Abstract
Strain FF8T (= CSUR P860 = DSM 28259) was isolated in Dakar, Senegal, from the urine of a 65-year-old man with acute cystitis. This strain shows a similarity of sequence of 16S rRNA of 98.38% with Weeksella virosa, and its GenBank accession numbers are HG931340 and CCMH00000000. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis had a poor score, ranging from 1.32 to 1.56, that did not allow identification of the bacterium. Using a polyphasic study made of phenotypic and genomic analyses, strain FF8T was a Gram-negative, aerobic rod and a member of the family Flavobacteriaceae. The sequenced genome is 2 562 781 bp with one chromosome but no plasmid. It exhibits a G + C content of 35.9% and contains 2390 protein-coding and 56 RNA genes, including a complete rRNA operon. On the basis of these data, we propose the creation of Weeksella massiliensis sp. nov.
Keywords: Culturomics, cystisis, genome, human, taxonogenomics, urine, Weeksella massiliensis
Introduction
The genus Weeksella (Holmes et al., 1986) was first described in 1986 [1]. To date, this genus includes one species, Weeksella virosa, which has been isolated from human clinical specimens [1], [2].
The current classification of prokaryotes relies on a polyphasic strategy combining phenotypic and genotypic characteristics [3], [4]. These include 16S rRNA sequence similarity, G + C content and DNA-DNA hybridization (DDH). However, these tools have significant drawbacks, notably that the recommended threshold values do not apply to all species or genera [5], [6].
Thanks to the progress made in sequencing technologies and their lowering costs, almost 40 000 bacterial genome sequences are currently available, covering many phyla [7]. Recently we proposed to integrate phenotypic characteristics, notably the MALDI-TOF spectrum, and genomic analysis and comparison in the taxonomic description of bacterial species [5], [8], [9]. We named this strategy taxonogenomics [5].
Strain FF8T (= CSUR P860 = DSM 28259) was isolated from the urine of a 65-year-old man treated at the Hôpital Principal de Dakar, Senegal. That is a Gram-negative bacterium, aerobic, indole negative, nonmotile and rod shaped. This bacterium was cultivated as part of the MALDI-TOF implementation at Hôpital Principal de Dakar aiming to improve the routine laboratory identification of microorganisms [10].
Here we present a summary classification and a set of features for Weeksella massiliensis sp. nov., together with the description of the complete genome sequencing and annotation. These characteristics support the circumscription of the species Weeksella massiliensis.
Organism Information
Classification and features
In July 2013, a urine sample was collected from a 65-year-old Senegalese man with acute cystitis. From this clinical sample strain FF8 (Table 1) was isolated by cultivation on 5% sheep's blood–enriched Columbia agar (bioMérieux, Marcy l’Etoile, France). When analysed by MALDI-TOF, no identification was obtained because the strain displayed low scores.
Table 1.
Classification and general features of Weeksella massiliensis strain FF8T[19]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain: Bacteria | TAS [34] | |
| Phylum: Bacteroidetes | TAS [11], [12] | ||
| Class: Flavobacteriia | TAS [35], [36] | ||
| Order: Flavobacteriales | TAS [35], [36], [37] | ||
| Family: Flavobacteriaceae | TAS [38] | ||
| Genus: Weeksella | TAS [1] | ||
| Species: Weeksella massiliensis | IDA | ||
| (Type) strain: FF8T | IDA | ||
| Gram stain | Negative | IDA | |
| Cell shape | Rod | IDA | |
| Motility | Not motile | IDA | |
| Sporulation | Non–spore forming | NAS | |
| Temperature range | Mesophile | IDA | |
| Optimum temperature | 37°C | IDA | |
| Optimum pH range | 7.2–7.4; 7.3 | ||
| Carbon source | Unknown | ||
| MIGS-6 | Habitat | Human | IDA |
| MIGS-6.3 | Salinity | Unknown | |
| MIGS-22 | Oxygen requirement | Aerobic | TAS |
| MIGS-15 | Biotic relationship | Free living | TAS |
| MIGS-14 | Pathogenicity | Unknown | |
| MIGS-4 | Geographic location | Dakar | TAS |
| MIGS-5 | Sample collection | November 28, 2013 | TAS |
| MIGS-4.1 | Latitude | 14.6937000 | TAS |
| MIGS-4.1 | Longitude | −17.4440600 | TAS |
| MIGS-4.4 | Altitude | 12 m above sea level | TAS |
MIGS, minimum information about a genome sequence.
Evidence codes are as follows: IDA, inferred from direct assay; TAS, traceable author statement (i.e., direct report exists in the literature); NAS, nontraceable author statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species or on anecdotal evidence). These evidence codes are from http://www.geneontology.org/GO.evidence.shtml of the Gene Ontology project [39]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
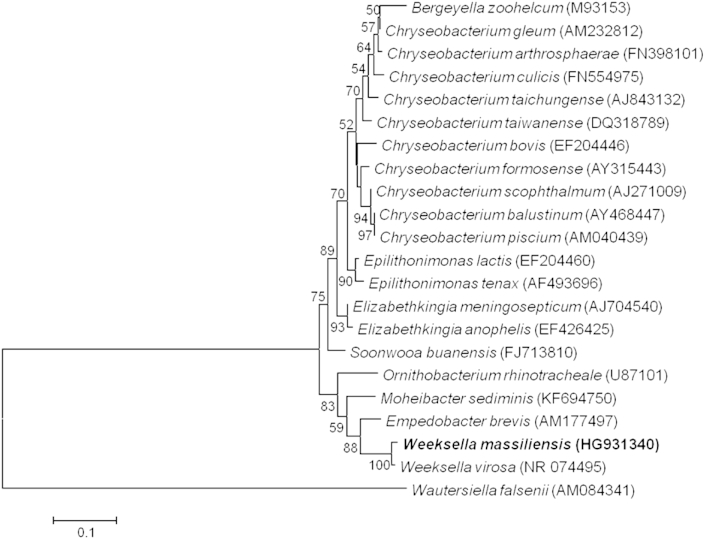
Strain FF8 exhibited a 98.38% 16S rRNA sequence similarity with Weeksella virosa strain DSM 16922T (GenBank accession number NR_074495), the phylogenetically closest bacterial species with standing in nomenclature. These values were lower than the 98.7% 16S rRNA gene sequence threshold recommended by Meier-Kolthoff et al., 2013, to delineate a new species within phylum Bacteroidetes [11], [12] without carrying out DDH [13]. A phylogenetic tree based on the 16S rRNA sequence highlights the position of Weeksella massiliensis strain FF8T among the family Flavobacteriaceae (Fig. 1). Different growth temperatures (25, 30, 37, 45 and 56°C) were tested. Growth was obtained between 25 and 37°C, with an optimal growth at 37°C. Growth of the strain was tested also under anaerobic and microaerophilic conditions using GENbag anaer and GENbag microaer systems, respectively (bioMérieux), and under aerobic conditions, with or without 5% CO2. Thus the optimal growth was observed under aerobic and microaerophilic conditions. No growth was observed under anaerobic conditions. The colonies were opaque, light yellow in color with a smooth surface, not haemolytic on 5% sheep's blood–enriched Columbia agar (bioMérieux) and approximately 2 mm in diameter. A motility test was negative. Cells are Gram-negative, non-spore-forming rods of regular shape with rounded ends (Fig. 2) and have mean diameter of 0.3 μm (range, 0.2–0.5 μm) and a mean length of 1.5 μm (range, 0.8–2.1 μm) (Fig. 3). Weeksella massiliensis strain FF8T does not grow on MacConkey agar [14].
Fig. 1.
Phylogenetic tree showing position of Weeksella massiliensis strain FF8T relative to other type strains within family Flavobacteriaceae. Strains and their corresponding 16S rRNA GenBank accession numbers are: Bergeyella zoohelcum strain ATCC 43767, M93153 (GA: AGYA00000000); Chryseobacterium gleum strain ATCC 35910, M58772 (GA: ACKQ00000000); Chryseobacterium arthrosphaerae strain CC-VM-7, FN398101; Chryseobacterium culicis strain R4-1A, FN554975; Chryseobacterium taichungense strain CC-TWGS1-8T, AJ843132; Chryseobacterium taiwanense strain BCRC 17412, DQ318789 (GA: JWTA00000000); Chryseobacterium bovis strain H9, EF204446; Chryseobacterium formosense strain CC-H3-2, AY315443 (GA: JPRP00000000); Chryseobacterium scophthalmum strain LMG 13028, AJ271009; Chryseobacterium balustinum strain LMG 8329, AY468447; Chryseobacterium piscium strain LMG 23089T, AM040439; Epilithonimonas lactis strain H1, EF204460 (GA: JPLY00000000); Epilithonimonas tenaxstrain DSM 16811, AF493696 (GA: AUAA00000000); Elizabethkingia meningoseptica strain ATCC 13253T, AJ704540 (GA: BARD00000000); Elizabethkingia anophelis strain Ag1, EF426426 (GA: AHHG00000000); Soonwooa buanensis strain HM0024T, FJ713810; Ornithobacterium rhinotracheale strain DSM 15997, NR102940 (GA: CP003283); Moheibacter sediminis strain M0116, KF694750; Empedobacter brevis strain NBRC 14943, NR112974 (GA: ARNT00000000); W. massilliensis strain FF8T, HG931340 (GA: CCMH00000000); Weeksella virosa strain DSM 16922T, NR074495 (GA: CP002455); and Empedobacter falsenii strain NF 993T, AM084341. Sequences were aligned using MUSCLE [42], and phylogenetic tree inferred by maximum likelihood method with Kimura 2-parameter model from MEGA 6 software [43]. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1000 times to generate majority consensus tree. E. falsenii was used as outgroup. Scale bar = rate of substitution per site of 0.1.
Fig. 2.
Gram staining of Weeksella massiliensis strain FF8T.
Fig. 3.
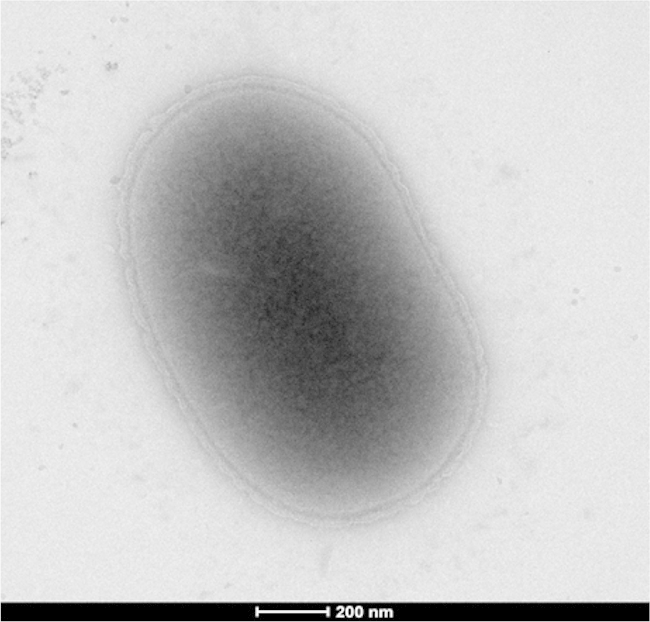
Transmission electron microscopy of Weeksella massiliensis strain FF8T. Cells were observed with Tecnai G20 transmission electron microscope operated at 200 keV. Scale bar = 200 nm.
Chemotaxonomic information
This bacterium possesses catalase and oxidase. Using an API ZYM strip (bioMérieux), positive reactions were observed for alkaline phosphatase, esterase, esterase–lipase, leucine arylamidase, acid phosphatase and naphthol-AS-BI-phosphohydrolase. Negative reactions were noted for α-chymotrypsin, cystine arylamidase, valine arylamidase, trypsin, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-mannosidase, α-fucosidase and N-acetyl-β-glucosaminidase. Strain FF8T is susceptible to ceftriaxone, amoxicillin/clavulanic acid, penicillin, imipenem, gentamicin and doxycycline but resistant to nitrofurantoin, vancomycin, trimethoprim/sulfamethoxazole and metronidazole. The minimum inhibitory concentrations (MICs) for some antibiotics tested by Weeksella massiliensis strain FF8T sp. nov. are listed in Table 2. A comparison of phenotypic characteristics with W. virosa, Bergeyella zoohelcum [15] and Moheibacter sediminis [16] is presented in Table 3.
Table 2.
Antimicrobial susceptibility and MICs of Weeksella massiliensis strain FF8T sp. nov.
| Antibiotic | MIC (mg/L) | Interpretation |
|---|---|---|
| Penicillin | 0.125 | Susceptible |
| Ceftriaxone | 0.06 | Susceptible |
| Amoxicillin/clavulanic acid | 0.16 | Susceptible |
| Imipenem | 0.15 | Susceptible |
| Gentamycin | 0.06 | Susceptible |
| Vancomycin | 3 | Resistant |
| Trimethoprim/sulfamethoxazole | 4 | Resistant |
MIC, minimum inhibitory concentration.
Table 3.
Differential characteristics of Weeksella massiliensis strain FF8T (data from this study), W. virosa strain DSM 16922T[1], Bergeyella zoohelcum strain D658 T[15], Moheibacter sediminis strain M0116T[16], Elizabethkingia meningoseptica[40] and Chryseobacterium bovis[41]
| Character | Weeksella massiliensis | Weeksella virosa | Bergeyella zoohelcum | Moheibacter sediminis | Elizabethkingia meningoseptica | Chryseobacterium bovis |
|---|---|---|---|---|---|---|
| Cell diameter (μm) | 0.2–0.5 | NA | NA | 0.2–0.3 | 0.5–1.0 | 0.5–0.9 |
| Oxygen requirement | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic | Aerobic |
| Gram stain | − | − | − | − | − | − |
| Motility | − | − | − | − | − | − |
| Endospore formation | − | − | − | − | − | − |
| Production of | ||||||
| Alkaline phosphatise | + | + | + | + | + | + |
| Acid phosphatise | + | + | + | + | + | + |
| Catalase | + | + | + | + | + | + |
| Oxidase | + | + | + | + | + | + |
| Nitrate reductase | − | − | − | − | − | − |
| Urease | − | − | + | − | − | − |
| α-Galactosidase | − | − | − | − | + | NA |
| β-Galactosidase | − | − | − | − | − | + |
| β-Glucuronidase | − | − | − | − | − | NA |
| α –Glucosidase | − | − | − | + | + | + |
| β-Glucosidase | − | − | − | + | − | NA |
| Esterase | + | + | − | + | − | + |
| Esterase lipase | + | + | NA | + | + | + |
| Naphthol-AS-BI-phosphohydrolase | + | + | + | + | + | + |
| N-acetyl-β-glucosaminidase | − | − | − | − | NA | + |
| Utilization of | ||||||
| 5-keto-gluconate | − | NA | NA | + | NA | NA |
| d-Xylose | + | − | + | − | + | − |
| d-Fructose | − | − | − | − | + | + |
| d-Glucose | − | − | − | − | + | + |
| d-Mannose | − | − | NA | − | NA | + |
| Habitat | Human | Human | Parasite saprophytic | Sediment | Human | Animal |
NA, data not available.
Extended features descriptions
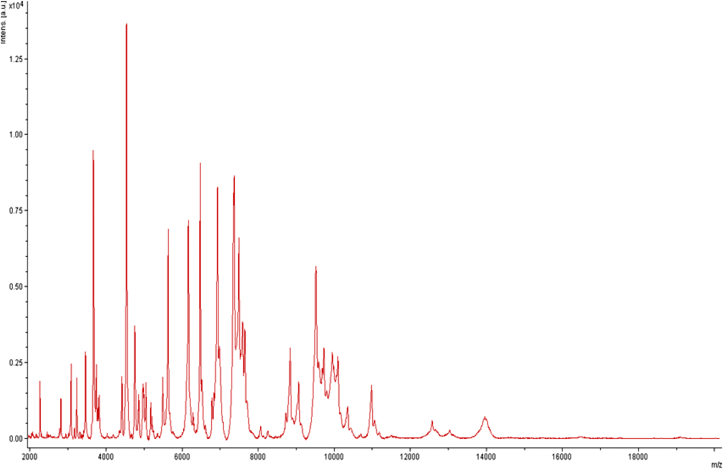
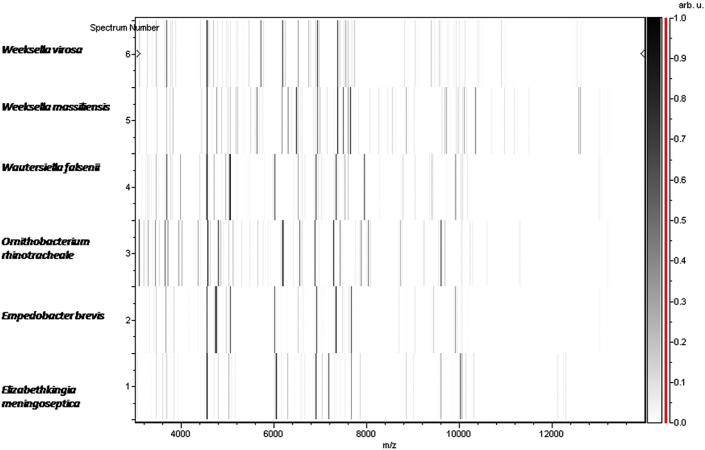
MALDI-TOF protein analysis was performed with a Microflex LT (Bruker Daltonics, Leipzig, Germany), as previously reported [17], [18]. The scores previously established by Bruker allowing validating (or not) the identification of species compared to the database of the instrument were applied. Briefly, a score of ≥ 2.000 with a species with a validly published name provided allows the identification at the species level; a score of ≥ 1.700 and < 2.000 allows the identification at the genus level; and a score of < 1.700 does not allow any identification. We performed 12 distinct deposits from 12 isolated colonies of strain FF8T. Two microliters of matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid) in 50% acetonitrile and 2.5% trifluoroacetic acid were distributed on each smear and submitted at air drying for 5 minutes. Then the spectra from the 12 different colonies were imported into the MALDI BioTyper software (version 2.0, Bruker) and analysed by standard pattern matching (with default parameter settings) against the main spectra of 6252 bacteria. Scores ranging from 1.32 to 1.56 were obtained for strain FF8T, suggesting that it was not a member of any known species. The reference mass spectrum from strain FF8T was incremented in our database (Fig. 4). The gel view highlighted spectrum differences with other Flaviobacteriaceae species (Fig. 5).
Fig. 4.
Reference mass spectrum from Weeksella massiliensis strain FF8T. This reference spectrum was generated by comparison of 12 individual colonies.
Fig. 5.
Gel view comparing Weeksella massiliensis strain FF8T to members of family Flavobacteriaceae. Gel view displays raw spectra of all loaded spectrum files arranged in pseudo-gel-like look. X-axis records m/z value. Left y-axis displays running spectrum number originating from subsequent spectra loading. Peak intensity expressed by grayscale scheme code. Color bar and right y-axis indicate relation between color peak; peak intensity displayed in arbitrary units. Displayed species are indicated at left.
Genome Sequencing Information
Genome project history
The organism was selected for sequencing on the basis of its phylogenetic position, 16S rRNA similarity and phenotypic differences with other members of the family Flavobacteriaceae. It was the second genome within Weeksella genus and the first genome of W. massiliensis sp. nov. The GenBank accession number is CCMH00000000 and consists of 54 contigs. Table 4 shows the project information and its association with minimum information about a genome sequence (MIGS, version 2.0) compliance [19]; associated MIGS records are also summarized in Supplemental Table S1.
Table 4.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | Paired-end 3.1 kb library |
| MIGS-29 | Sequencing platforms | Illumina MiSeq |
| MIGS-31.2 | Fold coverage | 30.88x |
| MIGS-30 | Assemblers | CLCGENOMICSWB4 |
| MIGS-32 | Gene calling method | Prodigal |
| Locus tag | Not indicated | |
| GenBank ID | CCMH00000000 | |
| GenBank date of release | August 22, 2014 | |
| GOLD ID | Gp0102101 | |
| BioProject ID | PRJEB5516 | |
| MIGS-13 | Source material identifier | DSM 28259 |
| Project relevance | MALDI-TOF implementation in Dakar |
MALDI-TOF, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MIGS, minimum information about a genome sequence.
Growth conditions and genomic DNA preparation
Weeksella massiliensis strain FF8T (= CSUR P860 = DSM 28259) was grown aerobically on 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C. Bacteria grown on four petri dishes were resuspended in 5 × 100 μL of Tris-EDTA buffer; 150 μL of this suspension was diluted in 350 μL Tris-EDTA buffer 10×, 25 μL proteinase K and 50 μL sodium dodecyl sulfate for lysis treatment. This preparation was incubated overnight at 56°C. Extracted DNA was then purified using 3 successive phenol–chloroform extractions (Thermo Fisher Scientific, Waltham, MA, USA) and ethanol precipitations at −20°C overnight. After centrifugation, the DNA was suspended in 65 μL elution buffer. The genomic DNA concentration was measured at 53.7 ng/μL using the Qubit assay with the high sensitivity kit (Life Technologies, Carlsbad, CA, USA).
Genome sequencing and assembly
Genomic DNA of Weeksella massiliensis FF8T was sequenced on a MiSeq sequencer (Illumina, San Diego, CA, USA) using the mate pair strategy. The genomic DNA (gDNA) was bar coded in order to be mixed with 11 other projects with the Nextera Mate-Pair sample prep kit (Illumina). The mate pair library was prepared with 1 μg of genomic DNA using the Nextera Mate-Pair Illumina guide. The gDNA sample was simultaneously fragmented and tagged with Mate-Pair junction adapters. The fragmentation pattern was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with a DNA 7500 lab chip. The DNA fragment sizes ranged from 1 to 10 kb with an optimal size at 3.1 kb. No size selection was performed, and only 143 ng of tagmented fragments were circularized. The circularized DNA was mechanically sheared to small fragments with an optimal at 645 bp on a Covaris device S2 in microtubes (Covaris, Woburn, MA, USA). The library profile was visualized on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies), and the final concentration library was measured at 6.03 nmol/L. The libraries were normalized at 2 nM and pooled. After a denaturation step and dilution at 15 pM, the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and sequencing were performed in a single 27-hour run in a 2 × 151 bp. Total information of 6 Gb was obtained from a 1158K/mm2 cluster density with a cluster passing quality control filters of 88.9% (21 834 000 clusters). Within this run, the index representation for Weeksella massiliensis was determined to 8.32%. The 1 613 495 paired reads were filtered according to the read qualities. These reads were trimmed then assembled thought CLCgenomicsWB4 software. Finally, the draft genome of Weeksella massiliensis consists of 17 scaffolds made of 54 contigs and generated a genome size of 2.56 Mb with G + C content of 35.9%.
Genome annotation
Open reading frames (ORFs) were predicted using Prodigal [20] with default parameters, but the predicted ORFs were excluded if they were spanning a sequencing gap region. The predicted bacterial protein sequences were searched against the GenBank database [21] and the Clusters of Orthologous Groups (COGs) database using BLASTP. The tRNAScanSE tool [22] was used to find tRNA genes, whereas ribosomal RNAs were found by using RNAmmer [23] and BLASTn against the GenBank database. Lipoprotein signal peptides and the number of transmembrane helices were predicted using SignalP [24] and TMHMM [25], respectively. ORFans were identified if their BLASTP E value was lower than 1e−03 for alignment length greater than 80 aa. If alignment lengths were smaller than 80 aa, we used an E value of 1e−05. Such parameter thresholds have already been used in previous works to define ORFans. Artemis [26] was used for data management, and DNA Plotter [27] was used for visualization of genomic features. The Mauve alignment tool (version 2.3.1) was used for multiple genomic sequence alignment [28]. To estimate the mean level of nucleotide sequence similarity at the genome level, we used calculated the average genomic identity of orthologous gene sequences (AGIOS) parameter using an in-lab pipeline named Marseille Average Genomic Identity (MAGi). Briefly, this software combines the Proteinortho software [29] for detecting orthologous proteins in pairwise comparisons of genomes, then retrieves the corresponding genes and determines the mean percentage of nucleotide sequence identity among orthologous ORFs using the Needleman-Wunsch global alignment algorithm. The script was created to calculate the average genomic identity of orthologous gene sequences (AGIOS) between genomes by MAGi. The MAGi script created to calculate AGIOS values written in Perl and BioPerl modules. Genome-to-genome distance (GGDC) analysis was also performed using the GGDC Web server (http://ggdc.dsmz.de/) as previously reported [30], [31].
Genome Properties
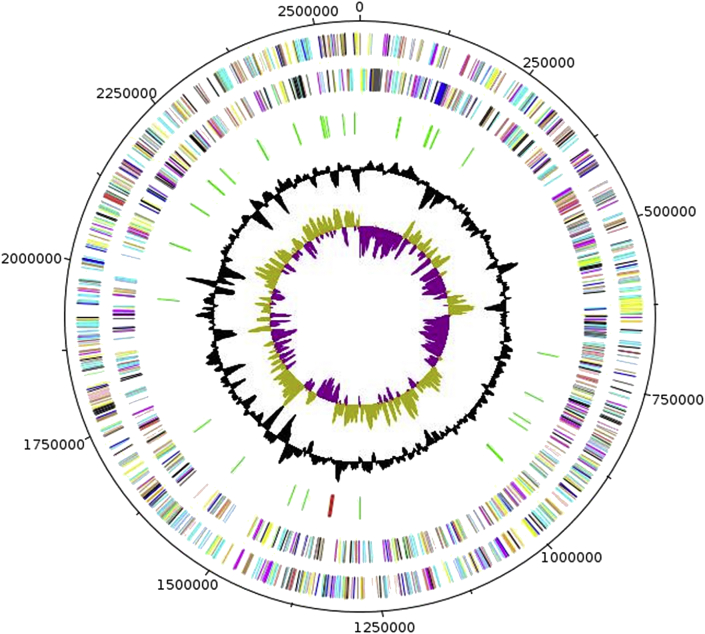
The genome of W. massiliensis strain FF8T is 2 562 781 bp long with a 35.9% G + C content (Fig. 6). Of the 2446 predicted genes, 2390 were protein-coding genes and 56 were RNAs genes. Three rRNA genes (one 16S rRNA, one 23S rRNA and one 5S rRNA) and 53 predicted tRNA genes were identified in the genome. A total of 1428 genes (58.38%) were assigned a putative function. Eighty genes were identified as ORFans (3.27%). The remaining genes were annotated as hypothetical proteins. The properties and the statistics of the genome are summarized in Table 5. The distribution of genes into COGs functional categories is presented in Table 6.
Fig. 6.
Graphical circular map of Weeksella massiliensis strain FF8T chromosome. From outside in, outer two circles show open reading frames oriented in forward (colored by COGs categories) and reverse (colored by COGs categories) directions, respectively. Third circle marks rRNA gene operon (red) and tRNA genes (green). Fourth circle shows G + C content plot. Innermost circle shows GC skew, with purple indicating negative values and olive positive values. COGs, Clusters of Orthologous Groups (COGs) database.
Table 5.
Nucleotide content and gene count levels of genome
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 2 562 781 | 100 |
| DNA coding (bp) | 413 280 | 16.12 |
| DNA G + C (bp) | 920 038 | 35.9 |
| DNA scaffolds | 07 | — |
| Total genes | 2446 | 100 |
| Protein coding genes | 2390 | 97.71 |
| RNA genes | 56 | 2.28 |
| Pseudo genes | ND | — |
| Gens in internal clusters | ND | — |
| Genes with function prediction | 1428 | 58.38 |
| Genes assigned to COGs | 1567 | 64.06 |
| Genes with Pfam domains | 1022 | 41.78 |
| Genes with signal peptides | 279 | 11.40 |
| Genes with transmembrane helices | 528 | 21.58 |
| ORFan genes | 80 | 3.27 |
| CRISPRs | 2 |
COGs, Clusters of Orthologous Groups (COGs) database; CRISPR, clustered regularly interspaced short palindromic repeat; ND, not determined.
Total is based on either size of genome (bp) or total number of protein-coding genes in annotated genome.
Table 6.
Number of genes associated with general COGs functional categoriesa
| Code | Value | % | Description |
|---|---|---|---|
| J | 139 | 5.82 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0 | RNA processing and modification |
| K | 83 | 3.47 | Transcription |
| L | 122 | 5.10 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 17 | 0.71 | Cell cycle control, cell division, chromosome partitioning |
| V | 43 | 1.80 | Defense mechanisms |
| T | 46 | 1.92 | Signal transduction mechanisms |
| M | 158 | 6.61 | Cell wall/membrane biogenesis |
| N | 4 | 0.17 | Cell motility |
| U | 27 | 1.13 | Intracellular trafficking and secretion |
| O | 86 | 3.60 | Posttranslational modification, protein turnover, chaperones |
| C | 106 | 4.44 | Energy production and conversion |
| G | 56 | 2.34 | Carbohydrate transport and metabolism |
| E | 148 | 6.19 | Amino acid transport and metabolism |
| F | 56 | 2.34 | Nucleotide transport and metabolism |
| H | 87 | 3.64 | Coenzyme transport and metabolism |
| I | 80 | 3.35 | Lipid transport and metabolism |
| P | 107 | 4.48 | Inorganic ion transport and metabolism |
| Q | 39 | 1.63 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 211 | 8.83 | General function prediction only |
| S | 116 | 4.85 | Function unknown |
| — | 823 | 33.64 | Not in COGs |
COGs, Clusters of Orthologous Groups (COGs) database.
Total is based on total number of protein-coding genes in annotated genome.
Insights From Genome Sequence
Here we compared the genome sequence of W. massiliensis strain FF8T with those of Weeksella virosa strain DSM 16922T (GenBank accession number CP002455), Chryseobacterium gleum strain ATCC 35910T (ACKQ00000000), Elizabethkingia meningoseptica ATCC 13253T (BARD00000000) and Empedobacter brevis strain NBRC 14943T (ARNT00000000). The draft genome of W. massiliensis has a larger size than that of W. virosa (2.56 Mb and 2.27 Mb, respectively) but smaller than those of C. gleum, E. meningoseptica and E. brevis (5.57 Mb, 3.96 Mb and 3.79 Mb, respectively). Weeksella massiliensis has a higher G + C content than E. brevis (35.89% and 32.7%, respectively) but lower than W. virosa, C. gleum and E. meningoseptica (35.9%, 36.8% and 36.4%, respectively). Because it has been suggested in the literature that the G + C content deviation is at most 1% within species, these data prove that this strain is a new taxon [32]. The gene content of W. massiliensis is larger than that of W. virosa (2446 and 2171, respectively) but smaller than those of C. gleum, E. meningoseptica and E. brevis (5369, 3423 and 3655, respectively). However, the distribution of genes into COGs categories was similar in all compared genomes. In addition, W. massiliensis shared 2390, 2049, 5289, 3369 and 3589 orthologous genes with W. virosa, C. gleum, E. meningoseptica and E. brevis, respectively.
Because only one genome was available for the genus Weeksella, we used the genomes from closely related species for the calculation of AGIOS values. The AGIOS values ranged from 66.45% to 73.15% between W. virosa and other members of the family Flavobacteriaceae (Table 7). W. massiliensis exhibited similar values when compared to representatives of other genera, from 66.65% to 72.25%, but a higher value when compared to W. virosa (87.85%). In addition, digital DDH similarities between the genomes were calculated by the GGDC Web server version 2.0, as recommended (Table 8) [30], [31], [32], [33].
Table 7.
Numbers of orthologous proteins shared between genomes (upper right) and AGIOS values obtained (lower left)
| Weeksella massiliensis | Weeksella virosa | Chryseobacterium gleum | Elizabethkingia meningoseptica | Empedobacter brevis | |
|---|---|---|---|---|---|
| W. massiliensis | 2390 | 1584 | 1421 | 1254 | 1647 |
| W. virosa | 87.85 | 2049 | 1297 | 1174 | 1551 |
| C. gleum | 66.69 | 66.45 | 5289 | 1523 | 1658 |
| E. meningoseptica | 66.65 | 66.54 | 73.15 | 3369 | 1523 |
| E. brevis | 72.25 | 71.80 | 67.39 | 67.42 | 3589 |
Bold indicates numbers of proteins per genome.
Table 8.
Pairwise comparison of W. massiliensis, W. virosa, C. gleum, E. meningoseptica and E. brevis using GGDC
| Weeksella massiliensis | Weeksella virosa | Chryseobacterium gleum | Elizabethkingia meningoseptica | Empedobacter brevis | |
|---|---|---|---|---|---|
| W. massiliensis | 100% | 32.9% | 22.1% | 19.7% | 19.7% |
| W. virosa | 100% | 26.7% | 22.6% | 18.9% | |
| C. gleum | 100% | 19.9% | 24.1% | ||
| E. meningoseptica | 100% | 16.8% | |||
| E. brevis | 100% |
GGDC, genome-to-genome distance.
Conclusions
On the basis of phenotypic, phylogenetic and genomic analyses, we formally propose the creation of Weeksella massiliensis sp. nov. that contains strain FF8T as type strain. The strain was isolated from the urine of a 65-year-old Senegalese man who sought care at the Hôpital Principal de Dakar, Senegal.
Description of Weeksella massiliensis sp. nov.
Weeksella massiliensis (mas.il.i.en'sis. L. gen. fem. n. massiliensis, of Massilia, the Latin name for Marseille, where strain FF8T was cultivated). Colonies were 2 mm in diameter and are opaque and light yellow with a smooth surface on 5% sheep's blood-enriched Columbia agar. It is not haemolytic on blood agar. Cells are Gram negative and not motile, with a mean diameter of 0.3 μm (range, 0.2–0.5 μm) and a mean length of 1.5 μm (range, 0.8–2.1 μm). Cells were catalase and oxidase positive. Positive reactions were observed for alkaline phosphatase, esterase, esterase–lipase, leucine arylamidase, phosphatase acid and naphthol-AS-BI-phosphohydrolase activities. Negative reactions were noted for α-chymotrypsin, cystine arylamidase, valine arylamidase, trypsin, α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, α-mannosidase, α-fucosidase and N-acetyl-β-glucosaminidase activities. Weeksella massiliensis strain FF8T is susceptible to ceftriaxone, amoxicillin/clavulanic acid, penicillin, imipenem, gentamicin and doxycycline but resistant to nitrofurantoin, vancomycin, trimethoprim–sulfamethoxazole and metronidazole. The G + C content of the genome is 35.9%. The 16S rRNA and genome sequences are deposited in GenBank under accession numbers HG931340 and CCMH00000000, respectively. The type strain FF8T (= CSUR P860 = DSM 28259) was isolated from the urine of a 65-year-old man with acute cystitis at Hôpital Principal de Dakar, Senegal.
Acknowledgements
The authors thank the Xegen Company (http://www.xegen.fr/) for automating the genomic annotation process. This study was funded by the Méditerranée-Infection Foundation. We also thank J.-P. Baudoin for creating the electron microscope photos.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.nmni.2015.09.013.
Conflict of Interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Holmes B., Steigerwalt A.G., Weaver R.E., Brenner D.J. Weeksella virosa gen. nov., sp. nov. (formerly group IIF) found in human clinical specimens. Syst Appl Microbiol. 1986;8:185–190. [Google Scholar]
- 2.Validation list no. 23 Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Bacteriol. 1987;37:179–180. [Google Scholar]
- 3.Tindall B.J., Rosselló-Móra R., Busse H.J., Ludwig W., Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 4.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 5.Ramasamy D., Mishra A.K., Lagier J.C., Padhmanabhan R., Rossi M., Sentausa E. A polyphasic strategy incorporating genomic data for the taxonomic description of new bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- 6.Sentausa E., Fournier P.E. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect. 2013;19:790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 7.Reddy T.B.K., Thomas A., Stamatis D., Bertsch J., Isbandi M., Jansson J. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta) genome project classification. Nucl Acids Res. 2014;43:1099–1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagier J.C., Armougom F., Mishra A.K., Nguyen T.T., Raoult D., Fournier P.E. Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci. 2012;6:315–324. doi: 10.4056/sigs.2685971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier J.C., Karkouri K.E., Mishra A.K., Robert C., Raoult D., Fournier P.E. Non contiguous-finished genome sequence and description of Enterobaceter massiliensis sp. nov. Stand Genomic Sci. 2013;7:399–412. doi: 10.4056/sigs.3396830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fall B., Lo C.I., Samb-Ba B., Perrot N., Diawara S., Gueye M.W. The ongoing revolution of MALDI-TOF mass spectrometry for microbiology reaches tropical Africa. Am J Trop Med Hyg. 2015;92:641–647. doi: 10.4269/ajtmh.14-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Validation list no. 143 Int J Syst Evol Microbiol. 2012;62:1–4. [Google Scholar]
- 12.Krieg N.R., Ludwig W., Euzéby J., Whitman W.B. Phylum XIV. Bacteroidetes phyl. nov. In: Krieg N.R., Staley J.T., Brown D.R., Hedlund B.P., Paster B.J., Ward N.L., editors. Bergey's manual of systematic bacteriology. 2nd ed. Springer; New York: 2011. pp. 4–25. [Google Scholar]
- 13.Meier-Kolthoff J.P., Göker M., Spröer C., Klenk H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol. 2013;195:413–418. doi: 10.1007/s00203-013-0888-4. [DOI] [PubMed] [Google Scholar]
- 14.Reina J., Gil J., Salva F., Gomez J., Alomar P. Microbiological characteristics of Weeksella virosa (formerly CDC group IIf) isolated from the human genitourinary tract. J Clin Microbiol. 1990;28:2357–2359. doi: 10.1128/jcm.28.10.2357-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandamme P., Bernardet J.F., Segers P., Kersters K., Holmes B. New perspectives in the classification of the Flavobacteriia: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int J Syst Bacteriol. 1994;44:827–831. [Google Scholar]
- 16.Zhang R.G., Tan X., Zhao X.M., Deng J., Lv J. Moheibacter sediminis gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from sediment, and emended descriptions of Empedobacter brevis, Empedobacter falsenii and Weeksella virosa. Int J Syst Evol Microbiol. 2014;64:1481–1487. doi: 10.1099/ijs.0.060178-0. [DOI] [PubMed] [Google Scholar]
- 17.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 18.Seng P., Rolain J.M., Fournier P.E., La Scola B., Drancourt M., Raoult D. MALDI-TOF–mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5:1733–1754. doi: 10.2217/fmb.10.127. [DOI] [PubMed] [Google Scholar]
- 19.Field D., Garrity G., Gray T., Morrison N., Selengut J., Sterk P. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benson D.A., Karsch-Mizrachi I., Clark K., Lipman D.J., Ostell J., Sayers E.W. GenBank. Nucleic Acids Res. 2012;40:48–53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowe T.M., Eddy S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 26.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 27.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechner M., Findeib S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Auch A.F., Klenk H.P., Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142–148. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meier-Kolthoff J.P., Klenk H.P., Göker M. Taxonomic use of DNA G + C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 33.Auch A.F., Von Jan M., Klenk H.P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernardet J.F. Class II. Flavobacteriia class. nov. In: Krieg N.R., Staley J.T., Brown D.R., Hedlund B.P., Paster B.J., Ward N.L., editors. Bergey's manual of systematic bacteriology. 2nd ed. Springer; New York: 2011. pp. 4–105. [Google Scholar]
- 36.List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2012;62:1017–1019. doi: 10.1099/ijsem.0.001149. [DOI] [PubMed] [Google Scholar]
- 37.Garrity G.M., Holt J.G. Taxonomic outline of the Archaea and Bacteria. In: Krieg N.R., Staley J.T., Brown D.R., Hedlund B.P., Paster B.J., Ward N.L., editors. Bergey's manual of systematic bacteriology. 2nd ed. Springer; New York: 2011. pp. 155–166. [Google Scholar]
- 38.Bernardet J.F., Nakagawa Y., Holmes B. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae, and emended description of the family. Int J Syst Evol Microbiol. 2002;52:1049–1070. doi: 10.1099/00207713-52-3-1049. [DOI] [PubMed] [Google Scholar]
- 39.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K.K., Kim M.K., Lim J.H., Park H.Y., Lee S.T. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int J Syst Evol Microbiol. 2005;55:1287–1293. doi: 10.1099/ijs.0.63541-0. [DOI] [PubMed] [Google Scholar]
- 41.Hantsis-Zacharov E., Senderovich Y., Halpern M. Chryseobacterium bovis sp. nov., isolated from raw cow’s milk. Int J Syst Evol Microbiol. 2008;58:1024–1028. doi: 10.1099/ijs.0.65500-0. [DOI] [PubMed] [Google Scholar]
- 42.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.