Abstract
Glucose is a ubiquitous energy source for most living organisms. Its uptake activity closely reflects cellular metabolic demand in various physiopathological conditions. Extensive efforts such as positron emission tomography, magnetic resonance imaging and fluorescence microscopy have been made to specifically image glucose uptake but all with limitations. Here, we report a new platform to visualize glucose uptake activity in live cells and tissues by performing stimulated Raman scattering on a novel glucose analogue labeled with a small alkyne moiety. Cancer cells with differing metabolic activities can be distinguished. Heterogeneous uptake patterns are observed with clear cell-cell variations in tumor xenograft tissues, neuronal culture and mouse brain tissues. Hence, by offering the distinct advantage of optical resolution yet without the undesirable influence of fluorophores, our method will be a valuable tool to study energy demands of living systems with subcellular resolution.
Keywords: alkyne tags, glucose uptake, imaging agents, Raman spectroscopy, SRS microscopy
Glucose is the primary energy source for virtually all living organisms, from bacteria to human. As a hydrophilic small molecule, the uptake of glucose is mediated by cell surface transporters to pass through the otherwise impermeable hydrophobic plasma membrane, during which the ATP-independent facilitated diffusion down a glucose concentration gradient is the major pathway. Since the uptake of glucose is biologically regulated and closely correlated to a wide range of metabolic transformations, several prominent techniques have been developed to selectively image glucose uptake activity. Among them, radioactive tracers of glucose analogues (Figure 1a, b) are widely applied in clinical positron emission tomography (PET) to image glucose uptake activity in vivo. Although these PET analogues cannot be metabolized like D-glucose in the glycolytic pathway and thus accumulate inside cells, their net flux across cell membrane still reflects the total exogenous cellular glucose use when imaged at steady-state conditions.[1] Indeed, imaging steady-state glucose uptake level has been proved by extensive PET studies to be a key diagnostic marker in many diseases. More recently, magnetic resonance imaging (MRI) has achieved glucose imaging in tumors through chemical exchange saturation transfer and hyperpolarization of 13C labeled D-glucose.[2] Unfortunately, both PET and MRI techniques have limited spatial resolution, and cannot visualize glucose uptake in single cells. It is well appreciated that glucose uptake and energy demands in many crucial physiological (e.g., brain activity) and pathological (e.g., tumor development) processes are intrinsically heterogeneous at the cellular level.[3] Hence, to visualize glucose uptake activity in single living cells is indispensable in evaluating the metabolic demand of individual cells and unveiling the underlying cell-to-cell variations.
Figure 1.
Structures of existing D-glucose analogues and synthesis of the new 3-O-propargyl-D-glucose. (a) [18F]2-Fluoro-2-deoxy-D-glucose ([18F]FDG). (b) [11C]3-O-methyl-D-glucose ([11C]3-OMG). (c) 2-NBD-2-deoxy-D-glucose (2-NBDG). (d) Synthetic route of 3-OPG 1. a) NaH, propargyl bromide, DMF, RT, 12 h, 90%; b) Dowex® 50WX8, H2O, 80°C, 20 h, 96%.
To monitor glucose uptake with cellular resolution, fluorophore-conjugated glucose analogues have been developed for fluorescence microscopy. 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]-2-deoxy-D-glucose (2-NBDG) (Figure 1c), which is also a glucose analogue that cannot be metabolized further in glycolysis, is one of the most popular probes and has been applied in a variety of cell types.[4] However, conjugation of glucose with fluorophores, which are mostly hydrophobic or charged, alters the hydrophilic property of natural glucose. Indeed, the water solubility of 2-NBDG is substantially reduced to ~10 mg/ml. Moreover, physical sizes of fluorophores are always larger than glucose itself, due to the requirement of extended conjugation for visible light absorption. Thus, most fluorescent glucose analogues including 2-NBDG exhibit non-specific interactions and retentions in cells and tissues,[5] which would bias the true glucose distribution to varying degrees.
Here we report a new modality to visualize glucose uptake activity at the subcellular level by stimulated Raman scattering (SRS) imaging of vibrationally labeled D-glucose, and demonstrate its utility in live cancer cells, tumor xenograft tissues, primary neurons and mouse brain tissues. We designed, synthesized and characterized a novel glucose analogue, named 3-O-propargyl-D-glucose (3-OPG, 1, Figure 1d). 3-OPG has a minor chemical modification comparable to the PET tracer [11C]3-OMG (Figure 1b) yet avoiding the use of radioactive isotopes and allowing for subcellular optical imaging. Compared with fluorophores, alkyne tag (C≡C) is physically much smaller and has been proven to be chemically inert and bioorthogonal in cells by numerous click-chemistry studies.[6] Thus, we expect 3-OPG to not only preserve the hydrophilic nature of glucose but also prevent its non-specific interactions in cells and tissues. Detection wise, alkyne tag exhibits a pronounced C ≡ C stretching Raman peak (~2100 cm−1) in the cell-silent spectral region.[7] Very recently alkyne tags have been coupled to SRS microscopy as a powerful vibrational label for metabolic imaging in live cells with high sensitivity, specificity and biocompatibility.[8] Therefore, by targeting the unique vibrational signature of alkyne tags, SRS imaging of 3-OPG uptake could be a valuable method to study energy demands and metabolic status in living systems with subcellular resolution and minimal perturbation.
We first synthesized 3-OPG by conjugating a propargyl group to glucose 3' OH group (Figure 1d). Briefly, commercially available 1,2:5,6-Di-O-isopropylidene-α-D-glucofuranose 2 is reacted with propargyl bromide to selectively introduce the alkyne moiety onto the 3’ position, which gives 3 in 90% yield. Deprotection of 3 in strong acids affords 3-OPG 1 with 96% yield. Comparing 3-OPG with 3-OMG (Figure 1), one can readily tell their structures are highly similar except for the replacement of a methyl group by a propargyl group, which is a common strategy to introduce the alkyne group and maintain the original function, such as in propargyl-choline[6b] and O-propargyl-puromycin.[6c]
Spontaneous Raman spectrum of 3-OPG is then characterized with a sharp and strong peak at 2129 cm−1 in the cell-silent region (Figure 2a, black), indicative of Raman-active mode of C ≡ C stretching. Negligible cellular background in 1800–2400 cm−1 is confirmed by both spontaneous Raman spectrum and SRS image at 2129 cm−1 of blank HeLa cells (Figure S1). Intrinsic Raman peaks of glucose, such as those from C-C and C-O bond vibrations, are also observed in the fingerprint region from 1000–1800 cm−1 (Figure 2a, black). Note that these peaks are not only much weaker than the alkyne peak but also overlapping with the cellular background (Figure 2a, red), which makes label-free Raman imaging of glucose in cells rather challenging.[9] Thus, introducing an exogenous alkyne tag not only promotes the sensitivity but also endows imaging specificity in the otherwise crowded cellular background without the need of spectral unmixing.
Figure 2.
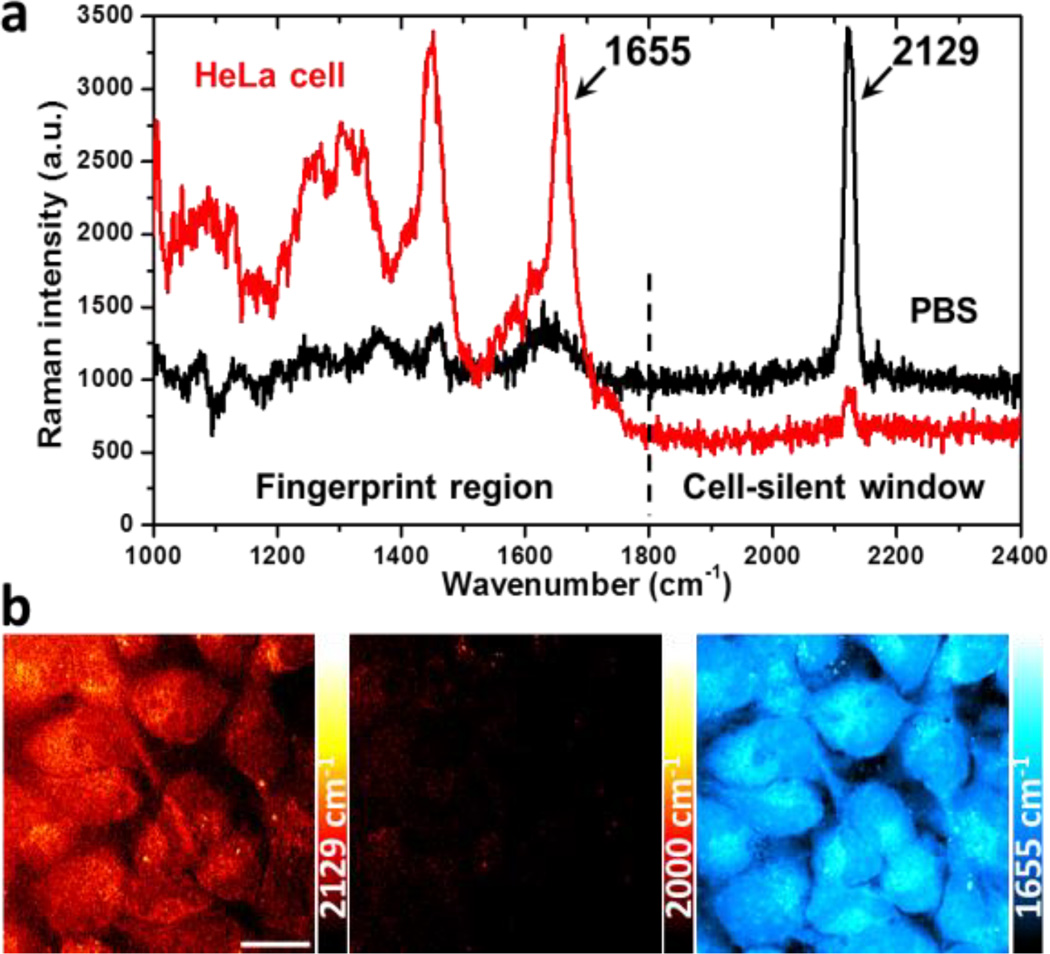
Uptake of 3-OPG in live mammalian cells. (a) Spontaneous Raman spectra of 25 mM 3-OPG solution in PBS (black) and HeLa cells incubated with 25 mM 3-OPG for 4 hours (red). (b) SRS imaging of 3-OPG uptake (2129 cm−1) in live HeLa cells incubated with 8 mM 3-OPG for 1 hour. The average intracellular concentration is estimated to be 3–5 mM. Images at 2000 cm−1 (off-resonance) and 1655 cm−1 (protein amide I) show the same area of cells. The 2129 cm−1 color bars in all images correspond to a linear 3-OPG concentration range of 0–26 mM, unless otherwise noted. Scale bar: 20 µm.
The uptake of 3-OPG is then tested in mammalian cell cultures. A distinct peak (2129 cm−1) emerges after incubating HeLa cells with 25 mM 3-OPG for 4 hours (Figure 2a, red), which exactly matches with that in the phosphate-buffered saline (PBS) solution spectrum of 3-OPG, confirming cellular uptake of 3-OPG. No cytotoxicity of 3-OPG is observed from cell viability assays (Figure S2). Cellular distribution of 3-OPG can be visualized with high sensitivity and speed by SRS imaging of the alkyne Raman peak at 2129 cm−1 in HeLa cells incubated with 8 mM 3-OPG for 1 hour (Figure 2b). The corresponding off-resonance image at 2000 cm−1 confirms the background-free nature of SRS. As a reference, SRS image at 1655 cm−1 shows cellular protein distribution by targeting at protein amide I band.
To quantify the intracellular uptake level, we correlated SRS intensity at 2129 cm−1 with known concentrations of 3-OPG solutions under identical conditions, which gives a detection sensitivity of 1.4 mM with 30 µs acquisition time per pixel and ~26 s/frame (Figure S3). For higher signal-to-noise ratio and more accurate quantifications, we chose 25 mM 3-OPG as incubation concentration for further biochemistry characterizations.
To validate that 3-OPG is transported into cells through glucose transporters, several control experiments are performed. First, uptake of 3-OPG is competed using D-glucose at different concentrations (Figure 3). As the concentration of D-glucose increases, cellular uptake of 3-OPG decreases accordingly, reaching 78% with 10 mM D-glucose and 53% with 50 mM D-glucose, consistent with the higher transporter affinity of D-glucose (Figure 3a–c). In contrast, with 50 mM L-glucose, which is not recognized by glucose transporters for its opposite chirality, 3-OPG uptake level is unaffected (Figure 3d).
Figure 3.
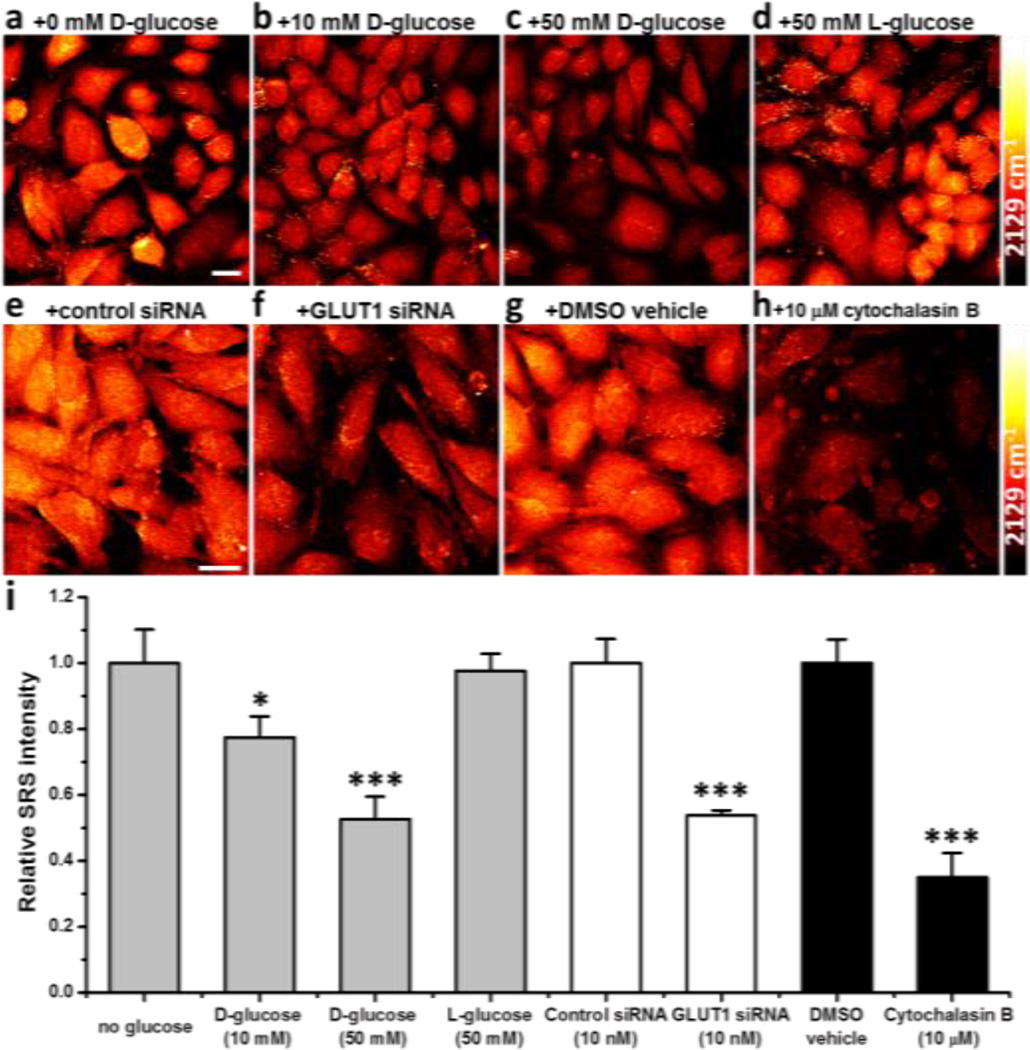
Glucose transporter dependence of 3-OPG uptake in mammalian cells. Representative SRS images of live HeLa cells incubated with 3-OPG in the presence of (a) 0 mM D-glucose, (b) 10 mM D-glucose, (c) 50 mM D-glucose, and (d) 50 mM L-glucose. (e, f) HeLa cells were transfected with 10 nM negative control siRNA (e) or GLUT1 siRNA (f) before incubation with 3-OPG. (g, h) HeLa cells were incubated with 3-OPG in the presence of DMSO vehicle (g) or 10 µM cytochalasin B (h). Scale bar: 20 µm. (i) Relative SRS intensity of 3-OPG uptake in HeLa cells under the conditions above. Data are shown as Mean + standard deviation (SD) (n≥3 replicates for each group). *, p<0.05; ***, p<0.001 by Student’s t-test. p<0.05 is considered statistically significant.
Four different transporter inhibition and stimulation assays are then conducted. Using RNA interference to suppress the expression of GLUT1 transporter, HeLa cells transfected with 10 nM GLUT1 siRNA for 48 hours show a much reduced uptake of 3-OPG (54%) compared to the control groups (Figure 3e, f). Moreover, HeLa cells incubated with 10 µM cytochalasin B, a potent inhibitor of glucose carriers, display a drastic reduction to 35% in 3-OPG uptake (Figure 3g, h). Efflux experiments of 3-OPG loaded HeLa cells were also performed with a rapid decay after 30 minutes and the addition of cytochalasin B significantly inhibits the efflux of 3-OPG (Figure S4), indicating the transport of 3-OPG in both directions is carrier-mediated. We further studied the effect of insulin stimulation on 3-OPG uptake by GLUT4 in rat muscle cells. With 1 µg/ml insulin for 24 hours, which stimulates GLUT4 translocation to plasma membrane, ~30% increase in 3-OPG uptake is observed in fused L6 cells (Figure S5), suggesting GLUT4-mediated uptake of 3-OPG. Taken together, these results further prove the glucose transporter dependence of 3-OPG uptake and its role as a D-glucose analogue to mammalian cells.
Uptake kinetics of 3-OPG is also examined (Figure S6 and S7). At 37 °C, buildup of 3-OPG inside HeLa cells reaches a plateau within 30 minutes, indicating fast uptake of 3-OPG at physiological temperature and allowing steady-state imaging. This equilibrium time is consistent with the reported size and weight dependence of transport kinetics for different glucose analogues.[10] Lowering to room temperature slows down 3-OPG uptake, as expected. Linear fitting of 3-OPG uptake at early times also reveals a ~3 folds decrease in initial transport rate from 37 °C to room temperature. Quantitatively, by plotting initial rates of 3-OPG uptake against different incubation concentrations (Figure S7), its Km and Vmax parameters are extracted in HeLa cells where GLUT1 is the most abundant isoform.[11] The fitted values of Km=26±4 mM and Vmax=0.71±0.05 mM/min of 3-OPG uptake turn out to be very close to that for 3-OMG uptake by human GLUT1 expressed in Xenopus oocytes (Km=13–20 mM and Vmax=0.74 mM/min assuming 106 oocytes/L).[12] Competition experiments using 3-OMG show a moderate reduction to 70% in 3-OPG uptake (Figure S8), which is again in agreement with their similar Km.
Further, important insights can be gained by simultaneously imaging cellular uptake of 3-OPG and 2-NBDG by the same set of cells (Figure S9). While 2-NBDG displays a heterogeneous pattern with stronger signal enriched in cellular structures surrounding the nucleus, 3-OPG is distributed more homogeneously throughout the cells including inside the nucleus. Such differential contrast between 3-OPG and 2-NBDG is likely due to non-specific retention of 2-NBDG in extra-nuclear organelles (such as endoplasmic reticulum) and its low permeability across nuclear membrane caused by the strong hydrophobicity and/or large size of the NBD fluorophores.[5] Hence we have obtained direct imaging evidence that 3-OPG could be a more truthful probe to report the genuine glucose uptake and distribution inside cells.
Tumors are well known to have aberrant metabolism with upregulated glucose uptake. Cancer cells of different phenotypes also have varied glucose uptake levels, which could serve as a marker for tumor malignancy and metastatic potential.[2a] We thus compare glucose uptake activity between HeLa, a human cervical cancer cell line and U-87 MG, a human primary glioblastoma cell line, using 3-OPG as imaging probe (Figure S10). As the highest-grade glioma (WHO grade IV), glioblastoma is the most aggressive form of malignant primary brain tumor. Incubated with 3-OPG, U-87 MG cells display a much higher alkyne peak than that of HeLa cells in Raman spectra (Figure S10a), suggesting a more pronounced glucose uptake activity. SRS images further show the whole intracellular distribution of 3-OPG in both cancer cells. Consistent with the spectroscopic results, U-87 MG cells indeed exhibit a brighter intracellular signal than HeLa cells, confirming much increased glucose uptake activity (Figure S10b). Quantitatively, 3-OPG uptake in U-87 MG cells is over 1.7 times higher than that of HeLa cells (Figure S10c). Thus SRS imaging of 3-OPG uptake could be a useful technique to distinguish cancer cells with varied metabolic characteristics.
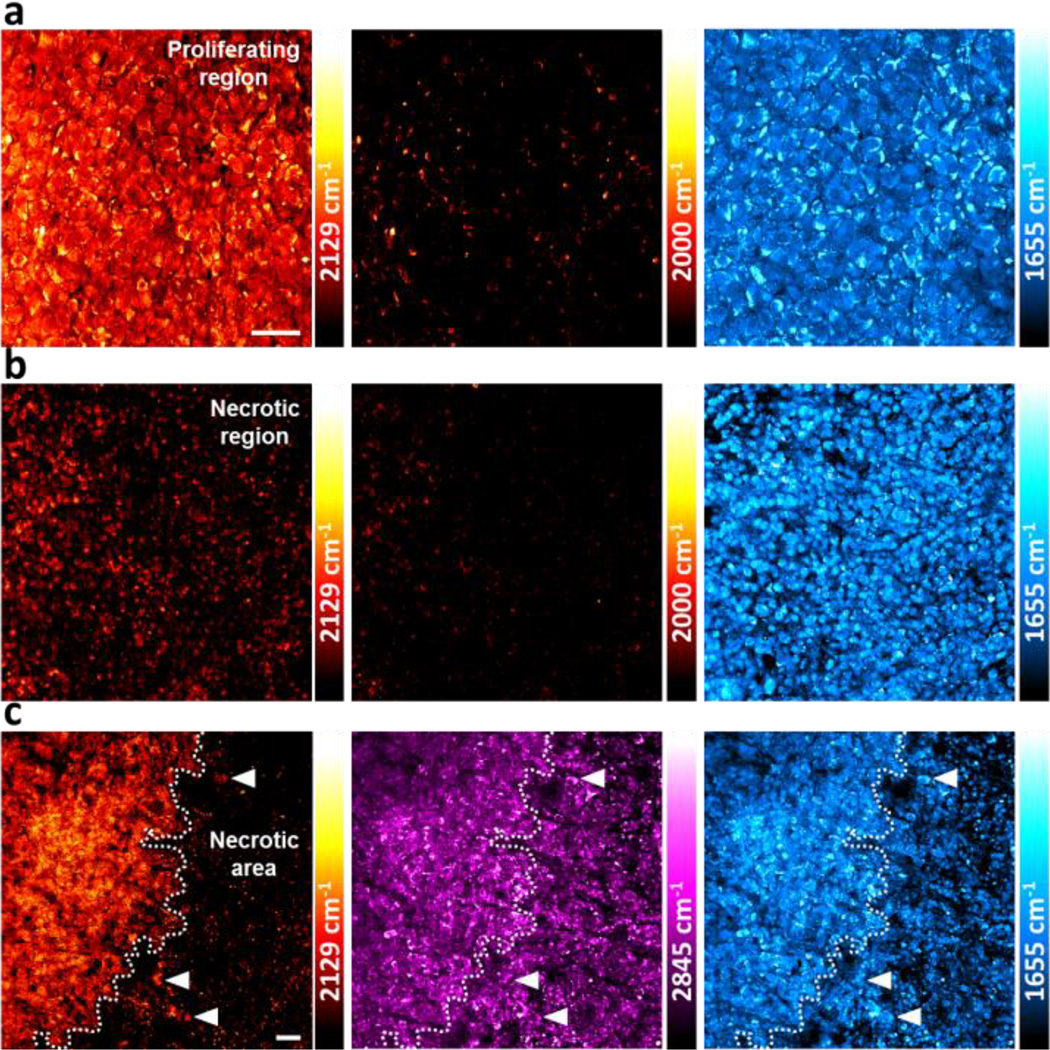
Uptake of 3-OPG is further visualized in mouse subcutaneous tumor xenograft model. Tumor regions with distinct glucose uptake levels are observed (Figure 4). In outer region of tumor, cells undergo fast proliferation and hence uptake glucose significantly (Figure 4a). In contrast, toward the center of tumor, cells display small and broken morphology (as shown in the amide channel at 1655 cm−1), suggesting necrosis and diminished cellular activity. Indeed, little 3-OPG uptake is seen in the SRS image with at least a 5 times lower signal than that in the tumor-propagating region (Figure 4b).
Figure 4.
SRS imaging of 3-OPG uptake in U-87 MG tumor xenograft tissues. (a) Strong uptake of 3-OPG in the proliferating region of tumor. (b) Little uptake is seen in the tumor region with necrotic cell morphology. The 2129 cm−1 color bars in (a, b) correspond to a 3-OPG concentration range of 0–53 mM. (c) Sharp contrast of 3-OPG uptake at the interface of two tumor regions. Scale bar: 40 µm.
The interface between the two tumor regions can be clearly distinguished by highlighting the 3-OPG uptake contrast at the single cell level, providing useful microscopic insights (Figure 4c). Necrosis is known to be a histological hallmark of glioblastoma and an important predictor for tumor prognosis in clinical glioma diagnostics and staging.[13] We discover that the metabolic contrast from 3-OPG uptake is much more pronounced than the label-free images of endogenous lipid (at 2845 cm−1) or protein (at 1655 cm−1) distributions. Most notably, a small population of tumor cells with high glucose uptake activity is identified within the region of diminished activity (Figure 4c, arrowhead), reflecting cell-cell variation of metabolic status. Therefore, this showcases the capability of vibrational imaging of glucose uptake activity using 3-OPG and provides opportunity for studying intratumoral heterogeneity in energy consumptions during brain tumor growth with superb contrast and cellular resolution.
Brain is the central control of activity in higher organisms and has a high demand of energy. Fast and adequate glucose supply is critical for maintaining the neural activity. We therefore demonstrate imaging 3-OPG uptake in both in vitro neuron culture and ex vivo mouse brain tissue (Figure 5). In cultured mouse primary hippocampal neurons, not only high level of 3-OPG is imaged inside the soma, but also it can be seen clearly in the neuronal processes (Figure 5a), reflecting energy need in axons and dendrites for neural signaling. Intercellularly, significant variation in 3-OPG uptake is observed among different neurons, with over 2 times difference from cells with low-glucose level to high-glucose demanding cells (Figure 5a, arrowhead). This observation from the 3-OPG channel, which is absent from the corresponding protein amide image, indicates heterogeneous energy requirement at single neuron level. Going beyond the neuron cultures, 3-OPG uptake is also visualized in the hippocampus of mouse brain tissue slice cultured ex vivo. Pronounced signal is present in both the granule cell layer and nearby hilar region of dentate gyrus which is known to exhibit high metabolic activity[14] (Figure 5b). Thus, 3-OPG is generally applicable as a vibrational imaging probe for glucose transport in both tumor and brain study with subcellular resolution.
Figure 5.
SRS imaging of 3-OPG uptake in neuronal systems. (a) SRS images of cultured mouse primary hippocampal neurons incubated with 3-OPG. Scale bar: 20 µm. (b) SRS images of mouse brain tissue slice cultured ex vivo with 3-OPG. Significant uptake of 3-OPG is seen at the granule cell layer of dentate gyrus. Scale bar: 40 µm.
In summary, we have designed, synthesized and characterized a novel glucose analogue with an alkyne vibrational tag (3-OPG). By coupling SRS microscopy to the alkyne tag, we have demonstrated vibrational imaging of glucose uptake activity in living cells and tissues with high sensitivity, specificity and biocompatibility. Our method can differentiate cancer cells with varied metabolic activities and reveal heterogeneous glucose uptake patterns in tumor xenograft tissues as well as in neuronal culture and mouse brain tissues at cellular resolution. Previous studies have been reported on glucose sensing in blood and metabolic incorporation using isotope-labeled glucose for imaging downstream products with Raman techniques.[15] However, they are either not in cellular environment or cannot visualize glucose uptake activity specifically. If compared to the classic PET tracers, 3-OPG can be synthesized in a highly cost-effective way without special handling of radioactive materials and endows optical imaging with subcellular resolution. If compared to the bulky fluorescent glucose analogues, 3-OPG has a minor structural modification without adding a significant size and weight to glucose, rendering better affinity to glucose transporters, less non-specific interaction and higher translocation efficiency (e.g., crossing the blood-brain barrier). With the use of near-infrared lasers for nonlinear excitation, SRS is capable of high-resolution and deep tissue imaging with minimal phototoxicity and essentially no photobleaching,[16] and has recently achieved video-rate imaging in live animals and humans with potential clinical applications.[17] We therefore expect that vibrational imaging of 3-OPG with SRS microscopy would become an attractive technique for studying glucose uptake activity at the cellular level, especially in brain and malignant tumors that are inherently in high demand of energy.
Supplementary Material
Footnotes
We thank Y. Shin for providing hippocampal neurons and J. Hirtz for assistance on mouse brain tissues. W. Min acknowledges support from NIH Director’s New Innovator Award, ARO MURI W911NF-12-1-0594 and Alfred P. Sloan Research Fellowship.
Supporting information for this article is available on the WWW under http://.
Contributor Information
Fanghao Hu, Department of Chemistry, Columbia University, New York, New York 10027 (USA).
Dr. Zhixing Chen, Department of Chemistry, Columbia University, New York, New York 10027 (USA)
Dr. Luyuan Zhang, Department of Chemistry, Columbia University, New York, New York 10027 (USA)
Yihui Shen, Department of Chemistry, Columbia University, New York, New York 10027 (USA).
Lu Wei, Department of Chemistry, Columbia University, New York, New York 10027 (USA).
Prof. Wei Min, Email: wm2256@columbia.edu, Department of Chemistry, Columbia University, New York, New York 10027 (USA); Kavli Institute for Brain Science, Columbia University, New York, New York 10027 (USA).
References
- 1.Plathow C, Weber WA. J. Nucl. Med. 2008;49:43S–63S. doi: 10.2967/jnumed.107.045930. [DOI] [PubMed] [Google Scholar]
- 2.a) Walker-Samuel S, Ramasawmy R, Torrealdea F, Rega M, Rajkumar V, Johnson SP, Richardson S, Goncalves M, Parkes HG, Arstad E, Thomas DL, Pedley RB, Lythgoe MF, Golay X. Nat. Med. 2013;19:1067–1072. doi: 10.1038/nm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rodrigues TB, Serrao EM, Kennedy BW, Hu DE, Kettunen MI, Brindle KM. Nat. Med. 2014;20:93–97. doi: 10.1038/nm.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Vaupel P, Kallinowski F, Okunieff P. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]; b) Okumoto S, Jones A, Frommer WB. Annu. Rev. Plant Biol. 2012;63:663–706. doi: 10.1146/annurev-arplant-042110-103745. [DOI] [PubMed] [Google Scholar]
- 4.a) Yamada K, Saito M, Matsuoka H, Inagaki N. Nat. Protoc. 2007;2:753–762. doi: 10.1038/nprot.2007.76. [DOI] [PubMed] [Google Scholar]; b) O'Neil RG, Wu L, Mullani N. Mol. Imaging Biol. 2005;7:388–392. doi: 10.1007/s11307-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 5.a) Hughes LD, Rawle RJ, Boxer SG. PloS one. 2014;9:e87649. doi: 10.1371/journal.pone.0087649. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zanetti-Domingues LC, Tynan CJ, Rolfe DJ, Clarke DT, Martin-Fernandez M. PloS one. 2013;8:e74200. doi: 10.1371/journal.pone.0074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Prescher JA, Bertozzi CR. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]; b) Jao CY, Roth M, Welti R, Salic A. P. Natl. Acad. Sci. USA. 2009;106:15332–15337. doi: 10.1073/pnas.0907864106. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Liu J, Xu Y, Stoleru D, Salic A. P. Natl. Acad. Sci. USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Grammel M, Hang HC. Nat. Chem. Biol. 2013;9:475–484. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Rouhanifard SH, Nordstrom LU, Zheng T, Wu P. Chem. Soc. Rev. 2013;42:4284–4296. doi: 10.1039/c2cs35416k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Yamakoshi H, Dodo K, Okada M, Ando J, Palonpon A, Fujita K, Kawata S, Sodeoka M. J. Am. Chem. Soc. 2011;133:6102–6105. doi: 10.1021/ja108404p. [DOI] [PubMed] [Google Scholar]; b) Yamakoshi H, Dodo K, Palonpon A, Ando J, Fujita K, Kawata S, Sodeoka M. J. Am. Chem. Soc. 2012;134:20681–20689. doi: 10.1021/ja308529n. [DOI] [PubMed] [Google Scholar]; c) Weeks T, Wachsmann-Hogiu S, Huser T. Opt. Express. 2009;17:17044–17051. doi: 10.1364/OE.17.017044. [DOI] [PubMed] [Google Scholar]
- 8.a) Wei L, Hu F, Shen Y, Chen Z, Yu Y, Lin CC, Wang MC, Min W. Nat. methods. 2014;11:410–412. doi: 10.1038/nmeth.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hong S, Chen T, Zhu Y, Li A, Huang Y, Chen X. Angew. Chem. Int. Ed. 2014;53:5827–5831. doi: 10.1002/anie.201400328. Angew. Chem. 2014, 126, 5937–5941. [DOI] [PubMed] [Google Scholar]; c) Lee HJ, Zhang W, Zhang D, Yang Y, Liu B, Barker EL, Buhman KK, Slipchenko LV, Dai M, Cheng JX. Sci. Rep. 2015;5:7930. doi: 10.1038/srep07930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akeson M, Brackmann C, Gustafsson L, Enejder A. J. Raman Spectrosc. 2010;41:1638–1644. [Google Scholar]
- 10.a) Aloj L, Caraco C, Jagoda E, Eckelman WC, Neumann RD. Cancer Res. 1999;59:4709–4714. [PubMed] [Google Scholar]; b) Speizer L, Haugland R, Kutchai H. Biochim. Biophys. Acta. 1985;815:75–84. doi: 10.1016/0005-2736(85)90476-6. [DOI] [PubMed] [Google Scholar]
- 11.Young CD, Lewis AS, Rudolph MC, Ruehle MD, Jackman MR, Yun UJ, Ilkun O, Pereira R, Abel ED, Anderson SM. PloS one. 2011;6:e23205. doi: 10.1371/journal.pone.0023205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Wang D, Pascual JM, Iserovich P, Yang H, Ma L, Kuang K, Zuniga FA, Sun RP, Swaroop KM, Fischbarg J, De Vivo DC. J. Biol. Chem. 2003;278:49015–49021. doi: 10.1074/jbc.M308765200. [DOI] [PubMed] [Google Scholar]; b) Wang D, Yang H, De Vivo D. Neurology. 2013;80 P02.092. [Google Scholar]
- 13.Raza SM, Lang FF, Aggarwal BB, Fuller GN, Wildrick DM, Sawaya R. Neurosurgery. 2002;51:2–12. doi: 10.1097/00006123-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Cho ZH, Son YD, Kim HK, Kim ST, Lee SY, Chi JG, Park CW, Kim YB. J. Nucl. Med. 2010;51:1545–1548. doi: 10.2967/jnumed.110.076182. [DOI] [PubMed] [Google Scholar]
- 15.a) Berger AJ, Itzkan I, Feld MS. Spectrochim. Acta Mol. Biomol. Spectrosc. 1997;53:287–292. doi: 10.1016/s1386-1425(96)01779-9. [DOI] [PubMed] [Google Scholar]; b) McNichols RJ, Cote GL. J. Biomed. Opt. 2000;5:5–16. doi: 10.1117/1.429962. [DOI] [PubMed] [Google Scholar]; c) Shafer-Peltier KE, Haynes CL, Glucksberg MR, Van Duyne RP. J. Am. Chem. Soc. 2003;125:588–593. doi: 10.1021/ja028255v. [DOI] [PubMed] [Google Scholar]; d) Weeks T, Schie IW, Wachsmann-Hogiu S, Huser T. J. Biophotonics. 2010;3:169–175. doi: 10.1002/jbio.200900068. [DOI] [PubMed] [Google Scholar]; e) Noothalapati Venkata HN, Shigeto S. Chem. Biol. 2012;19:1373–1380. doi: 10.1016/j.chembiol.2012.08.020. [DOI] [PubMed] [Google Scholar]; f) Li J, Cheng J-X. Sci. Rep. 2014;4:6807. doi: 10.1038/srep06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, He C, Tsai JC, Kang JX, Xie XS. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Alfonso-Garcia A, Mittal R, Lee ES, Potma EO. J. Biomed. Opt. 2014;19:71407. doi: 10.1117/1.JBO.19.7.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang D, Wang P, Slipchenko MN, Cheng JX. Acc. Chem. Res. 2014;47:2282–2290. doi: 10.1021/ar400331q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Saar BG, Freudiger CW, Reichman J, Stanley CM, Holtom GR, Xie XS. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ji M, Orringer DA, Freudiger CW, Ramkissoon S, Liu X, Lau D, Golby AJ, Norton I, Hayashi M, Agar NY, Young GS, Spino C, Santagata S, Camelo-Piragua S, Ligon KL, Sagher O, Xie XS. Sci. Transl. Med. 2013;5:201ra119. doi: 10.1126/scitranslmed.3005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.