Abstract
Francisella species are Gram-negative, nonmotile, pleomorphic coccobacilli, facultative intracellular fastidious bacteria. We report the isolation of a Francisella-like species from a blood culture collected from a 44-year-old bacteraemic patient in Perth, Western Australia. The organism was identified to species level by 16S rRNA sequencing and by fatty acid methyl esters analysis. The strain genotypically resembled Francisella hispaniensis, a species previously isolated from human blood in Spain.
Keywords: Fatty acids, Francisella, gene sequencing, sepsis, taxonomy
Case report
A 44-year-old woman was admitted for investigation of an acute febrile illness. She had a history of a liver transplant over a decade previously for Budd-Chiari syndrome and was currently receiving renal dialysis via an arteriovenous fistula. She was receiving prednisone and sirolimus before the onset of fever. Because she had a cough at admission, she was investigated for a possible lower respiratory infection and treated with intravenous amoxicillin and oral azithromycin. A single oral dose of ciprofloxacin was provided on the day of admission. A series of cultures was collected, but the only one to yield a significant result was her blood culture, which grew a Gram-negative bacillus on the third day after admission. A further single dose of ciprofloxacin was provided orally, and the azithromycin was stopped and both intravenous cefepime and oral metronidazole were started. The patient defervesced steadily and was consistently afebrile 2 days after admission. At admission, temperature was 37.7°C. It rose to 38.6°C, then fell, with subsequent daily peak readings of 37.7, 36.8 and 36.9°C. The patient reported a recent family holiday on the northwestern coast of the state, when she had had a minor abrasion from a fishhook, but no subsequent signs of local infection there.
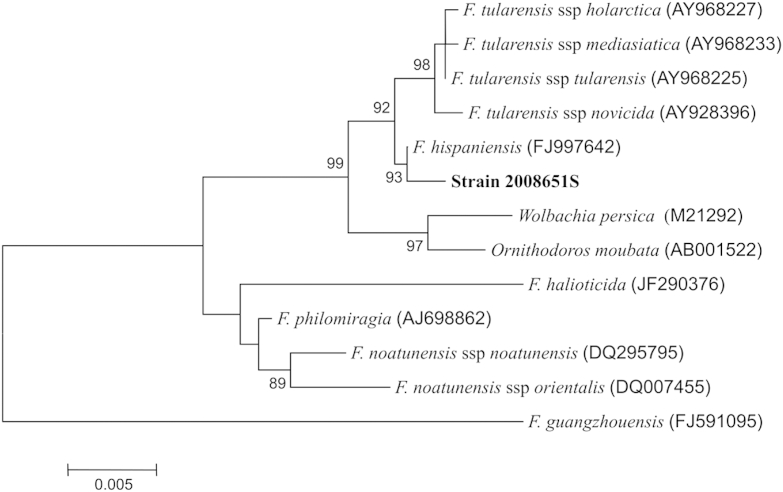
The isolate grew on chocolate and horse blood agars without additives. The morphology of the isolate suggested a Haemophilus-like microorganism, but no identification was obtained with the Vitek 2 NHI card or API NH ID commercial systems (bioMérieux, France). The fatty acid methyl esters (FAME) composition was determined by the MIDI-Inc ID system [1] which identified the isolate as F. tularaemia with a similarity index of 0.504. In this system, a similarity index of >0.500 suggests a good identification. The taxonomic position of strain 2008651S was further confirmed by high-throughput sequencing on an Ion Torrent PGM (Life Technologies, USA). Briefly, DNA was extracted from strain 2008651S by DNA mini kit (Qiagen, USA) before library preparation and sequencing using Ion Xpress Plus Fragment Library Kit, Ion PGM 200 Xpress Template Kit V2 and Ion PGM 200 sequencing kit on a Ion 318 sequencing chip according to manufacturer instructions. 16S rRNA sequence was assembled by mapping reads to complete 16S rRNA sequence from Francisella tularensis subsp. tularensis SCHU S4 (NC_006570), Francisella tularensis subsp. novicida U112 (NC_008601) and Francisella cf. novicida 3523 (NC_017449) [2], the last of which was more recently reclassified as Francisella hispaniensis [3]. All mapping runs produced identical consensus sequences for each target. A phylogenetic tree derived from assembled 16S rRNA sequences revealed that strain 2008651S indeed belonged to F. hispaniensis and that there was a 99.8% nucleotide identity between the 16S rRNA gene sequence of strain 2008651S and that of Francisella hispaniensis. These sequences were further aligned with the 16S rRNA sequences of F. tularensis spp. tularensis, F. tularensis ssp. holarctica, F. tularensis spp. mediasiatica, F. tularensis ssp. novicida, F. philomiragia, F. noatunensis ssp. noatunensis, F. noatunensis spp. orientalis, Wolbachia persica, Ornithodoros moubata and the recently proposed F. guangzhouensis [4] (Fig. 1) using the online Clustal Omega Software (http://www.ebi.ac.uk/Tools/msa/clustalo). Evolutionary distances were calculated by Kimura's two-parameter model [5] with the MEGA 3.1 software package (http://www.megasoftware.net) [6]. Phylogenetic trees were constructed using the neighbour-joining method [7].
Fig. 1.
Neighbour-joining phylogenetic tree derived from 16S rRNA sequences showing position of strain 2008651S (1419 nt). Bar = 0.005 substitutions per nucleotide.
Although strain 2008651S shared the unique FAME profiles of Francisella species, it differed from those described for F. hispaniensis FhSp1T [3]. For example, the percentage of decanoic acid (C10:0) was nearly three times larger (54.3%) in strain 2008651S compared to 17.2% detected in F. hispaniensis FhSp1T. Moreover, tetracosanoic (C24:0) and tetradecenoic (C24:1) acids were absent in strain 2008651S but were present in F. hispaniensis FhSp1T at 2.5% and 5.0%, respectively (Table 1). Furthermore, the metabolic fingerprinting determined for strain 2008651S by the Biolog GN2 MicroPlate (Fisher Scientific) according to manufacturer's instructions, differed from the fingerprinting obtained for F. hispaniensis FhSp1T [3]. The strain was tested in duplicate, and Escherichia coli ATCC 25922 was used as a control. Strain 2008651S was unreactive after overnight incubation at 35°C, while F. hispaniensis FhSp1T was remarkably active in this system; with the exception of maltose and succinamic acid, all other tests were positive [3]. These results suggest that although strain 2008651S is genotypically similar to F. hispaniensis FhSp1T, phenotypical differences exist between these two strains (Table 2).
Table 1.
FAME composition of strain 2008651S and Francisella hispaniensis ATCC 15482
| Fatty acid | Strain 2008651S (%) | F. hispaniensis ATCC 15482 |
|---|---|---|
| C10:0 | 54.3 | 17.2 |
| C10:0 2-OH | 1.31 | 2.7 |
| C14:0 | 14.0 | 11.2 |
| iso C15:0 | — | 1.1 |
| iso C15:0 2OH and/or C16:1 ωtc | — | 1.4 |
| anteiso C15:0 | — | 1.6 |
| C16:0 | 9.0 | 13.1 |
| C16:0 3-OH | 3.0 | 2.8 |
| C18:0 | 2.8 | 3.8 |
| C18:0 3-OH | 8.5 | 14.2 |
| C18:1 ω9c | 5.1 | 7.1 |
| C20:0 | — | 1.7 |
| C22:0 | — | 2.5 |
| C24:0 | — | 2.5 |
| C24:1 | — | 5.0 |
FAME, fatty acid methyl ester; t, trace (<1%).
Data for F. hispaniensis ATCC 15482 obtained from Huber et al.[3].
Table 2.
Biochemical characteristics of strain 2008651S and Francisella hispaniensis FhSp1
| Characteristic | Strain 2008651S | F. hispaniensisa FhSp1T |
|---|---|---|
| Catalase | − | (+) |
| Oxidase | + | + |
| Growth on MacConkey agar | − | − |
| Indole | − | − |
| H2S production | + | − |
| Vogues-Proskauer | + | − |
| Nitrate reduction | − | − |
| Growth on triple-sugar iron agar | + | − |
| Hydrolysis of: | ||
| Gelatin | − | − |
| Aesculin | − | − |
| Urease hydrolysis | − | − |
| Ornithine decarboxylase | +b | − |
| β-Galactosidase | − | − |
| Oxidation of: | ||
| Glucose | +b | + |
| Fructose | +b | + |
| Maltose | −c | − |
| Sucrose | − | + |
F. hispaniensis FhSp1T obtained from Huber et al.[3].
From API NH strip.
Positive reaction in NH Vitek 2 card.
The GenBank accession number for the partial 16S rRNA gene sequence of Francisella species strain 2008651S is KT281843.
Discussion
Francisella, the agent of tularaemia, a zoonotic infection that affects humans and animals, is highly infectious, while other species are considered less virulent. Traditionally, tularaemia has been classified as type A, associated with F. tularemia ssp. tularemia, and type B, caused by F. tularemia ssp. holarctica, with the latter considered to be endemic in the Northern Hemisphere [8]. Huber et al. [3] first described F. hispaniensis from the blood of a Spanish patient, which, together with this report, suggests that this species is a potential human pathogen. The presence of these organisms in air-conditioning systems [4], abalone [9] and fish [10] may also represent alternative reservoirs with the potential to cause infection in the human population. Though Legionella infections are well known to be associated with air-conditioning systems, there is less awareness of the importance of these environmental reservoirs to infections by other facultative intracellular bacteria. The patient's travel in the north of this state may indeed have led to an exposure to air-conditioning systems, and the history of a minor fishhook injury is intriguing. However, the patient's ongoing treatment with immunosuppressive therapy and her chronic comorbidities suggest that this specific Francisella isolate caused an opportunistic infection after undetermined exposure. Nevertheless, the preliminary identification of a presumptive Francisella species led to mandatory sample processing in a secure level 3 biosafety laboratory, which slowed down identification. Because the patient responded to empirical therapy, no antimicrobial susceptibility testing was performed.
Currently the genus comprises six species and six subspecies [4], [9], [10]. A review of the literature revealed that Francisella species have previously been isolated from other Australian states [11], [12]. The more recent report described the isolation of F. tularensis ssp. holarctica from a woman bitten by a wild ringtail possum in Tasmania. Coincidentally, our patient was also a 44-year-old woman [12]. It is possible that the patient in this study contracted the strain after being bitten while fishing. Jackson et al. [12] in 2011 suggested that infection of the Tasmanian patient was not the result of the importation of wildlife, changes in ecologic and climate conditions or in settings of postwar social disruption. It is highly likely that the same applies in this case report.
In conclusion, this is the first case of bacteraemia due to F. hispaniensis in Western Australia. We concur with Jackson et al. [12] that the isolation of Francisella spp. from human clinical material should alert clinicians and veterinarians of the possibility of human infection with these organisms. From the taxonomic point of view, the definitive position of Francisella, Wolbachia and Ornithodoros species is in need of revision.
Conflict of interest
None declared.
References
- 1.Paisley R. MIDI-Inc.; Newark, DE, USA: 1999. Training manual: MIS whole cell fatty acid analysis by gas chromatography. [Google Scholar]
- 2.Sjödin A., Svensson K., Ohrman C., Ahlinder J., Lindgren P., Duodo S. Genome characterisation of the genus Francisella reveals insight into similar volutionary paths in pathogens of mammals and fish. BMC Genomics. 2012;22:13–268. doi: 10.1186/1471-2164-13-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huber B., Escudero R., Busse H.J., Seibold E., Scholtz H.C., Anda P. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al. 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int J Syst Evol Microbiol. 2010;760:1887–1896. doi: 10.1099/ijs.0.015941-0. [DOI] [PubMed] [Google Scholar]
- 4.Qu P.H., Chen S.Y., Scholtz H.C., Busse H.J., Gu Q., Käempfer P. Francisella guangzhouensis sp. nov isolated from air-conditioning systems. Int J Syst Evol Microbiol. 2013;63:3628–3635. doi: 10.1099/ijs.0.049916-0. [DOI] [PubMed] [Google Scholar]
- 5.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 6.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 8.Keim P., Johansson A., Wagner D.M. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 9.Brevik Ø.J., Ottem K.F., Kamaishi T., Watanabe K., Nylund A. Francisella halioticida sp. nov., a pathogen of farmed giant abalone (Haliotis gigantean) in Japan. J Appl Microbiol. 2011;111:1044–1056. doi: 10.1111/j.1365-2672.2011.05133.x. [DOI] [PubMed] [Google Scholar]
- 10.Ottem K.F., Nylund A., Karlsbakk E., Friis-Møller A., Krossøy B., Knappskog D. New species in the genus Francisella (Gammaproteobacteria; Francisellae); Francisella piscicida sp. nov. isolated from cod (Gadus morhua) Arch Microbiol. 2007;188:547–550. doi: 10.1007/s00203-007-0274-1. [DOI] [PubMed] [Google Scholar]
- 11.Whipp M.J., Davis J.M., Lum G., de Boer J., Zhou Y., Bearden S.W. Characterization of a novicida-like subspecies of Francisella tularensis isolated in Australia. J Med Microbiol. 2003;52:839–842. doi: 10.1099/jmm.0.05245-0. [DOI] [PubMed] [Google Scholar]
- 12.Jackson J., McGregor A., Cooley L., Ng J., Brown M., Ong C.W. Francisella tularensis subspecies holarctica, Tasmania, Australia, 2011. Emerg Infect Dis. 2011;18:1484–1486. doi: 10.3201/eid1809.111856. [DOI] [PMC free article] [PubMed] [Google Scholar]