Abstract
Does small GTPase K-Ras4A have a single state or two states, one resembling K-Ras4B and the other N-Ras? A recent study of K-Ras4A made the remarkable observation that even in the absence of the palmitoyl K-Ras4A can be active at the plasma membrane. Importantly, this suggests that K-Ras4A may exist in two distinct signaling states. In state 1 K-Ras4A is only farnesylated, like K-Ras4B; in state 2 farnesylated and palmitoylated, like N-Ras. The K-Ras4A hypervariable region (HVR) sequence is positively charged, in-between K-Ras4B and N-Ras. Taken together, this raises the possibility that the farnesylated but nonpalmitoylated state 1, like K-Ras4B, binds calmodulin and is associated with colorectal and other adenocarcinomas like lung cancer and PDAC (pancreatic ductal adenocarcinoma). On the other hand, state 2 may be associated with melanoma and other cancers where N-Ras is a major contributor, such as acute myeloid leukemia (AML). Importantly, H-Ras has two – single and double – palmitoylated states that may also serve distinct functional roles. The multiple signaling states of palmitoylated Ras isoforms question the completeness of small GTPase Ras isoform statistics in different cancer types and call for reevaluation of concepts and protocols. They may also call for reconsideration of oncogenic Ras therapeutics.
Keywords: KRAS, HRAS, NRAS, KRAS4A, KRAS4B, K-Ras4A, K-Ras4B, calmodulin, calcium, tissue-specificity, lung cancer, colorectal cancer, pancreatic cancer, adenocarcinoma
Introduction
Numerous studies have focused on the genes and gene products of small GTPase Ras isoforms, HRAS, NRAS, and KRAS, the most frequently mutated isoform in human cancers (1, 2). The KRAS gene has two splice variants, K-Ras4A and K-Ras4B (Fig. 1a). Mutations in KRAS4B, which are also present in KRAS4A, are common in cancer. The expression level of K-Ras4A in tumors was believed to be substantially lower, retaining the attention of the community almost exclusively on the K-Ras4B variant. Recent observations by Philips and coworkers (3) may however lead to re-evaluation of our views. The quantitative RT-PCR assay for K-Ras4A and K-Ras4B message that the authors developed allowed them to measure the absolute amounts of the two transcripts. They observed that K-Ras4A was widely expressed in all their human cancer cell lines. In particular, in human colorectal tumors the amounts equaled those of K-Ras4B (Fig. 2). Their analysis revealed that the C-terminus of K-Ras4A contains the CAAX motif, a palmitoylation site, and a bipartite polybasic region (PBR) (Fig. 1b), and that unlike K-Ras4B, K-Ras4A does not bind to the cytosolic chaperone δ-subunit of cGMP phosphodiesterase type 6 (PDE6δ). They concluded that efforts to develop anti–K-Ras drugs that interfere with membrane trafficking should consider the distinct mechanisms of both K-Ras splice variants.
Figure 1.
K-Ras4A is in-between K-Ras4B and N-Ras. (a) The human KRAS gene encodes two splice variants of the K-Ras protein, K-Ras4A and K-Ras4B. K-Ras4A and K-Ras4B are identical in their first 150 amino acid residues, and subject to the same oncogenic mutations at positions 12, 13, and 61 (denoted in red). However, they differ in their C-terminal hypervariable region (HVR) and in four residues (151, 153, 165, and 166) at the edges of helix 5 in the catalytic domain, adjoining the membrane. The four residue substitutions between K-Ras4A and K-Ras4B are highlighted in maroon and blue respectively. (b) The HVR of the K-Ras4A isoform contains a bipartite polybasic region, PBR1 and PBR2. K-Ras4B’s HVR has a highly charged polybasic domain, with an additional charged patch between the polybasic regions. K-Ras4B is farnesylated, and K-Ras4A and N-Ras are farnesylated and palmitoylated. H-Ras is fanesylated and doubly palmitoylated. Basic residues are colored in blue, acidic residues in red, hydrophobic residues in black and polar and glycine residues in green. Farnesylated cysteine residues are denoted by an orange lipid group, whereas the reversibly palmitoylated cysteine residues are denoted by green lipid groups. (c) In the N-Ras-like state (left), the K-Ras4A hypervariable region (HVR) is both palmitoylated and farnesylated, whereas in the K-Ras4B-like state (right), the K-Ras4A HVR is only farnesylated. As denoted by the red lipid head groups (negatively charged phospholipids) the composition of the plasma membrane differs between them: zwitterionic, in the case of the N-Ras-like state and anionic liquid membrane in the case of the K-Ras4B-like state. Basic residues (blue stretches) in the K-Ras4A HVR interact more favorably with the negatively charged phospholipid head groups of the plasma membrane (on the right) - though not as favorably as the more positively charged HVR of K-Ras4B. The lower positive charge of K-Ras4A allows interaction with zwitterionic membranes (on the left) - though not as favorably as the neutral N-Ras. The four catalytic domain residues that differ between K-Ras4A and K-Ras4B are shown in stick form.
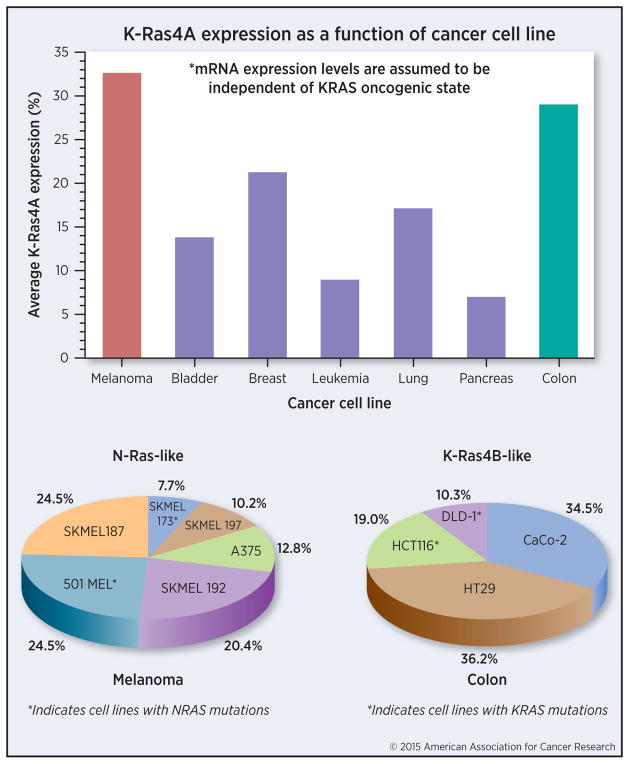
Figure 2.
The two states of K-Ras4A may contribute to different cancer types. We suggest that oncogenic K-Ras4A may mirror a K-Ras4B-like state in K-Ras4B-driven adenocarcinomas including colorectal, pancreatic, and lung cancers, whereas oncogenic K-Ras4A may reflect an N-Ras-like state in cancers where oncogenic N-Ras is frequent, such as melanoma and acute myeloid leukemia. The top panel provides estimated averages of K-Ras4A mRNA expression in different human cancer cell lines represented as a percentage of the total K-Ras expression, computed from the qPCR expression data in Fig. 1B of Tsai et al. (3). These unweighted averages were computed by measuring the percentages from the bar graphs in that figure, then summing the percentages from all cell lines of a given cancer type. The total for each cancer types was then divided by the number of cell lines reported for that cancer. This computation assumes that K-Ras4A expression is not dependent on whether the KRAS gene contains mutations. The bottom panel pie charts illustrate K-Ras4A expression in individual cell lines relative to all cell lines of that same tissue type, and suggest that significant variation exists even in the cancer types where K-Ras4A expression peaks. The percentages shown for each cell line were computed by taking the reported percentage expression for that cell line as a fraction of the sum of percentage expression levels for all cell lines of that cancer type. Here, the 4 colon and 6 melanoma cell lines reported in (3) are shown, with asterisks indicating those cell lines containing KRAS or NRAS mutations. We propose that the N-Ras-like state of K-Ras4A, in which it is palmitoylated and farnesylated, may contribute to its high expression levels in melanoma, whereas the K-Ras4B-like state of K-Ras4A, in which it is only farnesylated, may contribute to its high expression levels in colon cancer.
These remarkable observations couple with earlier pioneering studies on H-Ras (4) leading to important conclusions in oncogenic Ras research. These question the completeness of recorded statistics of isoforms in distinct cancer types (5, 6), and call for reassessment of procedures and protocols for tallying oncogenic mutations. In light of these observations, we contend that classifying Ras isoforms solely by their sequences may be erroneous; their signaling states in the tissue should also be catalogued. We note that even though our hypothesis is in agreement with all currently-available observations, to date it has not been directly tested.
K-Ras4A is in-between K-Ras4B and N-Ras
Sequence comparisons of H-Ras, N-Ras, K-Ras4A and K-Ras4B indicate that the catalytic domains (residues 1–166) are almost identical. K-Ras4A1–166 differs from its K-Ras4B1–166 counterpart in only four amino acids (Fig. 1a), located at the edges of helix 5, which is believed to adjoin the membrane. The HVRs of all isoforms at the C-terminal domain differ markedly (Fig. 1b) (7). Importantly, this results in distinct interactions of the HVRs with the membrane (Fig. 1c). Even though direct experimental evidence is lacking, current data suggest that this may differentially restrict Ras orientations (8, 9) thereby encoding selective effector recruitment and signaling. K-Ras4A is most similar to K-Ras4B. Their HVRs are highly positively charged, with that of the K-Ras4B containing an additional positively charged patch. The lysine residues in K-Ras4B form an almost continuous stretch; in K-Ras4A they are fewer and divided into regions, the common polybasic region 1 and 2 (PBR1, PBR2) (3). The lipid post-translational modification patterns at the C-terminal differ as well (Fig. 1b). H-Ras has two palmitoyl groups and a farnesyl, N-Ras one palmitoyl and farnesyl, K-Ras4A similarly one palmitoyl and farnesyl, and K-Ras4B only a farnesyl group. Thus, like K-Ras4B, K-Ras4A has a positively charged HVR – albeit to a lesser extent – and a farnesyl group. PBR2, closer to the C-terminal and just following the palmitoylated C180 and prior to the farnesylated C185, is more important to K-Ras4A membrane association than PBR1. Tsai et al. (3) observed that substitution of the basic residues in common polybasic region 1 and 2 (PBR1+2) with glutamine generated a K-Ras4A mutant that localized on the plasma membrane similarly to N-Ras. Of particular note, elimination of the palmitoylation through a C180S mutation mislocalized K-Ras4A in internal membranes. However, the mutant trafficked to the plasma membrane, unlike palmitoylation-deficient C181S N-Ras. Along similar lines, inhibiting palmitoylation of K-Ras4A with 2-bromopalmitate failed to block its expression on the plasma membrane, which was not the case with N-Ras.
Taken together, K-Ras4A may have two functional states (Fig. 1c): state 1 has polybasic regions and farnesyl; state 2 also has palmitoyl. Since the stoichiometry of the palmitoylation was not determined, the relative proportions of the two K-Ras4A states are unknown. Sequence-wise whereas K-Ras4A state 1 appears functionally similar to K-Ras4B, state 2 resembles N-Ras. In both K-Ras4A and K-Ras4B, the positively charged HVR is able to associate with a negatively charged inner leaflet of the plasma membrane. The palmitoyl, a saturated fatty acid, can insert easily into any lipid microdomain, including lipid rafts or liquid ordered phase membranes. The farnesyl prefers to insert into liquid ordered phase membranes, rich in unsaturated fatty acids. Farnesyl insertion is reversible, since its hydrocarbon chain is shorter and contains cis double bonds, which is not the case for the palmitoyl with a long and straightened lipid tail. The loosely-packed anionic membranes in the liquid ordered phase (10) can accommodate the inserted farnesyl between the lipid tails (8) (Figure 1c), easing its reversible slipping into- and out of- the membrane, while the positively charged K-Ras4B HVR retains Ras on the membrane. This in- and out- movement may be orchestrated by K-Ras complex formation (11) including regulators and effectors, such as Raf, and lead to dynamic signaling bursts. In contrast, our on-going simulations suggest that the fully hydrogenated compact palmitoyl is stably lodged at the interface of zwitterionic solid phase membrane rafts. The farnesyl moiety is a permanent attachment to the Ras protein, whereas the palmitoyl moiety is not (6). Therefore, this permanent/dynamic modification state is in direct contrast to the ability of these modifications to regulate Ras insertion into cell membranes.
Ras isoform-selective effector signaling is largely dictated by its HVR-membrane interactions and PTM modifications which determine its preferred orientation on the membrane and the flexibility, and thus effector accessibility (8, 9). From the conformational standpoint, with five additional aliphatic carbons and additional interactions, geranylgeranyl is likely to resemble the farnesyl; however, with a higher energetic cost for removal from the membrane. Although farnesylation cannot provide sufficient hydrophobicity to stably moor K-Ras4B or K-Ras4A in a lipid bilayer (12), the positively charged HVR patches provide the extra stabilization.
The two states of K-Ras4A may distribute differentially across cancer types
The similarity between K-Ras4A state 1 and K-Ras4B, and K-Ras4A state 2 and N-Ras, raises the question of whether the states are differentially distributed across cancer types. Based on data compiled from the Cancer Cell Line Encyclopedia (CCLE), the International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas Data Portal (TCGA), (C. Der, http://www.cancer.gov/researchandfunding/priorities/ras) the incidence of K-Ras activating mutations is particularly high in adenocarcinomas, with over 95% in pancreatic cancer (overall frequency of all RAS mutations 88%), 85% in colorectal cancer (52%) and 96% in lung adenocarcinoma (32%). N-Ras mutations are frequent in acute myeloid leukemia (AML), 59% (11%) and melanoma, 94% (28%). The chart presented by the Philips lab shows the relative distributions of K-Ras4B versus K-Ras4A in major cancers, strikingly indicating K-Ras4A expression peaking in melanoma and particularly in colorectal cancers (Fig. 2). This raises the question – is K-Ras4A state 1, with only the polybasic regions and the farnesyl, preferentially expressed in adenocarcinomas, including colorectal, pancreatic, and lung, whereas K-Ras4A state 2, with both farnesyl and palmitoyl, preferentially expressed in melanoma and in AML? That is, do these two states of K-Ras4A, like K-Ras4B and N-Ras, differentially distribute across cancer types? If so (13, 14), the clinical implications are significant. In line with this, studies from the Balmain laboratory revealed the unexpected importance of the oncogenic K-Ras4A isoform in lung cancers (15). We speculate that it is state 1 – rather than state 2 – that is important in their lung carcinogenesis model. However, measuring the relative contribution of palmitoylated (state 1) versus depalmitoylated (state 2) endogenous K-Ras4A in tumors or cell lines derived from tumors is challenging. Metabolic labeling shows palmitoylation qualitatively, not quantitatively. Since there are no good immunoprecipitating antibodies for K-Ras, pan-Ras antibodies would need to be used for metabolic labeling studies making it impossible to distinguish endogenous K-Ras4A from N-Ras or H-Ras. Obtaining such quantitative data would require developing an antibody that distinguishes between palmitoylated and depalmitoylated K-Ras4A and using it to determine the stoichiometry of the two species.
K-Ras4A state 1 – but not state 2 – may bind calmodulin
Calmodulin plays an important role in oncogenic K-Ras biology (16–19), and the race is on to obtain a crystal structure, so far without success. Calmodulin temporally down-regulates Raf’s activation and amplifies PI3Kα activation (19, 20). Calmodulin may selectively bind to oncogenic K-Ras4B (21), sequestering its farnesyl from the membrane. Current data point to the farnesyl docking into a hydrophobic pocket in calmodulin with the negatively charged calmodulin surface creating a favorable environment for the positively-charged HVR (22). While additional data and controls in studies of oncogenic K-Ras4B-calmodulin interaction need to be assembled and implemented, the observation by the Philips lab that K-Ras4A can associate with the membrane in the absence of the palmitoyl raises the question of whether calmodulin also interacts with K-Ras4A state 1. If this is indeed the case, as would be expected in its K-Ras4B-like state, it raises the possibility of therapeutic targeting of calmodulin/K-Ras4A/PI3Kα in adenocarcinoma signaling similar to calmodulin/K-Ras4B/PI3Kα (19). With a positively charged HVR and a farnesyl – and no sterically obstructing palmitoyl – K-Ras4A possesses the hallmarks of calmodulin binding, albeit with a lower affinity. Thus, K-Ras4A may bind calmodulin in K-Ras – but not N-Ras – driven cancers.
K-Ras4A may generate signaling redundancy in therapeutics
The observations made in the Tsai et al. paper (3) further raise the question of why K-Ras4A? As K-Ras4B-like (state 1), the positively charged HVR avidly interacts with the negatively-charged membrane – but not as avidly as K-Ras4B; as N-Ras-like (state 2), the lower HVR charge can have an advantage in neutral membranes – though with lesser avidity than N-Ras. The K-Ras4A HVR sequence places it between K-Ras4B and N-Ras, suggesting that its post-translational modification pathway in the cell (farnesyl-only or farnesyl+palmitoyl) decides its functional fate. This leads us to speculate that K-Ras4A is the oldest Ras species which could fulfill both functions. As to the question of why has it been retained by evolution, one reasonable possibility is to create redundant pathways (23). This functional redundancy is the problem we currently face when targeting K-Ras and N-Ras cancers (24).
Finally, this raises the question of why unlike K-Ras4B, K-Ras4A does not bind to the cytosolic chaperone δ-subunit of cGMP phosphodiesterase type 6 (PDE6δ) whereas the more dissimilar N-Ras HVR sequence does (3). We believe that this requires further examination.
H-Ras, with two palmitoyls and a farnesyl, also exists in two states
A decade ago a pioneering study (4) observed that H-Ras, with two palmitoyls and a farnesyl may also exist in two states – N-Ras-like singly-palmitoylated and doubly-palmitoylated H-Ras. This compelling discovery can explain the heterogeneity and the expression of oncogenic H-Ras isoforms in largely N-Ras driven cancers, such as AML (13, 14). The sequences of N-Ras and H-Ras do not appear to possess distinctive features. The palmitoylation half-life of Ras proteins vary, and has been shown to be shorter for oncogenic H-Ras12V than for WT H-Ras (7, 25, 26), with palmitoylation/depalmitoylation linked to the GTP/GDP cycle (25). Regulation of the cleavage of each of the two palmitoyl-protein thioesterase linkages of H-Ras (4) can take place through FKBP12-catalyzed prolyl isomerization, indicating that depalmitoylation is enzymatic (27). Thus, our thesis is that in the singly palmitoylated state, oncogenic H-Ras can substitute for N-Ras which is singly palmitoylated. The two distinct states of oncogenic H-Ras may explain its occurrence in AML; in this cancer the N-Ras-like singly palmitoylated state – but not the doubly palmitoylated state – may be expressed.
Implications for different types of cancers
Functional redundancy versus specificity has been an ongoing question in cellular signaling (23, 28, 29). Perhaps the most significant conclusion is the emerging higher functional complexity of Ras isoforms and splice variants than previously thought. Even though the frequency of Ras gene mutations varies across cancer types, the preferential occurrence of oncogenic K-Ras versus N-Ras in colorectal carcinoma cannot be explained solely on this basis. N-Ras (28) and H-Ras (30) are expressed in mouse colorectal cancer cells; despite this, K-Ras mutations are six times more frequent than N-Ras, and H-Ras mutations are not present. However, expression levels may not indicate the effective local concentration at the membrane, which also depend on the HVR post-translational states, local membrane composition, presence of certain scaffolding proteins such as galectin 1, etc. In light of the new findings, the current statistics of mutant occurrences in distinct cancers may need to be revised and broken down into tissue- and cell-specific Ras isoform states. The interactions, mechanisms, and signaling pathways may differ.
To date, in cancer cell line analysis, K-Ras4A has been taken as a homogeneous entity. Based on observations reported in the literature we point out that this common perception may be mistaken. K-Ras4A occurrence in oncogenic K-Ras4B-driven adenocarcinomas, like colorectal, pancreatic and lung cancers may mirror a K-Ras4B-like state (it is only farnesylated), whereas K-Ras4A occurrence in N-Ras-driven cancers, such as melanoma and acute myeloid leukemia may reflect the N-Ras-like state (farnesylated+palmitoylated). This is important because of the clinical and biological implications. It emphasizes that the recorded statistics of isoforms in distinct cancer types does not account for their functional states in specific tumors which may differ according to the tumor type. K-Ras4A in melanoma or acute myeloid leukemia (AML) may not be identical to K-Ras4A in pancreatic ductal adenocarcinoma (PDAC), lung or colorectal cancer (CRC). This calls for a reanalysis of procedures and mutational data, and re-evaluating treatments. This also holds for the less frequent farnesylated and doubly-palmitoylated H-Ras, which may also exist in two states – N-Ras-like singly-palmitoylated and doubly-palmitoylated H-Ras, although oncogenic H-Ras is not as frequent compared to mutational activation of N-Ras and K-Ras (1). Here our thesis is that isoform identification in cancer rests not only on its sequence, but on its possible tissue-specific functional state. How is the differential regulation between states 1 and 2 accomplished? Among the several possible mechanisms, we favor one whereby the two states preferentially act in distinct cancers, i.e., are tissue-dependent. However, there is no direct evidence either way, and this requires experimental studies.
Future Prospects
Nature often re-exploits useful mechanisms. Thus, the mechanism proposed here for accomplishing distinct isoform functions which are dictated by differing membrane trafficking motifs can be general. The recent elegant work of Nishimura and Linder (31), who described two states of processing for one of the Cdc42 HVR splice variants, that are also relevant to distinctions among other highly related cancer-associated small GTPases such as the RalA and RalB isoforms (32), may provide examples.
Direct experimental evidence at the cellular or biochemical level for different functions of any Ras isoform, including K-Ras4A versus K-Ras4B, is still lacking. The evidence for differential functions comes from isoform utilization in Ras driven cancer (where K-Ras mutations predominate) and from transgenic models where isoforms do not always substitute for each other, as for example observed in the differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon (28). Notwithstanding, the emerging picture conforms to the general principle adopted by evolution, where single genes are adapted for different functions by extending isoform heterogeneity.
Recent reports in the literature support the call for reevaluation of hypotheses and approaches used to calculate the statistics of the occurrence of Ras isoforms in different cancer types. Enhanced tumor isoform and mutational analysis protocols that are able to quantitatively distinguish among isoform species may better forecast cancer progression and guide platforms for therapeutics. Tsai et al. (3) utilized a newly generated antibody that detected K-Ras4A but not K-Ras4B. This emphasizes the importance of tool and protocol availability to identify the different isoforms and functional states of K-Ras. Thus challenges are posed to the community to develop methods to accurately and completely determine and decipher molecular data, and resolve the treatment implications (33). The critical importance of completeness emphasizes the enormity of the challenge. Improved methods will more accurately identify drug targets, advance innovative concepts, unveil mechanisms and decipher redundant pathways. Pursuing these has the potential of inspiring new research directions.
Acknowledgments
We thank Mark Philips for his comments on the manuscript. This project has been funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported [in part] by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Cox AD, Der CJ. Ras history: The saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fetics SK, Guterres H, Kearney BM, Buhrman G, Ma B, Nussinov R, et al. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure. 2015;23:505–16. doi: 10.1016/j.str.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A. 2015;112:779–84. doi: 10.1073/pnas.1412811112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, et al. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol. 2005;25:6722–33. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nature reviews Drug discovery. 2014;13:828–51. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox AD, Der CJ, Philips MR. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin Cancer Res. 2015;21:1819–27. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;121:421–7. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- 8.Jang H, Abraham SJ, Chavan TS, Hitchinson B, Khavrutskii L, Tarasova NI, et al. Mechanisms of Membrane Binding of Small GTPase K-Ras4B Farnesylated Hypervariable Region. J Biol Chem. 2015;290:9465–77. doi: 10.1074/jbc.M114.620724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abankwa D, Gorfe AA, Inder K, Hancock JF. Ras membrane orientation and nanodomain localization generate isoform diversity. Proc Natl Acad Sci U S A. 2010;107:1130–5. doi: 10.1073/pnas.0903907107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoemaker SD, Vanderlick TK. Intramembrane electrostatic interactions destabilize lipid vesicles. Biophys J. 2002;83:2007–14. doi: 10.1016/S0006-3495(02)73962-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, et al. GTP-Dependent K-Ras Dimerization. Structure. 2015;23:1325–35. doi: 10.1016/j.str.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–90. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 13.Bowen DT, Frew ME, Hills R, Gale RE, Wheatley K, Groves MJ, et al. RAS mutation in acute myeloid leukemia is associated with distinct cytogenetic subgroups but does not influence outcome in patients younger than 60 years. Blood. 2005;106:2113–9. doi: 10.1182/blood-2005-03-0867. [DOI] [PubMed] [Google Scholar]
- 14.Parikh C, Subrahmanyam R, Ren R. Oncogenic NRAS, KRAS, and HRAS exhibit different leukemogenic potentials in mice. Cancer Res. 2007;67:7139–46. doi: 10.1158/0008-5472.CAN-07-0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To MD, Wong CE, Karnezis AN, Del Rosario R, Di Lauro R, Balmain A. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat Genet. 2008;40:1240–4. doi: 10.1038/ng.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villalonga P, Lopez-Alcala C, Bosch M, Chiloeches A, Rocamora N, Gil J, et al. Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol Cell Biol. 2001;21:7345–54. doi: 10.1128/MCB.21.21.7345-7354.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Alcala C, Alvarez-Moya B, Villalonga P, Calvo M, Bachs O, Agell N. Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J Biol Chem. 2008;283:10621–31. doi: 10.1074/jbc.M706238200. [DOI] [PubMed] [Google Scholar]
- 18.Bosch M, Gil J, Bachs O, Agell N. Calmodulin inhibitor W13 induces sustained activation of ERK2 and expression of p21(cip1) J Biol Chem. 1998;273:22145–50. doi: 10.1074/jbc.273.34.22145. [DOI] [PubMed] [Google Scholar]
- 19.Nussinov R, Muratcioglu S, Tsai CJ, Jang H, Gursoy A, Keskin O. The key role of calmodulin in KRAS-driven adenocarcinomas. Mol Cancer Res. 2015;13:1265–73. doi: 10.1158/1541-7786.MCR-15-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joyal JL, Burks DJ, Pons S, Matter WF, Vlahos CJ, White MF, et al. Calmodulin activates phosphatidylinositol 3-kinase. J Biol Chem. 1997;272:28183–6. doi: 10.1074/jbc.272.45.28183. [DOI] [PubMed] [Google Scholar]
- 21.Liao J, Planchon SM, Wolfman JC, Wolfman A. Growth factor-dependent AKT activation and cell migration requires the function of c-K(B)-Ras versus other cellular ras isoforms. J Biol Chem. 2006;281:29730–8. doi: 10.1074/jbc.M600668200. [DOI] [PubMed] [Google Scholar]
- 22.Abraham SJ, Nolet RP, Calvert RJ, Anderson LM, Gaponenko V. The hypervariable region of K-Ras4B is responsible for its specific interactions with calmodulin. Biochemistry. 2009;48:7575–83. doi: 10.1021/bi900769j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drosten M, Lechuga CG, Barbacid M. Genetic analysis of Ras genes in epidermal development and tumorigenesis. Small GTPases. 2013;4:236–41. doi: 10.4161/sgtp.26905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nussinov R, Tsai CJ, Mattos C. ‘Pathway drug cocktail’: targeting Ras signaling based on structural pathways. Trends Mol Med. 2013;19:695–704. doi: 10.1016/j.molmed.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. Distinct rates of palmitate turnover on membrane-bound cellular and oncogenic H-ras. J Biol Chem. 2003;278:19292–300. doi: 10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- 26.Magee AI, Gutierrez L, McKay IA, Marshall CJ, Hall A. Dynamic fatty acylation of p21N-ras. The EMBO journal. 1987;6:3353–7. doi: 10.1002/j.1460-2075.1987.tb02656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, et al. FKBP12 binds to acylated H-ras and promotes depalmitoylation. Molecular cell. 2011;41:173–85. doi: 10.1016/j.molcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–8. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellano E, Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2:216–31. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuhmacher AJ, Guerra C, Sauzeau V, Canamero M, Bustelo XR, Barbacid M. A mouse model for Costello syndrome reveals an Ang II-mediated hypertensive condition. J Clin Invest. 2008;118:2169–79. doi: 10.1172/JCI34385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura A, Linder ME. Identification of a novel prenyl and palmitoyl modification at the CaaX motif of Cdc42 that regulates RhoGDI binding. Mol Cell Biol. 2013;33:1417–29. doi: 10.1128/MCB.01398-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentry LR, Nishimura A, Cox AD, Martin TD, Tsygankov D, Nishida M, et al. Divergent Roles of CAAX Motif-Signaled Posttranslational Modifications in the Regulation and Subcellular Localization of Ral GTPases. J Biol Chem. 2015 doi: 10.1074/jbc.M115.656710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nussinov R, Jang H, Tsai CJ. The structural basis for cancer treatment decisions. Oncotarget. 2014;5:7285–302. doi: 10.18632/oncotarget.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]