643–1651. doi: 10.1007/s13277-014-2763-6. [DOI] [PubMed] [Google Scholar]
- 34.Sigova AA, Mullen AC, Molinie B, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci U S A. 2013;110(8):2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan F, Yao J, Chen Y, et al. A novel long non-coding RNA FOXCUT and mRNA pair promote progression and predict poor prognosis esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(6):2838–2849. [PMC free article] [PubMed] [Google Scholar]
- 36.Chung S, Nakagawa H, Uemura M, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011;102(1):245–252. doi: 10.1111/j.1349-7006.2010.01737.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang CM, Wu QQ, Li SQ, et al. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59(3):591–597. doi: 10.1007/s10620-013-2956-7. [DOI] [PubMed] [Google Scholar]
- 38.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 39.Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 40.Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Li M, Wang Z, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290(7):3925–3935. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie HW, Wu QQ, Zhu B, et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumor Biol. 2014;35(8):7743–7754. doi: 10.1007/s13277-014-2013-y. [DOI] [PubMed] [Google Scholar]
- 43.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 44.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333(2):213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 45.Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumor Biol. 2014;35(9):9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 46.Tsang WP, Enders KO, Simon SM, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 47.Gao T, He B, Pan Y, et al. H19 DMR methylation correlates to the progression of esophageal squamous cell carcinoma through IGF2 imprinting pathway. Clin Transl Oncol. 2014;16(4):410–417. doi: 10.1007/s12094-013-1098-x. [DOI] [PubMed] [Google Scholar]
- 48.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(11):7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 49.Zang W, Wang T, Huang J, et al. Long noncoding RNA PEG10 regulates proliferation and invasion of esophageal cancer cells. Cancer Gene Ther. 2015;22(3):138–144. doi: 10.1038/cgt.2014.77. [DOI] [PubMed] [Google Scholar]
- 50.Wu W, Bhagat TD, Yang X, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144(5):956–966. doi: 10.1053/j.gastro.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Song JH, Cheng Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63(6):881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasic I1, Shlien A, Durbin AD, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70(1):160–171. doi: 10.1158/0008-5472.CAN-09-1902. [DOI] [PubMed] [Google Scholar]
- 53.Liu Q1, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41(9):4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi P, Xu MD, Ni SJ, et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu-suo Tong, Xi-lei Zhou, Xiao-wei Wang, et al. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med. 2014;12:233. doi: 10.1186/s12967-014-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]