Abstract
A total of 189 Acinetobacter baumannii isolates were collected in 2011 from a teaching hospital in Chongqing, China. Susceptibility data showed strains carrying integrons were significantly more resistant to all tested antibiotics that strains lacking integrons. Five types of gene cassettes belonging to class I integrons were identified in this study, and for the first time two types of gene cassettes belonging to class II integrons are reported. Most of the cassettes belong to a class I integron (136/144) encoding arr3, aacA4, dfrA17, aadA5, aadB, cat, blaOXA10, aadA1, aadA2, dfrA and aacC1. Isolates contained a class I gene cassette; AadA2-HP-dfrA was the prevalent strain in this hospital. A class II integron was detected in eight strains, which contained the type IV fimbriae expression regulatory gene pilR and sulfate adenylyltransferase, suggesting a possible role in multidrug resistance. The major epidemic strains from intensive care unit patients belong to international clone 2. In conclusion, the presence of integrons was significantly associated with multiple drug resistance of A. baumannii in this hospital, and class I integron isolates bearing AadA2-HP-dfrA were the prevalent strain in this hospital.
Keywords: Acinetobacter baumannii, drug resistance, integron, integron cassettes, Intensive Care Unit
Introduction
Acinetobacter baumannii is an important cause of nosocomial infection in intensive care units (ICUs) and is often resistant to multiple antibiotics [1], [2], [3]. Integrons are partly responsible for the multiantibiotic resistance of A. baumannii. Integrons are genetic elements that can integrate, by site-specific recombination, antibiotic-resistant gene cassettes [4]. Five main types of integrons have been described based the basis of the sequence of the encoded 5′ integrases (40–58% identity), including class I (Tn402 derivatives), class II (Tn7 derivatives), class III (located in a transposon), class IV (a subset of conjugative transposon in Vibrio cholerae) and class V (located in a compound transposon carried on a plasmid in Vibrio salmonicida) [5], [6]. All the integrons have a 5′ conserved segment, including an intI gene encoding an integrase and an attI recombination site, but have distinct 3′ conserved segments. Gene cassettes from class I integrons confer resistance to all known β-lactams, all aminoglycosides, chloramphenicol, trimethoprim, streptothricin, rifampin, erythromycin, fosfomycin, lincomycin and antiseptics of the quaternary–ammonium compound family [7], while fewer resistance cassettes (dfrA1, sat1, sat2, aadB, blaCARB-4, aadA1) have been found that were associated with class II integrons [8]. In China, integrons containing drug-resistant cassettes were considered to be a major resistant element [9], [10], with an integron detection ratio ranging from 52.3% to 69.6%.
Similar to reports from other hospitals in China, A. baumannii and its colonization have greatly increased at the Second Affiliated Hospital of Chongqing Medical University during the last several years. The mechanism by which it is doing this needs to be investigated. The aim of this study was to explore the roles, characteristics and frequency of integrons in A. baumannii drug resistance development at our hospital.
Materials and Methods
Isolates, antimicrobial agents susceptibility testing and data analysis
Between January 2011 and December 2011, a total of 189 nonrepetitive clinical isolates of Acinetobacter baumannii were collected from the laboratory of the Second Affiliated Hospital of Chongqing Medical University. A. baumannii multidrug-resistant isolates are resistant to more than two of the following five drug classes: penicillins (piperacillin, ampicillin/sulbactam or ticarcillin/clavulanic acid), cephalosporins (cefepime, ceftriaxone, cefotaxime or ceftazidime), carbapenems (imipenem), trimethoprim/sulfame–thoxazole, fluoroquinolones (ciprofloxacin, levofloxacin or gatifloxacin), and aminoglycosides (gentamicin, tobramycin or amikacin) [11]. Minimum inhibitory concentrations were determined by bioMérieux Vitek 2 AST-GN 13 cards (http://www.biomerieux-diagnostics.com/vitek-2-ast-cards) or MicroScan Dried Gram Negative Combo 31 panels (https://www.beckmancoulter.com/wsrportal/WSR/diagnostics/clinical-products/microbiology/microscan-panels/index.htm) with the relevant instruments. Our routine quality control method was in accordance with the manufacturer's guidelines. The breakpoints of various antibiotics to A. baumannii followed Clinical and Laboratory Standards Institute guidelines. The susceptibility differences among A. baumannii strains were analysed by the WHONET 5.6 program (World Health Organization; http://www.who.int/drugresistance/whonetsoftware/en/). The chi-square test was used to determine the significance of differences. A difference was considered statistically significant if the p value was less than 0.05.
Integron detection
Primers for class I, II and III integrons identification are listed in Table 1. PCR amplification parameters have been reported previously [12]. One or two cassette amplicons representative of each size were selected for DNA sequence characterization. The resulting DNA sequences were analysed by the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/).
Table 1.
Primers for integron identification
Molecular typing and multilocus sequence typing (MLST) analysis of ICU strains
For the 33 strains from patients in the ICU, repetitive PCR testing (rep-PCR) was carried out to analyse the endemic group and additional sporadic isolates [15]. MLST analyses were performed to characterize the epidemic features of ICU-isolated strains [16]. The c scheme and protocols are publicly available online (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html).
Results
Antimicrobial susceptibility testing
The proportion of isolates resistant to tested antimicrobial agents included piperacillin (92.7%), ampicillin/sulbactam (84.5%), ticarcillin/clavulanic acid (80.4%), ceftazidime (88.7%), ceftriaxone (90.2%), cefotaxime (92.2%), cefepime (93.8%), imipenem (72.2%), amikacin (78.8%), gentamicin (81.4%), tobramycin (76.8%), ciprofloxacin (89.7%), gatifloxacin (62.6%), levofloxacin (67%) and trimethoprim/sulfame–thoxazole (81.4%). The majority (93.3%) of the isolates in this study were multidrug resistant (resistant to more than two categories of antibiotic), which was higher than the reported ratio [17], and 38.7% of the isolates were resistant to all antibiotics used in our test.
Integron assessment of A. baumannii isolates
Integrons were widely distributed among clinically isolated A. baumannii strains in this teaching hospital in Chongqing, China. Among the 189 isolates analysed by PCR, 144 isolates (76.2%) were found to have the integron cassette; it was absent in the rest. Among the integron-positive isolates, 136 had a class I integron cassette and eight had a class II integron cassette. We did not detect class III integrons in isolates from this hospital.
Mapping of integrons
The cassette assortments in all the strains were characterized by sequencing the amplification products (Table 2). Among the 128 class I integron-carrying isolates, five different types of gene cassettes in segments of approximately 1.3, 1.6, 2, 2.5 and 3.1 kb between the 5′ conserved segment and the 3′ conserved segment regions were amplified by PCR. The five types of class I integron cassettes included: aadA2-HP-dfrA, aacC1-ORF1-ORF2-aadA1, arr-3-aacA4, dfrA17-aadA5 and aadB-cat-blaOXA10-aadA1. The cassettes of aadA2-HP-dfrA accounted for 83.9% of all integron-positive cassettes, making it was the most prevalent type in the integron-carrying A. baumannii isolate. There were two types of class II cassettes, comprising type 4 fimbriae expression regulatory protein pilR and sulfate adenylyltransferase.
Table 2.
Integron cassette types and contents of Acinetobacter baumannii clinical isolates
| Integron type | Coding length | No. of isolates | Function annotation |
|---|---|---|---|
| Class I | 1332 | 2 | Arr-3 and aacA4 |
| Class I | 1606 | 2 | DfrA17-aadA5 |
| Class I | 3124 | 2 | AadB-cat-blaOXA10-aadA1 |
| Class I | 1998 | 135 | AadA2-HP-dfrA |
| Class I | 2513 | 11 | aacC1-ORF1-ORF2- aadA1 |
| Class II | 480 | 5 | Type 4 fimbriae expression regulatory protein, pilR |
| Class II | 1480 | 4 | Sulfate adenylyltransferase |
dfrA12, dihydrofolate reductase; HP, hypothetical protein (HP); aadA2, adenyltransferase; rifampin ADP-ribosylating transferase (arr-3) and aminoglycoside 6′-N-acetyltransferase (aacA4) genes, aminoglycoside 2′-O-adenylyltransferase (aadB), chloramphenicol acetyltransferase (cat), oxacillinase (blaoxa-10), and aminoglycoside 3′- (9)-O-adenylyltransferase (aadA1) genes.
Correlation between integron and drug resistance
We compared susceptibility data from integron-positive and -negative A. baumannii isolates (Table 3). The chi-square test was used to calculate the p value in terms of resistant and susceptible numbers of integron-positive and -negative isolates. Integron presence was significantly associated with resistance to certain antibiotics. The resistance ratio to amikacin, ampicillin/sulbactam, trimethoprim/sulfame–thoxazole, ciprofloxacin, gatifloxacin, piperacillin, gentamicin, ticarcillin/clavulanic acid, cefepime, ceftriaxone, cefotaxime, ceftazidime, tobramycin, imipenem and levofloxacin were detected in integron-positive A. baumannii strains than in integron-negative strains (p < 0.01).
Table 3.
Association between antibiotic profile and integrons in Acinetobacter baumannii strains
| Antibiotic | Antibiotic susceptibility |
Integron-positive isolates |
Integron-negative isolates |
p | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| %R | %I | %S | %R | %I | %S | %R | %I | %S | ||
| AMK | 78.6 | 2.3 | 19.1 | 91 | 0.8 | 8.3 | 35.7 | 7.1 | 57.1 | <0.005 |
| SAM | 84 | 4.8 | 11.2 | 92.4 | 2.1 | 5.6 | 55.6 | 13.3 | 31.1 | <0.005 |
| SXT | 81.3 | 0 | 18.7 | 91 | 0 | 9 | 46.7 | 0 | 53.3 | <0.005 |
| CIP | 89.3 | 1.1 | 9.6 | 95.8 | 0 | 4.2 | 66.7 | 4.4 | 28.9 | <0.005 |
| GAT | 61.8 | 23.1 | 15 | 68.4 | 25.6 | 6 | 40.5 | 14.3 | 45.2 | <0.005 |
| PIP | 92.5 | 3.5 | 4 | 97.7 | 0.8 | 1.5 | 76.2 | 11.9 | 11.9 | <0.005 |
| GEN | 81.3 | 3.2 | 15.5 | 91 | 2.1 | 6.9 | 46.7 | 6.7 | 46.7 | <0.005 |
| TIM | 79.8 | 7.5 | 12.7 | 85.7 | 9 | 5.3 | 59.5 | 2.4 | 38.1 | <0.005 |
| FEP | 93.6 | 1.6 | 4.8 | 96.5 | 1.4 | 2.1 | 84.4 | 2.2 | 13.3 | <0.005 |
| CRO | 89.8 | 7.5 | 12.7 | 96.5 | 2.1 | 1.4 | 68.9 | 24.4 | 6.7 | <0.005 |
| CTX | 91.9 | 5.8 | 2.3 | 97 | 1.5 | 1.5 | 76.2 | 19 | 4.8 | <0.005 |
| CAZ | 88.8 | 2.7 | 8.6 | 95.8 | 0.7 | 3.5 | 66.7 | 8.9 | 24.4 | <0.005 |
| TOB | 76.5 | 2.1 | 21.4 | 86.8 | 1.4 | 11.8 | 40 | 4.4 | 55.6 | <0.005 |
| IPM | 71.1 | 1.1 | 27.8 | 75.7 | 0.7 | 23.6 | 55.6 | 2.2 | 42.2 | <0.01 |
| LVX | 65.8 | 20.9 | 13.4 | 68.1 | 25.7 | 6.2 | 45.7 | 10.4 | 43.9 | <0.005 |
%R, resistant rate; %I, intermediate rate; %S, sensitive rate; AMK, amikacin; CAZ, ceftazidime; CIP, ciprofloxacin; CRO, ceftriaxone; CTX, cefotaxime; FEP, cefepime; GAT, gatifloxacin; GEN, gentamicin; IPM, imipenem; LVX, levofloxacin; PIP, piperacillin; SAM, ampicillin/sulbactam; SXT, trimethoprim/sulfame–thoxazole; TIM, ticarcillin/clavulanic acid; TOB, tobramycin.
Molecular typing of ICU-isolated strains
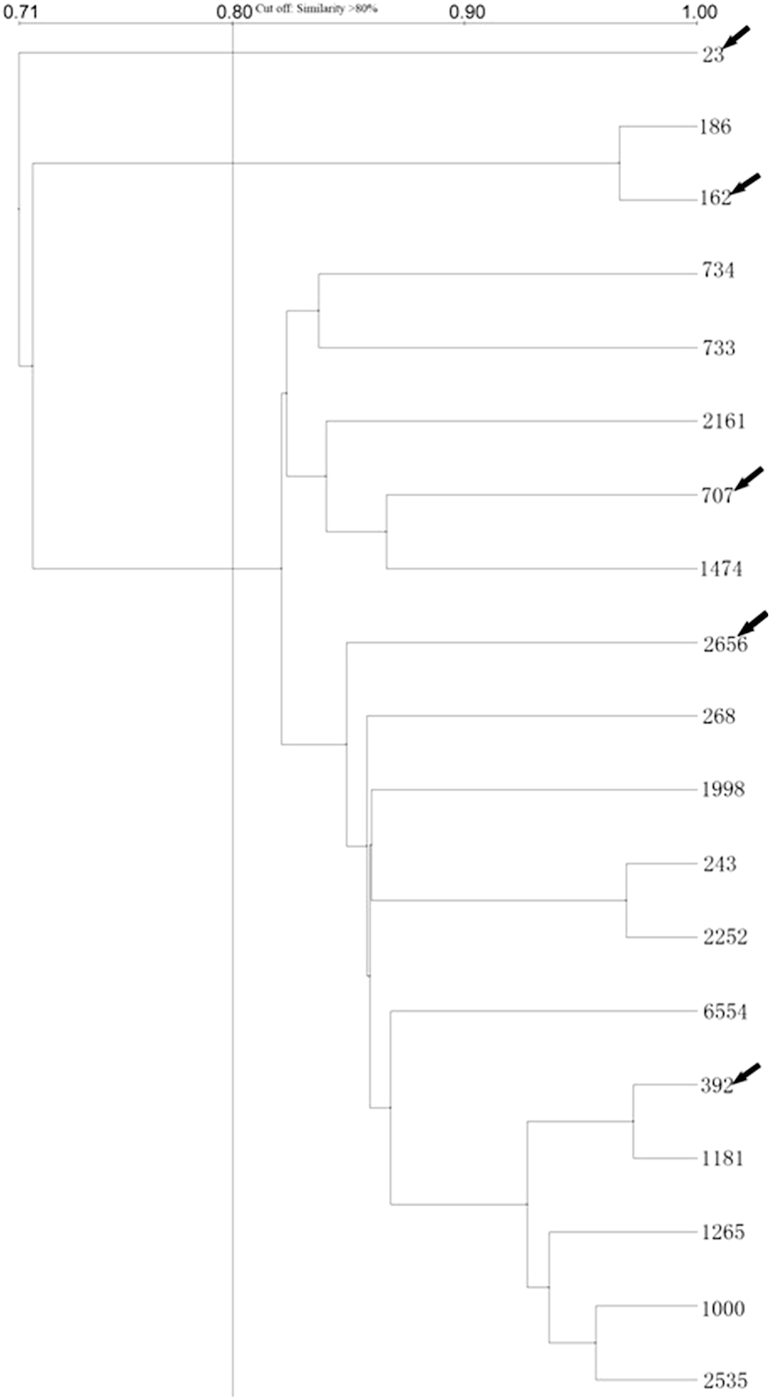
Because the ICU environment is the hardest hit with A. baumannii infection, molecular typing using rep-PCR and MLST were carried out in strains isolated from the ICU. Rep-PCR testing of ICU isolates showed that 16 of 19 isolated were closely related, with similarity scores of over 80% (Fig. 1). We assumed that isolates with closely linked rep-PCR results would be the same MLST type, so only five representative strains were processed for MLST typing. All five strains were consistent with sequence type (ST) 2, corresponding to European clone II [18].
Fig. 1.
Repetitive PCR analysis of isolates from intensive care unit. Isolates with more than 80% similarity were considered related. Arrow indicates isolates processed by multilocus sequence typing analysis to determine genotypes of Acinetobacter baumannii.
Discussion
Among the 189 clinical isolates in our study, resistance ratios to each tested antimicrobial agents were all above 60%. The susceptibility of clinical isolates to tobramycin (21.1%) and imipenem (26.8%) is more that 20%; only these two antimicrobial agents had a tolerable killing ability of clinical A. baumannii from this hospital. These data highlighted the severe challenge of drug-resistant A. baumannii nosocomial infection.
A total of 144 (76.2) of 189 strains were found to have an integron cassette, most of them belonging to a class I integron. The frequency of integron positivity in this hospital is higher than that in other countries and regions, such as 60% in the United Kingdom (A. baumannii) [19], 50% in the Netherlands (Enterobacteriaceae) [20], 59% in France (Enterobacteriaceae) [21], 52% in Taiwan (Escherichia coli) [22], 43% in Europe (Gram-negative isolates) [12], 41.5% in Brazil (Pseudomonas aeruginosa) [23] and 52.8% in Nanjing [10] and 40.7% in Zhejiang, China [24]. The class I integron-positive frequency (136/189, 72.0%) is similar to those in Guangzhou, China (74.9%), and Eastern China (69.6%) [9], [25].
AdaA2-HP-dfrA is the most prevalent integron cassette in our study. Recently a study in Southern China also revealed dfrXII-orfF-aadA2 to be the major integron cassette [9]. DfrA is a dihydrofolate reductase mediating the drug resistance to trimethoprim, and AadA2 is a aminoglycoside adenyltransferase involved in drug resistance to aminoglycoside. This integron cassette has been reported in other regions of China, including Nanjing and Zhejiang [10], [24], but the ratio of this cassette in other regions is lower than our data. Other drug-resistant genes were also present in these integron cassettes, including aacC1, aadA1, arr3, aacA4, dfrA17 and aadA5; and aadB, cat and blaOXA10 encoded resistance to aminoglycoside, rifampin, sulfanilamide, chloramphenicol and β-lactam, respectively. Most cassettes encoding various aminoglycoside resistance genes were consistent with previous studies [22], [26]. In addition, this study detected only two sulfanilamide resistance genes (dfrA12 and dfrA17) and one β-lactam resistance gene (blaOXA10) in A. baumannii integron, which seems insufficient to explain the significant correlations between integron positivity and these two types antimicrobial agents. Nearly all class I integrons characterized to date contain a 3′ conserved segment comprising a sulI resistance gene in addition to the qacEΔ1 gene; both of them mediated sulfanilamide resistance [27], and the β-lactam-resistant genes might locate outside of integrons or plasmids [12]. Integron positivity was also strongly correlated with fluoroquinolone resistance, although no fluoroquinolone-resistant genes existed in the integron. A previous study reported a clear association of integron structures with mutator plasmids, such as R46, in many Enterobacteriaceae [28], which can confer an increased baseline mutation rate in the host cell [29] and lead to an increase in the frequency of point mutations that give rise to fluoroquinolone-resistant phenotypes.
PilR is a member of the two-component regulatory system PilS/PilR that regulates the expression of type 4 fimbriae, which was first reported in Pseudomonas aeruginosa [30]. Type IV pili are essential for motility, biofilm formation, adherence to multiple surfaces and natural transformation and virulence [31], [32]. In general, the drug-resistant characters of class II integron-positive isolates and integron-negative isolates are not significantly different (except gatifloxacin; data not shown), so whether the two genes in the class II integrons are related to drug resistance needs further study.
Isolates from the ICU belonged to the ST2 genotype in this teaching hospital. The ST2 genotypic entity was also a predominated clonal lineage in multiple centers, such as Mediterranean countries [33] and Germany [34]. In addition, ICU-isolated strains also showed a relatively high carbapenem resistance (18 of 19 ICU isolates resistant to imipenem, for a resistance ratio of 95%; overall imipenem-resistant ratio is 71.1%), similar to that of ST2 strains isolated from other countries [35], [36].
Conclusion
This study demonstrated the wide distribution of integrons among A. baumannii within the Second Affiliated Hospital of Chongqing Medical University, Chongqing, China. A distinct feature of our findings is the prevalence of the major class I integron consisting of aadA2-HP-dfrA resistance genes in this hospital. Two novel class II integron cassettes also are reported. Regardless of the presence or absence of resistance genes within an integron, our data demonstrated a significant association between the presence of integrons and a reduced susceptibility to many antibiotics.
Conflict of Interest
None declared.
Acknowledgements
This work was funded by National Natural Science Foundation (81071316, 81271882, 81301394), New Century Excellent Talents in Universities (NCET-11-0703), the Fundamental Research Funds for the Central Universities (XDJK2012D007, XDJK2012D011, XDJK2013D003, XDJK2011C020) and Natural Science Foundation Project of CQ CSTC (2010BB5002, CSTC2013jcyjA10064).
References
- 1.Villers D., Espaze E., Coste-Burel M., Giauffret F., Ninin E., Nicolas F. Nosocomial Acinetobacter baumannii infections: microbiological and clinical epidemiology. Ann Intern Med. 1998;129(3):182–189. doi: 10.7326/0003-4819-129-3-199808010-00003. [DOI] [PubMed] [Google Scholar]
- 2.Gaynes R., Edwards J.R. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41(6):848–854. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 3.Dijkshoorn L., Nemec A., Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007;5(12):939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 4.Hall R.M., Collis C.M. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updat. 1998;1(2):109–119. doi: 10.1016/s1368-7646(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 5.Cambray G., Guerout A.M., Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 6.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4(8):608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 7.Partridge S.R., Tsafnat G., Coiera E., Iredell J.R. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33(4):757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 8.Xu Z., Li L., Shirtliff M.E., Alam M.J., Yamasaki S., Shi L. Occurrence and characteristics of class 1 and 2 integrons in Pseudomonas aeruginosa isolates from patients in southern China. J Clin Microbiol. 2009;47(1):230–234. doi: 10.1128/JCM.02027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Li H., Yang J., Zhan R., Chen A., Yan Y. Prevalence and characterization of integrons in multidrug resistant Acinetobacter baumannii in Eastern China: a multiple-hospital study. Int J Environ Res Public Health. 2015;12(8):10093–10105. doi: 10.3390/ijerph120810093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu B., Tong M., Zhao W., Liu G., Ning M., Pan S. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol. 2007;45(1):241–243. doi: 10.1128/JCM.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Freijo P., Fluit A.C., Schmitz F.J., Grek V.S., Verhoef J., Jones M.E. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998;42(6):689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- 13.Levesque C., Piche L., Larose C., Roy P.H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39(1):185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ploy M.C., Denis F., Courvalin P., Lambert T. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob Agents Chemother. 2000;44(10):2684–2688. doi: 10.1128/aac.44.10.2684-2688.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reboli A.C., Houston E.D., Monteforte J.S., Wood C.A., Hamill R.J. Discrimination of epidemic and sporadic isolates of Acinetobacter baumannii by repetitive element PCR-mediated DNA fingerprinting. J Clin Microbiol. 1994;32(11):2635–2640. doi: 10.1128/jcm.32.11.2635-2640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannouli M., Cuccurullo S., Crivaro V., Di Popolo A., Bernardo M., Tomasone F. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J Clin Microbiol. 2010;48(4):1223–1230. doi: 10.1128/JCM.02263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisenberg S.A., Schuetz A.N., Alexander E.A., Eiss B., Behta M., Saiman L. Endemic Acinetobacter baumannii in a New York hospital. PLoS One. 2011;6(12):e28566. doi: 10.1371/journal.pone.0028566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turton J.F., Gabriel S.N., Valderrey C., Kaufmann M.E., Pitt T.L. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13(8):807–815. doi: 10.1111/j.1469-0691.2007.01759.x. [DOI] [PubMed] [Google Scholar]
- 19.Turton J.F., Kaufmann M.E., Glover J., Coelho J.M., Warner M., Pike R. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J Clin Microbiol. 2005;43(7):3074–3082. doi: 10.1128/JCM.43.7.3074-3082.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones M.E., Peters E., Weersink A.M., Fluit A., Verhoef J. Widespread occurrence of integrons causing multiple antibiotic resistance in bacteria. Lancet. 1997;349(9067):1742–1743. doi: 10.1016/S0140-6736(05)62954-6. [DOI] [PubMed] [Google Scholar]
- 21.Sallen B., Rajoharison A., Desvarenne S., Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of enterobacteriaceae. Microb Drug Resist. 1995;1(3):195–202. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 22.Chang C.Y., Chang L.L., Chang Y.H., Lee T.M., Chang S.F. Characterisation of drug resistance gene cassettes associated with class 1 integrons in clinical isolates of Escherichia coli from Taiwan, ROC. J Med Microbiol. 2000;49(12):1097–1102. doi: 10.1099/0022-1317-49-12-1097. [DOI] [PubMed] [Google Scholar]
- 23.Fonseca E.L., Vieira V.V., Cipriano R., Vicente A.C. Class 1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol Med Microbiol. 2005;44(3):303–309. doi: 10.1016/j.femsim.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Xu H., Su Z., Wang S., Dai X., Chen J., Kong F. Four novel resistance integron gene-cassette occurrences in bacterial isolates from Zhenjiang, China. Curr Microbiol. 2009;59(2):113–117. doi: 10.1007/s00284-009-9405-z. [DOI] [PubMed] [Google Scholar]
- 25.Wu K., Wang F., Sun J., Wang Q., Chen Q., Yu S. Class 1 integron gene cassettes in multidrug-resistant Gram-negative bacteria in southern China. Int J Antimicrob Agents. 2012;40(3):264–267. doi: 10.1016/j.ijantimicag.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 26.White P.A., McIver C.J., Rawlinson W.D. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001;45(9):2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosser S.J., Young H.K. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44(1):11–18. doi: 10.1093/jac/44.1.11. [DOI] [PubMed] [Google Scholar]
- 28.Upton C., Pinney R.J. Expression of eight unrelated Muc+ plasmids in eleven DNA repair-deficient E. coli strains. Mutat Res. 1983;112(5):261–273. doi: 10.1016/0167-8817(83)90002-0. [DOI] [PubMed] [Google Scholar]
- 29.Ambler J.E., Pinney R.J. Positive R plasmid mutator effect on chromosomal mutation to nalidixic acid resistance in nalidixic acid-exposed cultures of Escherichia coli. J Antimicrob Chemother. 1995;35(5):603–609. doi: 10.1093/jac/35.5.603. [DOI] [PubMed] [Google Scholar]
- 30.Hobbs M., Collie E.S., Free P.D., Livingston S.P., Mattick J.S. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;7(5):669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Hao G., Galvani C.D., Meng Y., De La Fuente L., Hoch H.C. Type I and type IV pili of Xylella fastidiosa affect twitching motility, biofilm formation and cell-cell aggregation. Microbiology. 2007;153(Pt 3):719–726. doi: 10.1099/mic.0.2006/002311-0. [DOI] [PubMed] [Google Scholar]
- 32.Kang Y., Liu H., Genin S., Schell M.A., Denny T.P. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol Microbiol. 2002;46(2):427–437. doi: 10.1046/j.1365-2958.2002.03187.x. [DOI] [PubMed] [Google Scholar]
- 33.Di Popolo A., Giannouli M., Triassi M., Brisse S., Zarrilli R. Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin Microbiol Infect. 2011;17(2):197–201. doi: 10.1111/j.1469-0691.2010.03254.x. [DOI] [PubMed] [Google Scholar]
- 34.Schleicher X., Higgins P.G., Wisplinghoff H., Korber-Irrgang B., Kresken M., Seifert H. Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005–2009) Clin Microbiol Infect. 2013;19(8):737–742. doi: 10.1111/1469-0691.12026. [DOI] [PubMed] [Google Scholar]
- 35.Nemec A., Krizova L., Maixnerova M., Diancourt L., van der Reijden T.J., Brisse S. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J Antimicrob Chemother. 2008;62(3):484–489. doi: 10.1093/jac/dkn205. [DOI] [PubMed] [Google Scholar]
- 36.Adams-Haduch J.M., Onuoha E.O., Bogdanovich T., Tian G.B., Marschall J., Urban C.M. Molecular epidemiology of carbapenem-nonsusceptible Acinetobacter baumannii in the United States. J Clin Microbiol. 2011;49(11):3849–3854. doi: 10.1128/JCM.00619-11. [DOI] [PMC free article] [PubMed] [Google Scholar]