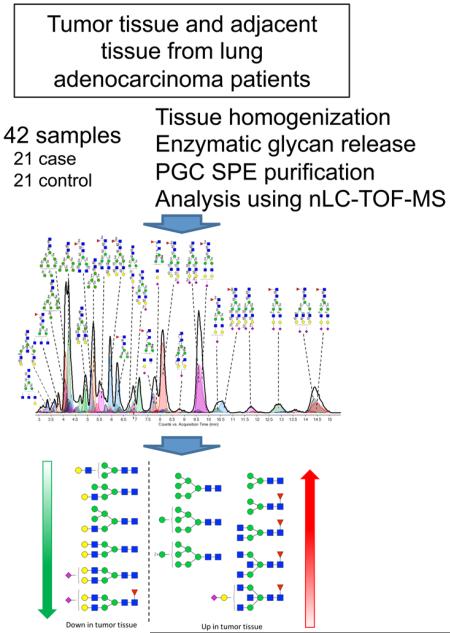
Abstract
To decrease the mortality of lung cancer, better screening and diagnostic tools as well as treatment options are needed. Protein glycosylation is one of the major post-translational modifications that is altered in cancer, but it is not exactly clear which glycan structures are affected. A better understanding of the glycan structures that are differentially regulated in lung tumor tissue is highly desirable and will allow us to gain greater insight into the underlying biological mechanisms of aberrant glycosylation in lung cancer. Here, we assess differential glycosylation patterns of lung tumor tissue and nonmalignant tissue at the level of individual glycan structures using nLC–chip–TOF–MS. Using tissue samples from 42 lung adenocarcinoma patients, 29 differentially expressed (FDR < 0.05) glycan structures were identified. The levels of several oligomannose type glycans were upregulated in tumor tissue. Furthermore, levels of fully galactosylated glycans, some of which were of the hybrid type and mostly without fucose, were decreased in cancerous tissue, whereas levels of non- or low-galactosylated glycans mostly with fucose were increased. To further assess the regulation of the altered glycosylation, the glycomics data was compared to publicly available gene expression data from lung adenocarcinoma tissue compared to nonmalignant lung tissue. The results are consistent with the possibility that the observed N-glycan changes have their origin in differentially expressed glycosyltransferases. These results will be used as a starting point for the further development of clinical glycan applications in the fields of imaging, drug targeting, and biomarkers for lung cancer.
Keywords: N-Glycosylation, NSCLC, tissue, nLC–MS profiling, gene expression
INTRODUCTION
Lung cancer is the leading cause of death for men and women in the United States and worldwide due to the inability to detect early stage disease and ineffective treatments for advanced disease.1 In a large randomized trial, low-dose spiral computerized tomography (LDCT) was recently shown to identify early stage lung cancer and, as a consequence, to reduce lung cancer mortality.2 LDCT is likely to become the first approved screening and early detection test for lung cancer, but it is plagued by a high false positive rate.2 There is a need to develop complementary screening and early detection tools based on molecular changes due to tumorigenesis. Given that our knowledge of the molecular biology of smoking-induced lung cancer has dramatically increased over the past few years, this approach is plausible. To date, this effort has been focused on the identification of genomic and/or proteomic signatures in a variety of different specimen types with limited success.3 A broader strategy that incorporates additional cancer characteristics is needed.
Protein glycosylation is one of the major post-translational modifications, and it has been proposed as a new paradigm in biomarker discovery and the search for drug targeting leads.4 Glycans are enzymatically synthesized by glycosyltransferases and, due to the different glycosidic linkages, have a large degree of structural heterogeneity. Glycosylation of proteins plays key roles in cell-cell and cell–matrix interactions as well as cellular differentiation and proliferation.5-7 It is not surprising, therefore, that, using numerous lectin-binding studies, glycans have been reported to be differentially expressed in tumor tissue compared to controls8 and that differential expression of glycosylation-related genes has been described.9 Furthermore, several of the proteins currently used as biomarkers for cancer in blood are highly glycosylated10 (e.g., CA125,11 PSA,12 CA19-9), indicating the potential importance of protein glycosylation in cancer and the potential of protein glycosylation analysis in disease identification and treatment.
Advances in technology, particularly the use of mass spectrometry for glycan profiling and identification,13-16 now allow for a more structure-specific strategy to be employed.17 Mass spectrometry in itself can determine only a glycan’s composition, i.e., the number and type of monosaccharides of which it consists, but does not allow the specific glycosidic linkages to be identified. The use of a complementary separation technique, such as HPLC, is needed to allow structure-specific analysis. A recent study addressed the glycosylation of colorectal cancer tissues compared to controls using mass spectrometric techniques.18 Among the differentially expressed glycan compositions observed were decreased levels of structures containing a bisecting GlcNAc, whereas levels of sulfated and paucimannosidic glycans as well as glycans containing sialylated Lewis epitopes were increased.18
The glycosylation pattern of the membrane fraction of colorectal cell lines has also been compared to the glycosylation of the membrane fraction of epithelial cells derived from colorectal tumors.19 It was observed that the membrane glycosylation of the cell lines differed from the glycosylation of the tumor cells, indicating that cell lines may not always provide biologically relevant glycosylation patterns. Therefore, further studies toward the characterization of differential glycosylation patterns of tumor tissue compared to non-malignant tissue are highly desirable. Elucidating disparate glycosylation patterns between malignant and nonmalignant lung tissue will allow us to gain greater insight into the underlying biological mechanisms of aberrant glycosylation in lung cancer. We anticipate that these results will then be used as a starting point for the further development of clinical glycan applications in the fields of imaging, drug targeting, and blood biomarkers.
In this study, we apply a nano HPLC coupled with time-of-flight mass spectrometry (nLC–TOF–MS)-based method with a porous graphitized carbon (PGC) stationary phase for the separation of N-glycans released from lung adenocarcinoma tissue samples and nonmalignant control tissue from the same individual. The method has been shown to be highly stable,20 with an average interday coefficient of variation of 4%, determined on log10-transformed integrals, and we previously determined the differential glycosylation profiles and a candidate biomarker panel for ovarian cancer using this method.21 The N-glycans are analyzed twice: once in unreduced form at the compositional level and once in reduced form at the level of individual glycan structures, which are identified using our in-house library for structural identification.22 Differential glycan compositions and structures are identified, and gene expression data from a published study23 is used to further address the underlying changes in the N-glycan biosynthetic pathway and to further understand lung adenocarcinoma tumor glycobiology.
EXPERIMENTAL SECTION
Human Samples
Forty-two deidentified malignant and adjacent normal lung tissue specimens were obtained from the New York University biorepository. All samples originated from patients who were current or former smokers with a diagnosis of lung adenocarcinoma (Stages I–IIIA). Informed consent was obtained from all 42 patients. The study was conducted in accordance with Institutional Ethics Committee approval at New York University and the University of California, Davis. Residual tumor and adjacent nonmalignant tissues were harvested from the resected lung after routine pathological protocols were completed. Two to three tissue pieces were aliquoted into a 1.5 mL Nunc vial, which was then immediately placed in liquid nitrogen. After transport in liquid nitrogen, each vial was barcoded and stored at −80 °C until it was analyzed. All specimens were clinically annotated for age, gender, race, histology, smoking status, pack-years, and stage of disease. Forty-one patients had stage I and 1 patient had stage IIIA disease. The average age of the cohort was 70.2 (±11.3) years and consisted of 13 males and 29 females. Six patients were current smokers, and 36 patients were former smokers; their average pack-year was 33.2 (±24.7).
Homogenization of Lung Cancer Tissue Specimens
All procedures were performed on dry ice to keep the samples cold. Depending on the weight of the tissue specimens, they were cut into either three (if >10.0 mg) or two pieces (if <10.0 mg), and all pieces from the same specimen were placed in a single 1.5 mL Eppendorf tube. Then samples were washed using 1 mL of cold PBS (Sigma-Aldrich, St. Louis, MO) by pipet mixing followed by subsequent aspiration to remove remaining blood. One-hundred microliters of HB buffer (0.25 M sucrose, 20 mM HEPES·KOH, and 10 μL of 1:10 protease inhibitor cocktail (Roche, Basel, Switzerland) per 10 mL of buffer, pH 7.4) was added to each of the samples, which were then homogenized using a Bullet Blender Storm for 3 min at speed 8 using prepacked tubes for hard tissues (Wisbiomed, San Mateo, CA).24
N-Glycan Sample Preparation
To assess the stability of the N-glycan sample preparation procedure, one standard serum sample (Sigma-Aldrich, St. Louis, MO) was included after every 10 tissue samples and as the first and last sample. The sample preparation procedure for serum is slightly different compared to that for tissue (see below), but all steps were performed in parallel for both the tissue and standard serum samples to ensure accurate sample preparation of the tissue samples.
Fifty microliters of 300 mM NH4HCO3 (Sigma-Aldrich, St. Louis, MO) with 15 mM dithiothreitol (DTT, Sigma-Aldrich, St. Louis, MO) was added to each of the tissue homogenates, whereas 25 μL of 200 mM NH4HCO3 with 10 mM DTT was added to 11 aliquots of 25 μL of a standard serum sample. Proteins were denatured by heat using four cycles of alternating between boiling water (100 °C) and room temperature (25 °C) water for 15 s each, and glycans subsequently were enzymatically released by addition of 2 and 1 μL of PNGaseF (New England Biolabs, Ipswich, MA) to the tissue and the serum samples, respectively, followed by 16 h incubation at 37 °C. Deglycosylated proteins were precipitated using 600 and 200 μL of ice-cold ethanol for the tissue and serum samples, respectively. After centrifugation, the supernatant containing the glycans was transferred to a 96-well plate according to the plate layout in Supporting Information Figure S1 and brought to dryness in vacuo.
N-Linked glycans released by PNGaseF were purified using graphitized carbon SPE plates with a 40 μL bed volume (Glygen, Columbia, MD), essentially as described earlier.20 Wells of the SPE plate were conditioned using 2 × 200 μL of 80% ACN containing 0.1% TFA (EMD chemicals, Gibbstown, NJ), followed by 3 × 200 μL of water. Glycan samples were reconstituted in 400 μL of water and subsequently loaded into the wells. Cartridges were washed using 7 × 200 μL of water, and N-glycans were eluted using 2 × 200 μL of 40% ACN containing 0.05% TFA. Samples were dried in vacuo and reconstituted in water prior to analysis. Fifty microliters of water was used for the standard serum samples and tissue samples <10.0 mg, whereas 100 μL of water was used for tissue samples >10.0 mg.
Glycan Reduction
Glycan reduction was essentially performed as described earlier.22 Briefly, 44 μL of a 2 M NaBH4 (Sigma-Aldrich, St. Louis, MO) solution in water was added to 44 μL of reconstituted unreduced glycan sample in a 96-well plate. The plate was incubated at 65 °C for 1 h, and the reduced glycans were then immediately purified using PGC SPE as described earlier. Samples were dried in vacuo and reconstituted in 50 μL of water prior to analysis.
nHPLC–chip–TOF–MS Analysis
N-Glycans were analyzed as described before20 using an Agilent (Santa Clara, CA) 6200 series nanoHPLC–chip–TOF–MS, consisting of an autosampler, which was maintained at 8 °C, a capillary loading pump, a nanopump, HPLC–chip–MS interface, and an Agilent 6210 time-of-flight mass spectrometer. The chip (Glycan Chip II, Agilent) contained a 9 × 0.075 mm i.d. enrichment column coupled to a 43 × 0.075 mm i.d. analytical column; both packed with 5 μm porous graphitized carbon (PGC). Upon injection of 1 μL of N-glycan sample, the sample was loaded onto the enrichment column using 3% ACN containing 0.1% formic acid (FA; Fluka, St. Louis, MO). Then, the analytical column was switched in-line so that the nanopump delivered a gradient of 3% ACN with 0.1% FA (solvent A) to 90% ACN with 1.0% FA (solvent B) over 17 min at a flow rate of 0.4 μL/min. Positive ions were generated using a capillary voltage of 1850 V with a nitrogen gas flow of 4 L/min at 325 °C. Mass spectra were acquired at a frequency of 0.63 spectra per second over a mass window of m/z 400 to 3000. The run order for the different samples is shown in Supporting Information Table S1.
Data Analysis
Data analysis was performed using Masshunter qualitative analysis (version B.03.01, Agilent) and Microsoft Excel for Mac 2011 (version 14.1.3, Microsoft), according to previous publications.20,25 Data was loaded into Masshunter qualitative analysis, and glycan features were identified and integrated using the Molecular Feature Extractor algorithm. First, signals above a signal-to-noise threshold of 5.0 were considered. Then, signals were deconvoluted using a tolerance of m/z 0.0025 ± 10 ppm. The resulting deconvoluted masses were subsequently annotated using a retrosynthetic theoretical glycan library that was previously developed26 and contained 331 possible N-glycan compositions. A 15 ppm mass error was allowed. Glycan compositions, retention times, and peak area were exported to csv-format for further evaluation.
Statistical Analysis
Prior to statistical analysis, raw peak areas were total-quantity-normalized based on the underlying assumption that the total amount of ionized glycans that reaches the detector is similar for different samples and glycan profiles for each data set. Glycans detected in fewer than 70% of samples were discarded from downstream analysis to reduce the bias that could be induced by imputation for missing not at random. Unobserved values for any remaining undetected glycans below the predefined detection limit were imputed as one-half of the glycan-specific minimum of the observed values. Finally, the normalized data were log2-transformed to reduce the influence of extreme values and to meet homogeneity of variance assumptions. Statistical analyses were conducted in R 3.0.1 language and environment.
We conducted a partial least-squares regression with linear discriminant analysis (PLS-LDA) to assess whether glycomic profiles could separate malignant from nonmalignant tissue samples. To adjust for covariates, we regressed intensity values on age, gender, and smoking history and used the residuals, further scaled to a variance of one, in the PLS-LDA. Leave-one-out cross-validation was used to estimate the classification accuracy, sensitivity, and specificity of the PLS-LDA using 1–10 latent components.
We conducted a differential analysis to identify specific glycans significantly differentially regulated between cancer tissue and control samples. Two approaches to the differential analysis were used. First, we used paired sample t-tests for univariate analysis without adjustments for covariates. Second, for multivariate analysis, we used a mixed effect model in order to take into account age, gender, and smoking history as covariates. For the mixed effect analysis, a random effect was included for each patient to account for correlation of glycan intensities in tissue samples from the same patient. Cancer status of the tissue sample (malignant vs nonmalignant), age, gender, and smoking history (pack-years) were included as fixed effects. A chi-squared test was used to determine significance of differences in intensity values between malignant and nonmalignant tissue samples. For both analyses, false discovery rates (FDR) were calculated to account for multiple testing.
Gene Expression
Landi et al. previously performed gene expression analysis using HG-U133A Affymetrix chips on fresh frozen tissue samples of adenocarcinoma and paired noninvolved lung tissue from current, former, and never smokers.23 Preprocessed expression data for 27 glycosyltransferases and glycosidases were extracted from the GENT database.27 Only matched pairs of current and former smokers were analyzed to mimic the inclusion criteria of the glycomics study. This resulted in a data set originating from paired malignant and nonmalignant samples from 22 patients that were current (12) or former (10) smokers. The gene expression data was analyzed using the same statistical methods for differential analysis as described above.
RESULTS
To assess the differential glycosylation profile of lung adenocarcinoma tissue relative to nondiseased lung tissue, samples were obtained from 42 individuals. Samples from both nonmalignant and malignant lung tissue, obtained from the same patient, were analyzed. Standard samples, which were included to assess sample preparation as well as instrument errors, showed consistent results, as shown in Supporting Information Figure S2, indicating that the data obtained was of good quality. The glycosylation profiles obtained from three samples showed very low intensities (Supporting Information Figure S3) compared to that of the other samples during the assessment of data quality and were excluded from all further statistical analyses. Because these samples were low in both the reduced and reduced analyses, it is not likely that these low intensities were due to instrument failure but, rather, sample preparation difficulties. An overview of the patient characteristics of the 39 remaining individuals is provided in Table 1.
Table 1. Sample Characteristics of the Sample Set Used for the Glycomics Analysis in This Study.
| variable | LC patients (n = 39) |
|---|---|
| total sample size, N | 39 |
| age, mean ± SD | 69.90 ± 11.50 |
| female, no. (%) | 26 (66.67%) |
| current smoker, no. (%) | 6 (15.38%) |
| pack/year, mean (±SD) | 33.73 (±24.81) |
Differential Glycan Compositions in Cancer Tissue Compared to Controls
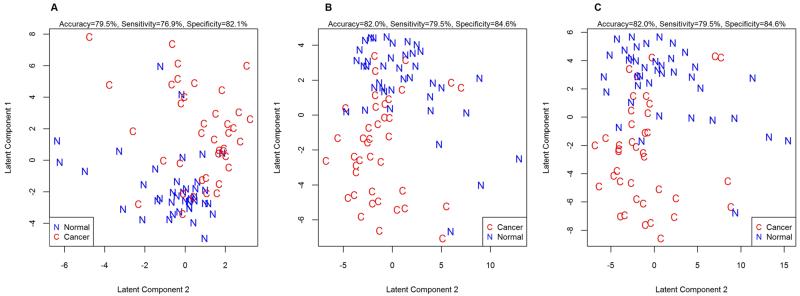
First, a glycan compositional analysis approach was used to assess whether the glycosylation of proteins is altered in adenocarcinoma tissue as compared to that in control tissue. In this method, glycans were not reduced, resulting in two signals for each glycan due to the separation of reducing-end anomers. Intensities of all signal peaks from a single composition were added to obtain a single representative measure for that composition prior to statistical analysis. Forty-five glycan compositions were consistently (>70% of samples) observed in the tissue samples. These compositions were used to conduct a PLS-LDA analysis to assess whether the global glycomic profile could separate adenocarcinoma tissue from control. The results are shown in Figure 1a. While complete separation between the cancerous tissue and the control tissue is not achieved, a clear distinction is observed, indicating changes in glycosylation associated with adenocarcinoma.
Figure 1.
Lung adenocarcinoma tissues can be separated from the nonmalignant tissue from the same individual based on their N-glycosylation pattern. Score plots of the PLS-LDA analysis are shown for (A) unreduced glycan compositions, (B) reduced glycan compositions, and (C) N-glycan structures. Using leave-one-out cross-validation, accurate classification rates were calculated to be 79.5% for the unreduced glycan compositions using 3 latent components, 82.0% for the reduced glycan compositions using 4 latent components, and 82.0 for the N-glycan structures using 3 latent components.
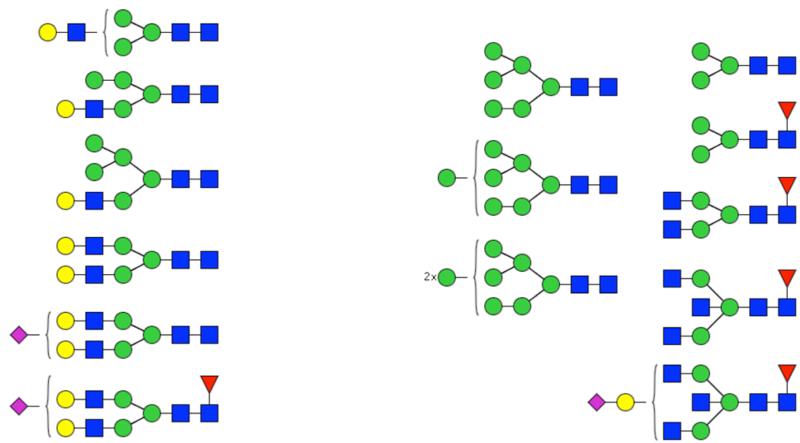
Differential analysis was performed at the level of individual glycan compositions to identify which glycan compositions contribute to the segregation between the cancerous and control tissues. Independent of whether the values were adjusted for the covariates, 14 glycan compositions were identified to be differentially expressed (FDR < 0.05) in cancerous tissue compared to controls (Table 2). One additional glycan composition (Hex5HexNAc4NeuAc2) was shown to be significant when adjusted for the covariates. Interestingly, the levels of several oligomannose type glycans were upregulated. This is concordant with previous studies in human colorectal tissue18 and mouse breast cancer tissue.28 Furthermore, levels of fully galactosylated glycans, some which were of the hybrid type and mostly without fucose, were decreased in cancerous tissue, whereas levels of non- or low-galactosylated glycans mostly with fucose were increased. These differences, which are further illustrated in Figure 2, have thus far not been reported in lung cancer or other cancer tissue studies.
Table 2. Glycan Compositions That Are Differentially Expressed in Adenocarcinoma Tissue.
| GlycanID | Unreduced analysis | Reduced analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor Meana |
Normal Meana |
Ratiob | Unadjusted for covariates |
Adjusted for covariates | Tumor Meana |
Normal Meana |
Ratiob | Unadjusted for covariates |
Adjusted for covaraites | |||||
| P-value | FDR | P-value | FDR | P-value | FDR | P-value | FDR | |||||||
| H3N2F0S0 | 401031 | 243979 | 1.64 | 0.001 | 0.004 | <0.001 | <0.001 | 632462 | 437396 | 1.45 | 0.009 | 0.032 | 0.004 | 0.018 |
| H3N2F1S0 | 143561 | 60381 | 2.38 | 0.002 | 0.010 | 0.002 | 0.008 | 368115 | 163315 | 2.25 | 0.007 | 0.032 | 0.002 | 0.012 |
| H3N4F1S0 | 282347 | 162840 | 1.73 | <0.001 | 0.003 | <0.001 | 0.001 | 336161 | 281400 | 1.19 | 0.268 | 0.391 | 0.259 | 0.378 |
| H3N5F1S0 | 157177 | 82413 | 1.91 | 0.015 | 0.048 | 0.013 | 0.040 | 283220 | 155035 | 1.83 | 0.002 | 0.015 | 0.001 | 0.012 |
| H3N6F1S0 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 25853 | 16755 | 1.54 | 0.008 | 0.032 | 0.007 | 0.025 |
| H4N2F1S0 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 23247 | 8457 | 2.75 | 0.001 | 0.008 | <0.001 | 0.002 |
| H4N3F0S0 | 69856 | 101298 | 0.69 | <0.001 | 0.003 | <0.001 | 0.001 | 183849 | 238680 | 0.77 | 0.178 | 0.319 | 0.170 | 0.304 |
| H4N4F0S1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 45527 | 70586 | 0.64 | 0.009 | 0.032 | 0.008 | 0.027 |
| H4N5F1S1 | 44795 | 18954 | 2.36 | 0.001 | 0.005 | 0.001 | 0.003 | 148386 | 100637 | 1.47 | 0.064 | 0.152 | 0.053 | 0.133 |
| H4N6F1S0 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 77475 | 45157 | 1.72 | 0.001 | 0.008 | <0.001 | 0.005 |
| H5N3F0S0 | 62297 | 85280 | 0.73 | 0.010 | 0.036 | 0.009 | 0.031 | 120491 | 165439 | 0.73 | 0.046 | 0.124 | 0.042 | 0.113 |
| H5N4F0S0 | 277677 | 405445 | 0.68 | <0.001 | 0.001 | <0.001 | <0.001 | 548738 | 938311 | 0.58 | <0.001 | 0.002 | <0.001 | 0.002 |
| H5N4F0S1 | 588178 | 809015 | 0.73 | <0.001 | 0.003 | <0.001 | 0.001 | 1076923 | 1515725 | 0.71 | 0.008 | 0.032 | 0.007 | 0.025 |
| H5N4F0S2 | 772633 | 999662 | 0.77 | 0.019 | 0.056 | 0.013 | 0.040 | 1162964 | 1586227 | 0.73 | 0.069 | 0.152 | 0.063 | 0.137 |
| H5N4F1S0 | 826507 | 870090 | 0.95 | 0.336 | 0.409 | 0.327 | 0.387 | 1309307 | 2004782 | 0.65 | 0.001 | 0.010 | 0.001 | 0.008 |
| H5N4F1S1 | 1349183 | 1697040 | 0.80 | 0.001 | 0.004 | <0.001 | 0.001 | 2379749 | 3133061 | 0.76 | 0.063 | 0.152 | 0.058 | 0.137 |
| H5N5F0S1 | 24276 | 38241 | 0.63 | 0.044 | 0.122 | 0.040 | 0.111 | 33159 | 66889 | 0.50 | 0.003 | 0.019 | 0.003 | 0.013 |
| H5N5F1S2 | 37578 | 53028 | 0.71 | 0.176 | 0.269 | 0.168 | 0.252 | 49067 | 100225 | 0.49 | 0.001 | 0.008 | 0.001 | 0.006 |
| H5N5F2S0 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 36795 | 51803 | 0.71 | 0.003 | 0.019 | 0.002 | 0.012 |
| H6N2F0S0 | 658421 | 482979 | 1.36 | <0.001 | 0.003 | <0.001 | <0.001 | 1026670 | 723347 | 1.42 | 0.010 | 0.032 | 0.005 | 0.020 |
| H6N3F0S0 | 89282 | 120565 | 0.74 | 0.007 | 0.028 | 0.005 | 0.017 | 180604 | 247451 | 0.73 | 0.008 | 0.032 | 0.007 | 0.025 |
| H6N4F0S0 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 55478 | 151490 | 0.37 | <0.001 | <0.001 | <0.001 | <0.001 |
| H6N4F0S1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 14364 | 44239 | 0.32 | <0.001 | 0.008 | <0.001 | 0.002 |
| H6N4F1S0 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 30500 | 61178 | 0.50 | 0.003 | 0.019 | 0.002 | 0.013 |
| H6N6F0S0 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | 28979 | 50194 | 0.58 | 0.004 | 0.022 | 0.003 | 0.013 |
| H7N2F0S0 | 510228 | 357363 | 1.43 | 0.001 | 0.004 | <0.001 | 0.001 | 622269 | 460219 | 1.35 | 0.006 | 0.027 | 0.002 | 0.012 |
| H7N6F1S2 | 20567 | 30991 | 0.66 | 0.140 | 0.242 | 0.132 | 0.221 | 25926 | 48286 | 0.54 content-type="color:#0f7f0f" | 0.002 | 0.017 | 0.002 | 0.012 |
| H8N2F0S0 | 592137 | 406776 | 1.46 | 0.001 | 0.004 | <0.001 | 0.001 | 685608 | 500068 | 1.37 | 0.005 | 0.026 | 0.002 | 0.012 |
Mean ion intensities are reported for each of the individual glycan compositions.
Ratio between the initial (control) and the final (case) state. Ratios color-coded in green represent glycan compositions of which levels are decreased in lung adenocarcinoma, whereas those in red represent glycan compositions of which levels are increased in lung adenocarcinoma. N.D. indicates that the glycan composition was not determined in the analysis.
Figure 2.
Differential expression of glycan compositions in lung adenocarcinoma tissue compared to controls. Putative structures are shown for the glycans that are present at significantly different (FDR < 0.05) levels in the unreduced analysis. Glycans of which the levels are decreased in malignant tissue are shown on the left, whereas glycans of which the levels are increased in malignant tissue are shown on the right. Symbol key: blue square is N-acetylglucosamine, green ball is mannose, yellow ball is galactose, red triangle is fucose, and purple diamond is N-acetylneuraminic acid.
Differential Glycan Composition with Chemical Reduction of Glycans
The preceding results showed that glycan compositions were clearly differential in lung cancer tissue compared to controls; however, these compositions are often composed of several glycan structures (isomers). To further elaborate the glycan structures that are differentially expressed in lung adenocarcinoma tissue, the glycans were reduced to avoid separation of the reducing end anomers and subsequently reanalyzed using nLC–PGC–chip–TOF–MS. Unexpectedly, more glycan compositions were consistently observed in the reduced samples compared to those in the unreduced set (73 vs 45 compositions, respectively). The reason for the increase in composition is not yet clear. Unreduced glycans separate according to the anomeric character of the reducing end. Each compound is then separated into two forms. Reduction leaves the compound with a single structure, thereby decreasing the total number of peaks. The differences could be issues associated with the peak capacity. Further exploration is needed, but this does not affect the results.
The data set composed of reduced glycans was assessed at the level of glycan compositions to evaluate whether similar differential results were obtained as compared to the data set comprising unreduced glycans. The reduced glycan compositions were used to conduct a PLS-LDA analysis to assess whether the global glycomic profile could separate adenocarcinoma tissue from control. The results are summarized in Figure 1b. As with the unreduced analysis, separation between the cancer and control tissues is observed, indicating the differentiating potential of the reduced glycan compositions in adenocarcinoma.
The differential analysis results with and without adjustment for the covariates are shown in Table 2. Again, glycan compositions were observed to be significantly (FDR < 0.05) altered in cancerous tissue compared to controls in higher numbers compared to the unreduced samples (22 compositions vs 15 compositions, respectively). However, the results are essentially the same: The levels of several oligomannose type glycans were upregulated. Decreased levels of some hybrid-type glycans as well as fully galactosylated glycans mostly without fucose were observed, and levels of the N-glycan core (Hex3HexNAc2) with and without fucose as well as two fucosylated, low-galactosylated tetraantennary glycans were increased. These results show the similarities of the unreduced and reduced sets and indicate that the analysis of reduced glycans is feasible, thus allowing for glycan structure-specific determination of differential glycosylation in adenocarcinoma.
Differential Glycan Structures in Cancer Compared to Controls
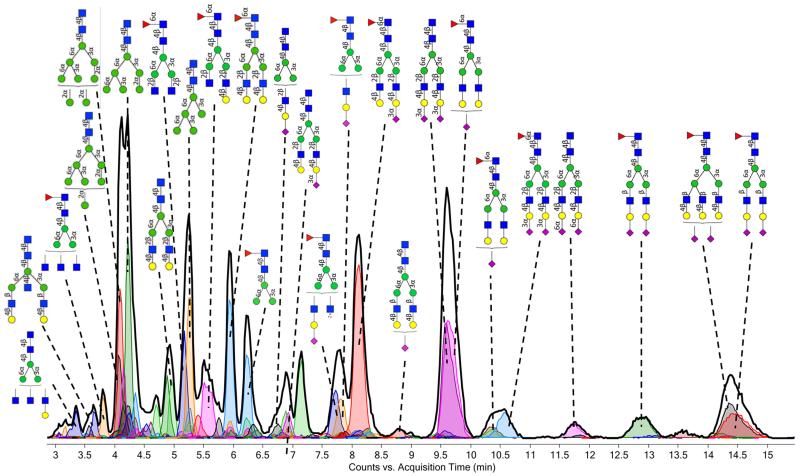
With the glycans reduced, each glycan structure is represented by one signal. This allows for the annotation of actual structures to each of the signals. Structural identification was performed by comparing retention times and accurate masses to an N-glycan database. Here, we used our in-house build library, which is based on serum N-glycans22 for the annotation of the tissue chromatograms. A typical chromatogram of a cancerous tissue with annotation of the higher-abundance glycan structures is shown in Figure 3. Although many of the glycan structures are similar to serum glycans, their relative abundances vary from those in serum. For example, high-mannose-type glycans are much more abundant in the tissue samples (both cancerous and control) compared to serum.21,25,29
Figure 3.
PGC–LC–MS chromatogram of reduced glycans from a malignant lung tissue sample. The extracted glycan chromatogram (chromatogram of all glycans summed) is shown in black, whereas extracted ion chromatograms of individual glycans are shown in color. The chromatograms have been annotated with actual structures, as obtained by comparing the retention times to our in-house built serum N-glycan library.22 For symbol key, see Figure 2.
A total of 115 glycan structures were consistently observed in the tissue samples. A PLS-LDA was performed using these structures. There was a clear, but not complete, separation between cancerous and control tissues (Figure 1c). The separation is similar to what was obtained using glycan compositions. This is further illustrated by an accuracy of classification of 82.0% using the glycan structures, whereas a 79.5% accuracy was obtained using the unreduced glycan compositions
A differential analysis was also preformed to identify individual glycan structures that show differential glycosylation patterns associated with adenocarcinoma. The results of the differential analyses are shown in Table 3. Of the 115 glycans consistently observed in more than 70% of the samples, 29 glycans were shown to be differentially expressed in cancerous tissue compared to controls regardless of adjusting for the covariates. Again, several oligomannose structures as well as structures with low levels of galactose were upregulated in cancerous tissue, whereas galactosylated structures of hybrid and complex type were downregulated. For several glycan compositions, multiple structures were shown to be significant. While the structures of the same composition were typically all altered in the same direction, the two glycans with composition Hex4HexNAc3Fuc1NeuAc1 were differentially expressed in opposite directions. Because of the opposite expression, composition Hex4HexNAc3Fuc1NeuAc1 was not significant in the compositional analysis, and the differential expression of these glycan structures would thus be missed in a compositional analysis. These results, therefore, show the potential of glycan structure-specific differentiation. The specific structures of the two differentially expressed glycans of composition Hex4HexNAc3Fuc1NeuAc1 are not yet known. Therefore, further structure elucidation studies will be necessary to evaluate the full biological effects of the different glycan isomers.
Table 3. Individual Glycan Structures That Are Differentially Expressed in Lung Adenocarcinoma Tissue.
| Composition | Average RT |
Structurea | Library entry |
Observed mass [da) |
theoretical mass (da) |
Delta M (ppm) |
Tumor Meanb |
Normal Meanb |
Ratioc | Unadjusted for covariates |
Adjusted for covariates |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P-value | FDR | P-value | FDR | ||||||||||
| H3N2F0S0 | 4.86 |
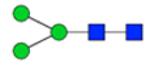
|
912.344 | 912.343 | 1.1 | 287152 | 164841 | 1.74 | 0.008 | 0.034 | 0.004 | 0.016 | |
| H3N2F0S0 | 3.64 |
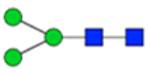
|
912.345 | 912.343 | 1.2 | 72518 | 28613 | 2.53 | <0.001 | 0.002 | <0.001 | <0.001 | |
| H3N2F1S0 | 6.19 |
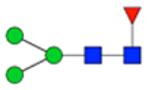
|
1058.401 | 1058.401 | −0.7 | 345819 | 150017 | 2.31 | 0.001 | 0.008 | <0.001 | 0.003 | |
| H4N2F0S0 | 4.51 |
|
1074.393 | 1074.396 | −3.4 | 54582 | 39713 | 1.37 | 0.004 | 0.019 | 0.002 | 0.012 | |
| H4N3F1S1 | 7.70 |
|
1714.632 | 1714.629 | 2.0 | 171879 | 113106 | 1.52 | 0.005 | 0.026 | 0.002 | 0.012 | |
| H4N3F1S1 | 11.07 |
|
1714.622 | 1714.629 | −4.0 | 15486 | 31287 | 0.49 | 0.001 | 0.006 | <0.001 | 0.003 | |
| H4N4F1S1 | 9.22 |
|
1917.710 | 1917.708 | 0.8 | 51559 | 98025 | 0.53 | 0.001 | 0.008 | 0.001 | 0.004 | |
| H4N6F1S0 | 5.86 |
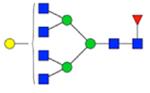
|
2032.739 | 2032.772 | −16.2 | 75633 | 43708 | 1.73 | 0.001 | 0.006 | <0.001 | 0.003 | |
| H5N3F0S0 | 4.14 |
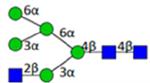
|
1439.529 | 1439.528 | 0.5 | 101534 | 137078 | 0.74 | 0.008 | 0.034 | 0.007 | 0.028 | |
| H5N4F0S0 | 3.15 |
|
N5400e | 1642.599 | 1642.608 | −5.5 | 83080 | 185213 | 0.45 | <0.001 | 0.001 | <0.001 | 0.001 |
| H5N4F0S0 | 3.50 |
|
1642.596 | 1642.608 | −6.9 | 44284 | 99390 | 0.45 | <0.001 | 0.001 | <0.001 | 0.001 | |
| H5N4F0S0 | 4.93 |
|
N5400a | 1642.609 | 1642.608 | 0.5 | 330335 | 485218 | 0.68 | 0.001 | 0.006 | 0.001 | 0.004 |
| H5N4F0S0 | 5.91 |
|
N5400C | 1642.613 | 1642.608 | 3.4 | 58345 | 101557 | 0.57 | <0.001 | 0.001 | <0.001 | 0.001 |
| H5N4F0S1 | 8.62 |
|
N5401d | 1933.715 | 1933.703 | 5.9 | 171516 | 356734 | 0.48 | 0.001 | 0.006 | <0.001 | 0.004 |
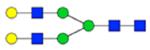
| H5N4F1S0 | 4.28 |
|
1788.675 | 1788.666 | 5.2 | 19550 | 38441 | 0.51 | <0.001 | 0.002 | <0.001 | 0.001 | |
| H5N4F1S0 | 5.89 |
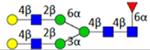
|
N5410a | 1788.672 | 1788.666 | 3.2 | 1059476 | 1623115 | 0.65 | 0.001 | 0.006 | 0.001 | 0.004 |
| H5N4F1S1 | 9.50 |
|
N5411e | 2079.761 | 2079.761 | −0.2 | 1359270 | 1981449 | 0.69 | 0.007 | 0.030 | 0.006 | 0.024 |
| H5N4F2S0 | 5.55 |
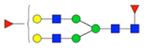
|
1934.746 | 1934.724 | 11.5 | 24148 | 12770 | 1.89 | 0.002 | 0.009 | 0.001 | 0.007 | |
| H5N4F2S1 | 8.59 |

|
2225.809 | 2225.819 | −4.5 | 22456 | 57641 | 0.39 | <0.001 | 0.002 | <0.001 | <0.001 | |
| H5N5F2S0 | 5.10 |
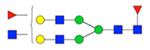
|
2137.816 | 2137.803 | 6.2 | 7940 | 24773 | 0.32 | <0.001 | 0.001 | <0.001 | <0.001 | |
| H6N2F0S0 | 4.21 |
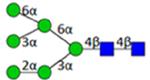
|
N6200a | 1398.502 | 1398.502 | 0.1 | 928055 | 655873 | 1.41 | 0.001 | 0.008 | <0.001 | 0.004 |
| H6N3F0S0 | 4.66 |
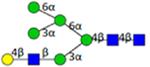
|
N6300a | 1601.582 | 1601.581 | 0.3 | 177960 | 240773 | 0.74 | 0.010 | 0.039 | 0.009 | 0.033 |
| H6N3F0S1 | 8.45 |

|
N6301C | 1892.682 | 1892.677 | 3.0 | 46392 | 92370 | 0.50 | 0.002 | 0.010 | 0.001 | 0.008 |
| H6N4F0S0 | 3.28 |
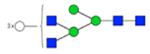
|
1804.659 | 1804.661 | −1.0 | 26884 | 67267 | 0.40 | <0.001 | 0.001 | <0.001 | 0.001 | |
| H6N4F0S0 | 3.81 |
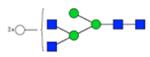
|
1804.665 | 1804.661 | 2.3 | 28505 | 79545 | 0.36 | <0.001 | <0.001 | <0.001 | <0.001 | |
| H6N5F0S0 | 5.37 |
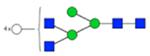
|
2007.740 | 2007.740 | 0.1 | 8220 | 12298 | 0.67 | 0.010 | 0.038 | 0.008 | 0.032 | |
| H6N5F1S0 | 7.55 |
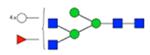
|
2153.797 | 2153.798 | −0.2 | 34884 | 61256 | 0.57 | 0.005 | 0.025 | 0.004 | 0.019 | |
| H7N2F0S0 | 4.07 |
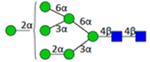
|
N7200a | 1560.551 | 1560.555 | −2.4 | 544399 | 403047 | 1.35 | 0.012 | 0.043 | 0.006 | 0.025 |
| H8N2F0S0 | 4.07 |
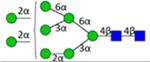
|
N8200a | 1722.601 | 1722.608 | −3.7 | 647154 | 475956 | 1.36 | 0.006 | 0.027 | 0.002 | 0.011 |
Symbol key: blue square is N-acetylglucosamine, green ball is mannose, yellow ball is galactose, red triangle is fucose, purple diamond is N-acetylneuraminic acid, and open ball is undefined hexose.
Mean ion intensities are reported for each individual glycan.
Ratio between the initial (control) and final (case) states. Ratios color-coded in green represent glycan compositions of which levels are decreased in lung adenocarcinoma, whereas those in red represent glycan compositions of which levels are increased in lung adenocarcinoma.
Expression of Glycosylation-Related Genes in Adenocarcinoma
Glycans are the product of glycosyltransferases; therefore, it is to be expected that differential glycan patterns are reflected in differential glycosyltransferase expression. To further assess this relationship, gene expression profiles from literature were compared to the glycomics analysis. Landi et al. previously performed gene expression analysis using HG-U133A Affymetrix chips on fresh frozen tissue samples of adenocarcinoma and paired noninvolved lung tissue.23 Data from 27 glycosyltransferases were extracted, which contained all enzymes relevant to N-glycan terminal processing that were available. Because Landi et al. presented no statistical analysis of the glycosyltransferases, we performed differential expression analysis. Of the 27 genes examined, 15 genes (MAN1A1, MAN1A2, MAN1C1, MAN2A1, MAN2A2, MGAT1, MGAT2, MGAT3, MGAT4B, B4GALT2, FUT1, FUT2, FUT3, FUT6, and FUT8) were found to be differentially expressed (FDR < 0.05) in the differential analyses, with and without adjustment for age and gender as covariates (Table 4).
Table 4. Differential Analysis of Gene Expression of N-Glycan Differentiating Genes in Paired Adenocarcinoma and Nonmalignant Tissue Samples.
| GENE | Unadjusted for covariates |
Adjusted for covariates | Means | Ratioa | |||
|---|---|---|---|---|---|---|---|
| P-Value | FDR | P-Value | FDR | Cancer | Normal | ||
| MAN1A1 | 0.005 | 0.009 | 0.003 | 0.006 | 365 | 527 | 0.69 |
| MAN1A2 | <0.001 | <0.001 | <0.001 | <0.001 | 98 | 68 | 1.43 |
| MAN1B1 | 0.264 | 0.264 | 0.247 | 0.247 | 223 | 197 | 1.13 |
| MAN1C1 | <0.001 | <0.001 | <0.001 | <0.001 | 273 | 414 | 0.66 |
| MAN2A1 | 0.011 | 0.014 | 0.004 | 0.006 | 881 | 610 | 1.44 |
| MAN2A2 | 0.011 | 0.014 | 0.005 | 0.006 | 195 | 248 | 0.79 |
| MGAT1 | <0.001 | 0.001 | <0.001 | <0.001 | 819 | 1064 | 0.77 |
| MGAT2 | <0.001 | <0.001 | <0.001 | <0.001 | 461 | 320 | 1.44 |
| MGAT3 | <0.001 | <0.001 | <0.001 | <0.001 | 57 | 91 | 0.62 |
| MGAT4A | 0.032 | 0.056 | 0.028 | 0.049 | 118 | 98 | 1.21 |
| MGAT4B | <0.001 | <0.001 | <0.001 | <0.001 | 466 | 334 | 1.40 |
| MGAT4C | 0.681 | 0.715 | 0.674 | 0.708 | 23 | 22 | 1.06 |
| MGAT5 | 0.593 | 0.655 | 0.584 | 0.646 | 16 | 18 | 0.90 |
| B4GALT1 | 0.145 | 0.203 | 0.136 | 0.190 | 154 | 127 | 1.21 |
| B4GALT2 | <0.001 | <0.001 | <0.001 | <0.001 | 308 | 191 | 1.61 |
| FUT1 | <0.001 | <0.001 | <0.001 | <0.001 | 141 | 223 | 0.63 |
| FUT2 | <0.001 | <0.001 | <0.001 | <0.001 | 183 | 87 | 2.12 |
| FUT3 | <0.001 | <0.001 | <0.001 | <0.001 | 253 | 90 | 2.83 |
| FUT4 | 0.287 | 0.344 | 0.276 | 0.331 | 107 | 117 | 0.92 |
| FUT5 | 0.295 | 0.344 | 0.284 | 0.331 | 7 | 6 | 1.20 |
| FUT6 | <0.001 | <0.001 | <0.001 | <0.001 | 206 | 138 | 1.49 |
| FUT7 | 0.109 | 0.163 | 0.101 | 0.151 | 20 | 15 | 1.28 |
| FUT8 | <0.001 | <0.001 | <0.001 | <0.001 | 650 | 229 | 2.84 |
| FUT9 | 0.026 | 0.050 | 0.023 | 0.044 | 26 | 19 | 1.37 |
| ST3GAL1 | 0.158 | 0.208 | 0.149 | 0.195 | 79 | 97 | 0.82 |
| ST3GAL2 | 0.946 | 0.946 | 0.944 | 0.944 | 53 | 54 | 0.99 |
| ST6GAL1 | 0.065 | 0.105 | 0.059 | 0.095 | 384 | 336 | 1.14 |
Ratio between the initial (control) and final (case) states. Ratios color-coded in green represent glycan compositions of which levels are decreased in lung adenocarcinoma, whereas those in red represent glycan compositions of which levels are increased in lung adenocarcinoma.
DISCUSSION
This article describes a detailed analysis of the differential N-glycosylation profile of early stage lung adenocarcinoma tissue compared to paired nonmalignant lung control tissue. Samples were chosen to evaluate the early indications of glycosylation changes due to tumorigenesis. Glycosylation profiles were first obtained from the native, unreduced glycans, but to allow for structural identification, a second analysis was performed upon reduction. This yielded 29 differentially expressed glycan structures. To further assess the regulation of the altered glycosylation, the glycomics data was compared to gene expression data from lung adenocarcinoma tissue compared to nonmalignant lung tissue, which was publicly available.23 While it would have been more ideal to have gene expression data on the same tissue samples, we did not have more tissue material available.
By analyzing unreduced glycans, we have previously been able to identify differential glycosylation patterns in the serum of ovarian cancer patients,21,30 lung cancer patients,29 and prostate cancer patients.25 However, structural identification is difficult when N-glycans are not reduced due to the separation of the reducing end anomers. In this study, we showed that the analysis of reduced N-glycans yields very similar, though extended, results, as more compositions can be observed. This resulted in more significantly different compositions (Table 2). Furthermore, the separation of reduced N-glycans using a PGC stationary phase in combination with our in-house N-glycan library allowed us to identify, for the first time, 29 glycan structures that were differentially expressed in lung cancer tissue. Notably, two glycans of composition Hex4HexNAc3Fuc1NeuAc1 were shown to be differentially expressed in the opposite direction (Table 3). The level of the overall composition was, therefore, not significantly altered (Table 2). This indicates the value of structure-specific analysis for the identification of differentiating glycans. However, when the predictive value of the three data sets (unreduced N-glycan compositions, reduced N-glycan compositions, and reduced N-glycan structures) was compared, they each yielded very similar results, indicating that for differential profiling (i.e., biomarker) studies a composition specific analysis is likely sufficient.
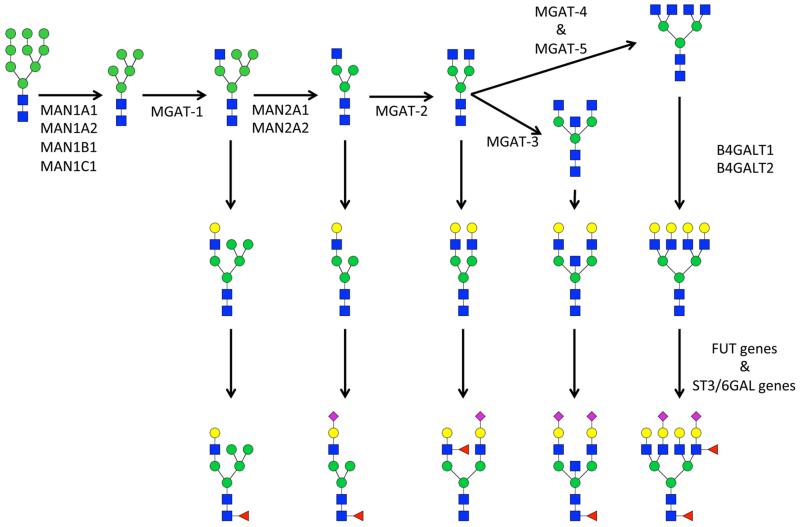
The glycomics analysis indicated that high-mannose-type glycans are consistently increased in lung adenocarcinoma tissue (Tables 2 and 3). Potentially, this could be caused by decreased levels of MGAT1, which indicate lower transformation of high-mannose-type glycans to hybrid-type glycans (Table 4 and Figure 4). Increased levels of high-mannose-type glycans have been previously associated with breast cancer progression.28 Levels of high-mannose-type glycans were also enhanced upon TGF-β-induced epithelial–mesenchymal transition in mouse mammary gland epithelial cells,31 indicating a possible role in cancer progression. However, in serum samples of ovarian cancer patients, high-mannose-type glycans were shown to be decreased.21,32 This is likely a reflection of the largely different protein profile of serum compared to tissue.
Figure 4.
Schematic overview of N-glycan processing catalyzed by carbohydrate acting enzymes in the Golgi. Enzymes involved in glycan processing have been included, but sugar nucleotide donors have not been included in this figure. For symbol key, see Figure 2.
The level of fucosylation, especially core fucosylation, was also increased in cancer compared to that in nonmalignant tissue (Table 2 and Figure 2). Comparison to previously established gene expression data indicated a possible regulatory role for FUT 8, as increased expression of FUT8, the fucosyltransferase that is known to catalyze the addition of the α1-6-linked fucose to the core GlcNAc,33 was observed (Figure 4 and Table 4). Increased levels of core fucosylation have previously been observed in sera of prostate cancer patients.34 Furthermore, increased gene expression levels of FUT8 have been observed in several cancers including lung,35 ovarian,36 thyroid,37 and colorectal,38 and that was associated with poor prognosis in colorectal cancer.38 Recently, a mechanism for the upregulation of FUT8 during the transition from epithelial to mesenchymal type cells was proposed,35 and it is anticipated that FUT8 or core fucosylated cancer-specific antigens could be excellent drug targets. Indeed, core fucosylated proteins have been proposed for use as biomarkers for hepatocellular carcinoma (HCC);39-41 however, a recent study did not find increased levels of core fucosylation in HCC tissue.42
In this study, we found levels of hybrid-type glycans to be decreased in lung adenocarcinoma tissue. This finding was supported by the decreased expression of MGAT1 and the increased expression of MGAT2 in adenocarcinoma tissue (Table 4), which would theoretically result in decreased levels of hybrid-type glycans (Figure 4). We previously reported decreased levels of four hybrid-type glycans in sera of ovarian cancer patients,21 but, to our knowledge, hybrid-type glycans have not typically been identified to be differentially expressed in cancer.
Galactosylation is well-known to be affected in serum of individuals with several inflammatory diseases; in particular, the galactosylation of immunoglobulin G is typically decreased.43-46 Here, we report decreased levels of galactosylation in the tissue samples of adenocarcinoma patients. The gene expression of one of the enzymes catalyzing the addition of a galactose to a GlcNAc residue was significantly increased (B4GALT2, see Table 4), whereas the expression of the other gene with that functionality was not altered (B4GALT1), and previous studies indicate that both transferases were upregulated in breast cancer tissue.9 However, the gene expression analysis was performed on different samples, which might explain this discrepancy. Gene expression levels do not always correlate with protein expression and protein activity levels, which could be a reason for the observed increase in gene expression and decrease of the final product. Another explanation might be that residual blood was left in the tissue even after thorough washing. It is well-known that lung tissue is well-perfused and that residual blood might remain. The decreased levels of galactosylated glycans found in the tissue samples could be a reflection of the decreased galactosylation previously reported on IgG.43-46 Furthermore, normal lung tissue is typically better perfused than tumor lung tissue, which might also contribute to the observation of decreased levels of typically high-abundant biantennary galactosylated glycans in blood.
Overall, this study describes, for the first time, actual glycan structures, in addition to compositions, that are differentially expressed in adenocarcinoma tissue compared to nonmalignant tissue from the same individual. These tissues not only consisted of tumor cells but also contained the surrounding stroma and other components of the tumor cell microenvironment. Glycomics changes may, therefore, partially reflect glycoproteins from the tumor microenvironment. Further studies will be necessary to identify proteins to which the glycans are attached. Such studies could be similar in nature to a recent report on the identification of the peptide moieties of differentially expressed glycopeptides in pancreatic tissue47 or could directly target glycopeptides. This will allow for a greater understanding of possible roles and functions of different glycosylation patterns in the development and progression of cancer. Glycan profiling of blood samples from these same lung cancer patients and in a large cohort of early stage patients with matched controls is ongoing. Data obtained from this study will be valuable in understanding the glycan composition in the blood and our quest to develop a biomarker for the detection of early stage lung cancer.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge financial support from the LUNGevity Foundation (no. 201118739) and the Tobacco Related Disease Research Program (no. 20PT0034). Funding provided by NIH grant nos. R01 GM049077 and U24 DK097154 is also gratefully acknowledged. The project described was further supported by the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), through grant no. UL1 TR000002.
ABBREVIATIONS
- FDR
false discovery rate
- GENT
gene expression database of normal and tumor tissues
- HCC
hepatocellular carcinoma
- LDCT
low dose spiral computerized tomography
- PGC
porous graphitized carbon
- PLS-LDA
partial least-squares regression with linear discriminant analysis
- PNGaseF
peptide-N-glycosidase F
- TOF
time-of-flight
- SPE
solid-phase extraction
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b00255.
Run order of the lung tissue samples on the nLC–TOF–MS (Table S1); plate layout used for the glycan analysis from lung adenocarcinoma tissue samples (Figure S1); quality control of the nLC–TOF–MS run showing the consistency during the MS runs (Figure S2); total ion chromatograms of the three low-intensity samples and three typical samples (Figure S3) (PDF).
The authors declare no conflict of interest.
The authors declare no competing financial interest.
REFERENCES
- (1).Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. Ca-Cancer J. Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- (2).The National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Pass HI, Beer DG, Joseph S, Massion P. Biomarkers and molecular testing for early detection, diagnosis, and therapeutic prediction of lung cancer. Thorac Surg Clin. 2013;23(2):211–24. doi: 10.1016/j.thorsurg.2013.01.002. [DOI] [PubMed] [Google Scholar]
- (4).Packer NH, von der Lieth CW, Aoki-Kinoshita KF, Lebrilla CB, Paulson JC, Raman R, Rudd P, Sasisekharan R, Taniguchi N, York WS. Frontiers in glycomics: bioinformatics and biomarkers in disease. An NIH white paper prepared from discussions by the focus groups at a workshop on the NIH campus, Bethesda MD (September 11-13, 2006) Proteomics. 2008;8(1):8–20. doi: 10.1002/pmic.200700917. [DOI] [PubMed] [Google Scholar]
- (5).Plzak J, Holikova Z, Smetana K, Jr., Dvorankova B, Hercogova J, Kaltner H, Motlik J, Gabius HJ. Differentiation-dependent glycosylation of cells in squamous cell epithelia detected by a mammalian lectin. Cells Tissues Organs. 2002;171(2-3):135–44. doi: 10.1159/000063707. [DOI] [PubMed] [Google Scholar]
- (6).Cheray M, Petit D, Forestier L, Karayan-Tapon L, Maftah A, Jauberteau MO, Battu S, Gallet FP, Lalloue F. Glycosylation-related gene expression is linked to differentiation status in glioblastomas undifferentiated cells. Cancer Lett. 2011;312(1):24–32. doi: 10.1016/j.canlet.2011.07.027. [DOI] [PubMed] [Google Scholar]
- (7).Wu YM, Liu CH, Huang MJ, Lai HS, Lee PH, Hu RH, Huang MC. C1GALT1 enhances proliferation of hepatocellular carcinoma cells via modulating MET glycosylation and dimerization. Cancer Res. 2013;73(17):5580–90. doi: 10.1158/0008-5472.CAN-13-0869. [DOI] [PubMed] [Google Scholar]
- (8).Hakomori S. Tumor-associated carbohydrate antigens defining tumor malignancy: basis for development of anti-cancer vaccines. Adv. Exp. Med. Biol. 2001;491:369–402. doi: 10.1007/978-1-4615-1267-7_24. [DOI] [PubMed] [Google Scholar]
- (9).Potapenko IO, Haakensen VD, Luders T, Helland A, Bukholm I, Sorlie T, Kristensen VN, Lingjaerde OC, Borresen-Dale AL. Glycan gene expression signatures in normal and malignant breast tissue; possible role in diagnosis and progression. Mol. Oncol. 2010;4(2):98–118. doi: 10.1016/j.molonc.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin. Chem. 2010;56(2):223–36. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Saldova R, Struwe WB, Wynne K, Elia G, Duffy MJ, Rudd PM. Exploring the glycosylation of serum CA125. Int. J. Mol. Sci. 2013;14(8):15636–54. doi: 10.3390/ijms140815636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Gilgunn S, Conroy PJ, Saldova R, Rudd PM, O’Kennedy RJ. Aberrant PSA glycosylation-a sweet predictor of prostate cancer. Nat. Rev. Urol. 2013;10(2):99–107. doi: 10.1038/nrurol.2012.258. [DOI] [PubMed] [Google Scholar]
- (13).Ruhaak LR, Miyamoto S, Lebrilla CB. Developments in the identification of glycan biomarkers for the detection of cancer. Mol. Cell. Proteomics. 2013;12(4):846–55. doi: 10.1074/mcp.R112.026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mechref Y, Hu Y, Garcia A, Hussein A. Identifying cancer biomarkers by mass spectrometry-based glycomics. Electrophoresis. 2012;33(12):1755–67. doi: 10.1002/elps.201100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wuhrer M. Glycomics using mass spectrometry. Glycoconjugate J. 2013;30(1):11–22. doi: 10.1007/s10719-012-9376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tharmalingam T, Adamczyk B, Doherty MA, Royle L, Rudd PM. Strategies for the profiling, characterisation and detailed structural analysis of N-linked oligosaccharides. Glycoconjugate J. 2013;30(2):137–46. doi: 10.1007/s10719-012-9443-9. [DOI] [PubMed] [Google Scholar]
- (17).Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14(4-5):525–46. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- (18).Balog CI, Stavenhagen K, Fung WL, Koeleman CA, McDonnell LA, Verhoeven A, Mesker WE, Tollenaar RA, Deelder AM, Wuhrer M. N-glycosylation of colorectal cancer tissues: a liquid chromatography and mass spectrometry-based investigation. Mol. Cell. Proteomics. 2012;11(9):571–85. doi: 10.1074/mcp.M111.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Chik JH, Zhou J, Moh ES, Christopherson R, Clarke SJ, Molloy MP, Packer NH. Comprehensive glycomics comparison between colon cancer cell cultures and tumours: Implications for biomarker studies. J. Proteomics. 2014;108C:146–162. doi: 10.1016/j.jprot.2014.05.002. [DOI] [PubMed] [Google Scholar]
- (20).Ruhaak LR, Taylor SL, Miyamoto S, Kelly K, Leiserowitz GS, Gandara D, Lebrilla CB, Kim K. Chip-based nLC-TOF-MS is a highly stable technology for large-scale high-throughput analyses. Anal. Bioanal. Chem. 2013;405(14):4953–8. doi: 10.1007/s00216-013-6908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kim K, Ruhaak LR, Nguyen UT, Taylor SL, Dimapasoc L, Williams C, Stroble C, Ozcan S, Miyamoto S, Lebrilla CB, Leiserowitz GS. Evaluation of glycomic profiling as a diagnostic biomarker for epithelial ovarian cancer. Cancer Epidemiol., Biomarkers Prev. 2014;23(4):611–21. doi: 10.1158/1055-9965.EPI-13-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Aldredge D, An HJ, Tang N, Waddell K, Lebrilla CB. Annotation of a serum N-glycan library for rapid identification of structures. J. Proteome Res. 2012;11(3):1958–68. doi: 10.1021/pr2011439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, Murphy SE, Yang P, Pesatori AC, Consonni D, Bertazzi PA, Wacholder S, Shih JH, Caporaso NE, Jen J. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lu J, Grenache DG. High-throughput tissue homogenization method and tissue-based quality control materials for a clinical assay of the intestinal disaccharidases. Clin. Chim. Acta. 2010;411(9-10):754–7. doi: 10.1016/j.cca.2010.02.009. [DOI] [PubMed] [Google Scholar]
- (25).Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136(18):3663–71. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kronewitter SR, An HJ, de Leoz ML, Lebrilla CB, Miyamoto S, Leiserowitz GS. The development of retrosynthetic glycan libraries to profile and classify the human serum N-linked glycome. Proteomics. 2009;9(11):2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–57. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).de Leoz ML, Young LJ, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB. High-mannose glycans are elevated during breast cancer progression. Mol. Cell Proteomics. 2011;10(1) doi: 10.1074/mcp.M110.002717. M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ruhaak LR, Nguyen UT, Stroble C, Taylor SL, Taguchi A, Hanash SM, Lebrilla CB, Kim K, Miyamoto S. Enrichment strategies in glycomics-based lung cancer biomarker development. Proteomics Clin Appl. 2013;7:664–76. doi: 10.1002/prca.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hua S, Williams CC, Dimapasoc LM, Ro GS, Ozcan S, Miyamoto S, Lebrilla CB, An HJ, Leiserowitz GS. Isomer-specific chromatographic profiling yields highly sensitive and specific potential N-glycan biomarkers for epithelial ovarian cancer. J. Chromatogr A. 2013;1279:58–67. doi: 10.1016/j.chroma.2012.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Tan Z, Lu W, Li X, Yang G, Guo J, Yu H, Li Z, Guan F. Altered N-Glycan Expression Profile in Epithelial-to-Mesenchymal Transition of NMuMG Cells Revealed by an Integrated Strategy Using Mass Spectrometry and Glycogene and Lectin Microarray Analysis. J. Proteome Res. 2014;13(6):2783–95. doi: 10.1021/pr401185z. [DOI] [PubMed] [Google Scholar]
- (32).Kronewitter SR, De Leoz ML, Strum JS, An HJ, Dimapasoc LM, Guerrero A, Miyamoto S, Lebrilla CB, Leiserowitz GS. The glycolyzer: Automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics. 2012;12(15-16):2523–38. doi: 10.1002/pmic.201100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Yanagidani S, Uozumi N, Ihara Y, Miyoshi E, Yamaguchi N, Taniguchi N. Purification and cDNA cloning of GDP-L-Fuc:N-acetyl-beta-D-glucosaminide:alpha1–6 fucosyltransferase (alpha1–6 FucT) from human gastric cancer MKN45 cells. J. Biochem. 1997;121(3):626–32. doi: 10.1093/oxfordjournals.jbchem.a021631. [DOI] [PubMed] [Google Scholar]
- (34).Saldova R, Fan Y, Fitzpatrick JM, Watson RW, Rudd PM. Core fucosylation and alpha2–3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology. 2011;21(2):195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- (35).Chen CY, Jan YH, Juan YH, Yang CJ, Huang MS, Yu CJ, Yang PC, Hsiao M, Hsu TL, Wong CH. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2013;110(2):630–5. doi: 10.1073/pnas.1220425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Takahashi T, Ikeda Y, Miyoshi E, Yaginuma Y, Ishikawa M, Taniguchi N. alpha1,6fucosyltransferase is highly and specifically expressed in human ovarian serous adenocarcinomas. Int. J. Cancer. 2000;88(6):914–9. doi: 10.1002/1097-0215(20001215)88:6<914::aid-ijc12>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- (37).Ito Y, Miyauchi A, Yoshida H, Uruno T, Nakano K, Takamura Y, Miya A, Kobayashi K, Yokozawa T, Matsuzuka F, Taniguchi N, Matsuura N, Kuma K, Miyoshi E. Expression of alpha1,6-fucosyltransferase (FUT8) in papillary carcinoma of the thyroid: its linkage to biological aggressiveness and anaplastic transformation. Cancer Lett. 2003;200(2):167–72. doi: 10.1016/s0304-3835(03)00383-5. [DOI] [PubMed] [Google Scholar]
- (38).Muinelo-Romay L, Vazquez-Martin C, Villar-Portela S, Cuevas E, Gil-Martin E, Fernandez-Briera A. Expression and enzyme activity of alpha(1,6)fucosyltransferase in human colorectal cancer. Int. J. Cancer. 2008;123(3):641–6. doi: 10.1002/ijc.23521. [DOI] [PubMed] [Google Scholar]
- (39).Comunale MA, Rodemich-Betesh L, Hafner J, Wang M, Norton P, Di Bisceglie AM, Block T, Mehta A. Linkage specific fucosylation of alpha-1-antitrypsin in liver cirrhosis and cancer patients: implications for a biomarker of hepatocellular carcinoma. PLoS One. 2010;5(8):e12419. doi: 10.1371/journal.pone.0012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Comunale MA, Wang M, Hafner J, Krakover J, Rodemich L, Kopenhaver B, Long RE, Junaidi O, Bisceglie AM, Block TM, Mehta AS. Identification and development of fucosylated glycoproteins as biomarkers of primary hepatocellular carcinoma. J. Proteome Res. 2009;8(2):595–602. doi: 10.1021/pr800752c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, Block TM, Mehta AS. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol., Biomarkers Prev. 2009;18(6):1914–21. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mehta A, Norton P, Liang H, Comunale MA, Wang M, Rodemich-Betesh L, Koszycki A, Noda K, Miyoshi E, Block T. Increased Levels of Tetra-antennary N-Linked Glycan but Not Core Fucosylation Are Associated with Hepatocellular Carcinoma Tissue. Cancer Epidemiol., Biomarkers Prev. 2012;21(6):925–33. doi: 10.1158/1055-9965.EPI-11-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Parekh RB, Dwek RA, Sutton BJ, Fernandes DL, Leung A, Stanworth D, Rademacher TW, Mizuochi T, Taniguchi T, Matsuta K, et al. Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature. 1985;316(6027):452–7. doi: 10.1038/316452a0. [DOI] [PubMed] [Google Scholar]
- (44).Huhn C, Selman MH, Ruhaak LR, Deelder AM, Wuhrer M. IgG glycosylation analysis. Proteomics. 2009;9(4):882–913. doi: 10.1002/pmic.200800715. [DOI] [PubMed] [Google Scholar]
- (45).Selman MHJ, Niks EH, Titulaer MJ, Verschuuren JJGM, Wuhrer M, Deelder AM. IgG Fc N-Glycosylation Changes in Lambed-Eaton Myasthenic Syndrome and Myasthenia Gravis. J. Proteome Res. 2011;10(1):143–152. doi: 10.1021/pr1004373. [DOI] [PubMed] [Google Scholar]
- (46).Bondt A, Selman MH, Deelder AM, Hazes JM, Willemsen SP, Wuhrer M, Dolhain RJ. Association between galactosylation of immunoglobulin G and improvement of rheumatoid arthritis during pregnancy is independent of sialylation. J. Proteome Res. 2013;12(10):4522–31. doi: 10.1021/pr400589m. [DOI] [PubMed] [Google Scholar]
- (47).Pan S, Chen R, Tamura Y, Crispin DA, Lai LA, May DH, McIntosh MW, Goodlett DR, Brentnall TA. Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J. Proteome Res. 2014;13(3):1293–306. doi: 10.1021/pr4010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.