Abstract
The ability of bacteria to use cGMP as a second messenger has been controversial for decades. Recently, nucleotide cyclases from Rhodospirillum centenum, GcyA, and Xanthomonas campestris, GuaX, have been shown to possess guanylate cyclase activities. Enzymatic activities of these guanylate cyclases measured in vitro were low, which makes interpretation of the assays ambiguous. Protein sequence analysis at present is insufficient to distinguish between bacterial adenylate and guanylate cyclases, both of which belong to nucleotide cyclases of type III. We developed a simple method for discriminating between guanylate and adenylate cyclase activities in a physiologically relevant bacterial system. The method relies on the use of a mutant cAMP receptor protein, CRPG, constructed here. While wild-type CRP is activated exclusively by cAMP,_CRPG can be activated by either cAMP or cGMP. Using CRP- and CRPG-dependent lacZ expression in two E. coli strains, we verified that R. centenum GcyA and X. campestris GuaX have primarily guanylate cyclase activities. Among two other bacterial nucleotide cyclases tested, one, GuaA from Azospillrillum sp. B510, proved to have guanylate cyclase activity, while the other one, Bradyrhizobium japonicum CyaA, turned out to function as an adenylate cyclase. The results obtained with this reporter system were in excellent agreement with direct measurements of cyclic nucleotides secreted by E. coli expressing nucleotide cyclase genes. The simple genetic screen developed here is expected to facilitate identification of bacterial guanylate cyclases and engineering of guanylate cyclases with desired properties.
Keywords: cGMP, cAMP, nucleotide cyclase, second messenger, signal transduction, Escherichia coli
INTRODUCTION
Cyclic adenosine 3’,5’-monophosphate (cAMP) and cyclic guanosine 3’,5’-monophosphate (cGMP) are the most important nucleotide messengers in eukaryotes. Bacteria also use cAMP to regulate diverse processes that include utilization of alternative sugars (catabolite repression), motility, virulence, etc. In contrast to cAMP, the existence and role of cGMP in bacteria has remained controversial for decades (1,2). Several studies claimed validity to the presence of cGMP signaling in bacteria, only to be rejected by subsequent rigorous testing.
Two recent reports have provided evidence of guanylate cyclases (cGMP synthases) in the proteobacterial species Rhodospillium centenum and Xanthomonas campestris (3,4). These reports documented intracellular cGMP and identified protein targets of cGMP-dependent signaling. In addition, in both studies, guanylate cyclases were overexpressed in E. coli, purified, and their activities were measured in vitro. However, enzymatic activities were low, which raised questions about the functionality of these enzymes in vivo. R. centenum GcyA and X. campestris GuaX are large multidomain proteins. At present, conditions required for their full activities are not yet known. Further, protein sequences of the substrate-binding sites in these bacterial guanylate cyclases are different from the sequences of eukaryotic guanylate cyclases (3,4), which suggests that these classes of enzymes have evolved from adenylate cyclases in parallel pathways. However, the low number of bacterial guanylate cyclases does not yet allow reliable distinction between guanylate and adenylate cyclases based on protein sequences.
To facilitate the identification of new bacterial guanylate cyclases, we developed an E. coli-based reporter system that does not require protein purification. The key component of the system is an engineered mutant cAMP receptor protein, CRP (also known as catabolite activator protein, CAP) (5,6) that can be activated by either cAMP or cGMP. This mutant protein, designated CRPG, along with the wild-type CRP, allows one to monitor guanylate cyclase activity in E. coli via CRP- and CRPG-dependent lacZ gene expression. The heterologous nucleotide cyclases expressed in the E. coli mutant lacking its native adenylate cyclase, Cya, can be assigned to adenylate cyclases if they activate lacZ expression via both CRP and CRPG. If they activate lacZ expression via CRPG only, they represent guanylate cyclases. This system is expected not only to facilitate characterization of cGMP-dependent signal transduction in bacteria but also to provide an E. coli-based screening platform for engineering guanylate cyclases with desired properties (for example, light-activated guanylate cyclases for optogenetic applications [7,8]). In addition, we show in this study that E. coli secretes most cAMP as well as cGMP, therefore direct measurements of cyclic nucleotides secreted by E. coli (e.g., by ELISA) presents yet another method for assessing the substrate specificity of heterologous nucleotide cyclases.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown at 37°C in LB medium. Antibiotics were used at the following concentrations (μg/mL): ampicillin (Ap), 100; kanamycin (Km), 25; spectinomycin (Sp), 100; tetracycline (Tc), 12. Cyclic nucleotides, cAMP or cGMP (Sigma), were added to the LB agar medium to a final concentration of 0.1 mM.
Table 1.
Strains and plasmids used in this study.
| Strain | Description | Source or Reference |
|---|---|---|
| DH5α | Strain used for cloning | New England Biolabs |
| UQ3811 | Strain used for CRP protein purification | 10 |
| BL21 [DE3] cya crp |
fhuA2 [lon] ompT gal (λDE3) [dcm] ΔhsdS cpr∷Kmr cyaA∷Spr |
New England Biolabs |
| Plasmid | ||
| pET23a(+) | Bacterial expression vector, Apr | Invitrogen |
| pETcyaA | pET23a(+) encoding B. japonicum USDA6 CyaA | This study |
| pETcyaB1 | pET23a(+) encoding aa 591-857 (catalytic domain) of Nostoc sp. CyaB1 | This study |
| pETgcyA | pET23a(+) encoding R. centenum GcyA | This study |
| pETguaA | pET23a(+) encoding Azospillrillum sp. B510 GuaA | This study |
| pETguaX | pET23a(+) encoding X. campestris GuaX | This study |
| pHY26-1 | Vector expressing His-tagged E. coli CRP | 10 |
| pHYC445 | Vector expressing His-tagged CRP triple (R123N, T127C, S128T) mutant, CRPG | This study |
| pRK415 | Broad-host range vector, IncP, TcR | 11 |
| pRK-CRP | pRK415 encoding E. coli CRP under the lac promoter | This study |
| pRK-CRPG | pRK415 encoding CRPG under the lac promoter | This study |
The BL21 [DE3] cya crp double mutant was constructed by a one-step gene inactivation method (9) using the BL21 [DE3] cya strain described earlier (8). lacZ expression in BL21 [DE3] cya crp expressing CRP or CRPG was assessed on LB plates supplemented with Ap, Tc, X-gal (40 μg/mL) and IPTG (50 μM) after 36 h incubation at 37 °C.
Plasmid construction
The triple, R123N T127C S128T, CRP mutant (designated CRPG) was constructed by site-directed mutagenesis using the QuikChange kit (Agilent Technologies). Plasmid pHY26-1 (10) containing the wild-type crp gene was used as template to yield pHYC445 that contains crpG. The crp and crpG genes were PCR-amplified from pHY26-1 and pHYC445, respectively, and cloned into XbaI and EcoRI sites of the vector pRK415 (11). The resulting plasmids pRKcrp and pRKcrpG express, respectively, crp and crpG from the lac promoter.
All nucleotide cyclases tested here were cloned into pET23a(+). The adenylate cyclase domain (aa 591-857) of the CyaB1 protein from Nostoc sp. PCC 7120 and the full-length guaB gene were synthesized by GeneArt Strings (Life Technologies) and assembled with pET23(+) using Gibson assembly (NEB). The gcyA gene (rc1-3783) from Rhodospirillium centenum was amplified by PCR from the plasmid kindly provided by Dr. C. Bauer (3). The guaA (AZL_d00190) and XC_0250 (hereby designated guaX) genes were amplified from genomic DNA of Azospillrillum sp. B510 and Xanthomonas campestris kindly provided by Drs. S. Sato and R. Ryan, respectively.
Overexpression and purification of CRP proteins
The His-tagged CRP and CRPG were overexpressed in strain UQ3811 and purified using a nickel-nitrilotriacetate column according to the manufacturer’s instruction (Novagen), as described earlier (10). The final purity of both proteins was >95% (based on SDS-PAGE).
Responsiveness of CRP and CRPG to cAMP and cGMP
To determine how effectively the CRP and CRPG proteins responded to cGMP and cAMP, the in vitro DNA-binding activities of CRP and CRPG were measured with varying concentrations of each cyclic nucleotide as described previously (12,13). In vitro DNA-binding assays were carried out using a fluorescence polarization method. The target DNA was a 26-mer CRP consensus sequence (5′-GTAAATGTGATGTACATCACATGGAT-3′) labeled with a fluorescent dye, Texas Red, as described previously (10). When the DNA-protein interaction occurs, fluorescence polarization is increased, which reflects slowed Brownian motion of the labeled DNA by the bound protein. Assays were performed in 50 mM Tris-HCl (pH 8.0), 150 mM KCl, and 1 mM EDTA with a probe concentration of 5 nM in the presence of 6.4 μM salmon sperm DNA (nonspecific competitor). The concentration of each protein was 150 nM.
Detection of secreted cyclic nucleotides
cAMP and cGMP production in the BL21[DE3] mutants that express the respective nucleotide cyclases were assessed by specific cAMP and cGMP ELISA kits (Enzo Life Sciences) according to the instructions of the manufacturer. Briefly, overnight cultures were transferred to fresh M9 minimal medium at an A600 of 0.02 and protein expression was induced by the addition of 0.1 mM IPTG. Growth was continued at 37°C until the cells reached stationary phase (A600 , 3~4) and cell-free supernatants were collected by centrifugation at 4°C. Since E. coli secretes most cAMP and cGMP into the medium (7,14), extracellular cyclic nucleotide levels were determined in cell-free supernatants.
RESULTS AND DISCUSSION
Construction and properties of the cAMP- and cGMP-responsive CRP mutant
To construct the CRP mutant that can be activated by cGMP, we combined three amino acid substitutions (R123N, T127C and S128T) in the cAMP-binding pocket, each of which has been shown earlier to improve cGMP responsiveness (13,15,16). We expected that mutations in all three residues would have a cumulative effect. R123N was suggested to help properly position cGMP in the binding pocket by enabling a new H-bond to Asn (15). The exact role of T127C and S128T replacements, which increased CRP responsiveness to cGMP, is unknown (16). It is noteworthy that the residue corresponding to Thr128 of CRP in the Pseudomonas aeruginosa CRP homolog, Vfr, is also important for cGMP responsiveness (13).
We overexpressed and purified, using nickel affinity resin, the triple mutant (R123N, T127C, S128T), designated CRPG. Figure 1 shows that the DNA binding by CRPG was stimulated equally well by cAMP and cGMP (Fig. 1). This is in contrast to DNA binding by wild-type CRP, which was increased only by cAMP but not cGMP. Further, the affinity of CRPG to DNA in the presence of cAMP and cGMP was approximately the same as the affinity to DNA of CRP in the presence of cAMP (Fig. 1). Therefore, in accord with our expectation, the three individual substitutions in the cAMP-binding pocket had cumulative effect on improving cGMP-dependent DNA binding by CRP.
Figure 1.
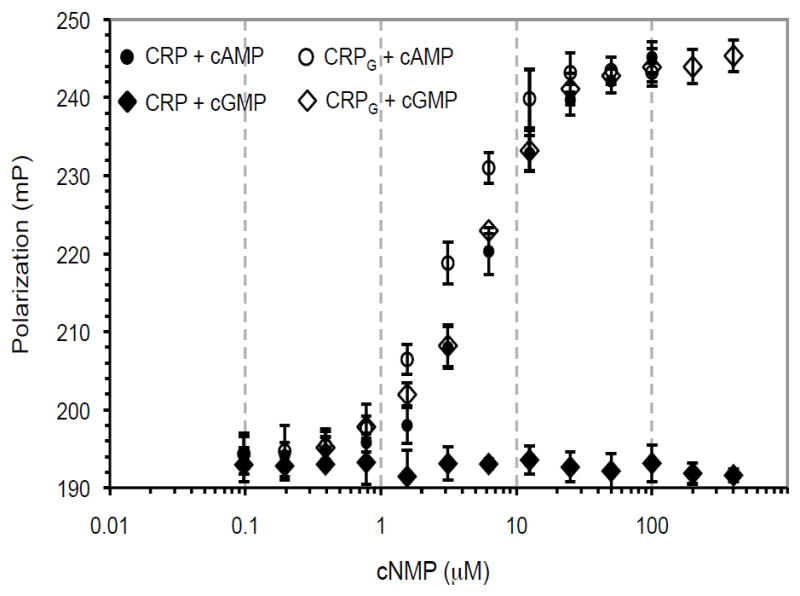
DNA-binding of CRP and CRPG at various concentrations of cAMP and cGMP. In vitro DNA binding was measured by fluorescence anisotropy.
Discrimination between cAMP and cGMP by wild-type CRP is complex because both molecules are known to bind with approximately equal affinities (17,18), yet, unlike cAMP, which stimulates DNA binding, cGMP inhibits it (19). Interestingly, cAMP binds to CRP in the anti conformation (20,21), whereas cGMP binds in the syn conformation (12,19,21). The structure of the cGMP-binding protein XC_0249 from Xanthomonas campestris shows that it too binds cGMP in the syn conformation (4,22). These observations are consistent with the dominant conformations of cAMP and cGMP in aqueous solutions in vitro, which are 95:5 syn/anti for cGMP and 30:70 syn/anti for cAMP (23). Based on these considerations, we hypothesize that CRPG binds cGMP in the syn conformation as does wild-type CRP, but the syn-cGMP binding of CRPG leads to an active form that may be locally different but globally similar to the active form of the wild-type CRP in the presence of cAMP. A crystal structure of the CRPG protein in the presence of cGMP is needed to understand the cGMP-activation of DNA binding by CRPG. However, regardless of the activation mechanism, equal responsiveness of CRPG to cAMP and cGMP presented an opportunity to develop an E. coli-based system that can discriminate between adenylate and guanylate cyclase activities.
CRP/CRPG reporter-based discrimination of adenylate and guanylate cyclases
To design a system for discriminating between adenylate and guanylate cyclase activities, we constructed a BL21 [DE3] cya crp double mutant that lacks genes encoding a native adenylate cyclase, Cya, and CRP. The crp and crpG genes were cloned in vector pRK415 downstream of a lac promoter and introduced into this double mutant. First, we tested the ability of the crp and crpG genes to activate the chromosomal lacZ gene expression in the presence of exogenously added cAMP or cGMP. As expected, CRP activated lacZ expression (judged by the appearance of blue color) in the presence of cAMP but not cGMP, whereas CRPG activated lacZ expression in the presence of both cGMP and, to a lesser extent, cAMP (Fig. 2A). We do not understand the apparent preference of CRPG for cGMP versus cAMP in E. coli in light of the approximately equal affinities for these cyclic nucleotides in vitro (Fig. 1). However, this fortunate circumstance only improves the discriminating capacity of the CRPG/CRP system.
Figure 2.
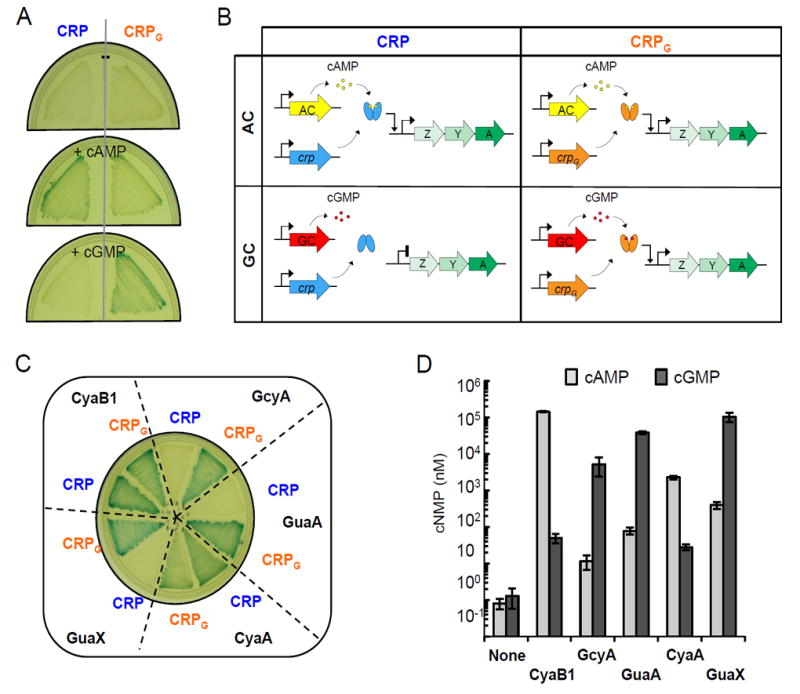
The CRP/CRPG reporter system for detecting adenylate and guanylate cyclase activities. A, The BL21 [DE3] cya crp strains expressing wild-type CRP from pRKcrp (A) or CRPG from pRKcrpG (G) were grown with or without the addition of cAMP or cGMP. The strain expressing CRP developed blue color only in the presence of cAMP whereas the strain expressing CRPG developed blue color in the presence of either cAMP or cGMP. B, Heterologous nucleotide cyclases were introduced into the BL21 [DE3] cya crp mutants expressing CRP or CRPG. The CyaB1 (catalytic domain) and CyaA (full-length) produced blue color in both strains, while GcyA, GuaA and GuaX produced blue colonies only in the presence of CRPG. The medium contained 50 μM IPTG. C, Analysis of secreted cNMPs from the E. coli strains expressing nucleotide cyclases. The same strains as in panel B were used. Cyclic nucleotide levels were measured by ELISA using cAMP- and cGMP-specific antibodies. Average data and standard deviations from three independent experiments are shown.
Next, we tested known adenylate and guanylate cyclases. The adenylate cyclase domain of CyaB1 from Nostoc sp. is known to possess adenylate cyclase activity (24). We introduced this domain into the BL21 [DE3] cya crp strains carrying either CRP or CRPG and observed that it activated lacZ expression in both test strains (Fig. 2B). In contrast, the full-length proteins R. centenum GcyA and X. campestris GuaX activated lacZ expression only in the presence of CRPG but not CRP (Fig. 2B). This indicates that E. coli cells expressing GcyA or GuaX accumulated mostly cGMP but not cAMP, which is consistent with the original reports (3,4).
To further test the utility of the CRP/CRPG system, we cloned two genes encoding putative nucleotide cyclases of unknown substrate specificity. One of these, AZL_d00190, hereby designated guaA, originates from plasmid pAB510d present in Azospillrillum sp. B510. This protein was suggested to have guanylate cyclase activity (3). Another enzyme, hereby designated CyaA, from Bradyrhizobium japonicum USDA 6 (YUI_RS0121825) had unknown substrate preference. The CRP/CRPG test showed that GuaA is a guanylate cyclase, while CyaA is an adenylate cyclase (Fig. 2B). Note that in our assay, expression of nucleotide cyclase and CRP/CRPG are controlled by IPTG concentration, which can be increased to enhance assay sensitivity (in case of low activity nucleotide cyclases) or decreased (to better discriminate between adenylate and guanylate cyclase activities of super-active cyclases).
Measurements of cNMPs secreted by E. coli expressing nucleotide cyclases
In search of an independent means for verifying the performance of the CRP/CRPG reporter system, we exploited the fact that both cAMP (14) and cGMP (7) are actively secreted by E. coli into the medium, although secretion efficiencies appear to differ dependent on which nucleotide is secreted (7). We measured secreted cNMPs in the BL21 [DE3] cya crp mutant expressing the nucleotide cyclases tested by the CRPG/CRP system. The results of analysis of secreted nucleotides via ELISA that involves the cAMP- and cGMP-specific antibodies (Table 2 and Fig. 2C) were in excellent agreement with the results of the E. coli CRP/CRPG reporter system (Fig. 2B). All strains expressing guanylate cyclases secreted >102-fold higher amounts of cGMP than cAMP, while strains expressing adenylate cyclases secreted 10-102-fold higher amounts of cAMP than cGMP. Such large differences make the distinction between these enzymes unambiguous. Importantly, the absolute levels of cNMPs secreted by cells expressing different cyclases varied widely, by orders of magnitude. The absolute amounts of secreted cNMPs may reflect differences in the expression levels, solubility of the heterologous cyclases as well as differences in their relative enzymatic activities. Despite such variation, the distinction between two classes of nucleotide cyclases was clear in the ELISA test as well as in the CRPG/CRP system reporter system.
Table 2.
Cyclic nucleotide production by nucleotide cyclases in strain BL21[DE3] cya crp.
| Protein | None | CyaB1 | GcyA | GuaA | CyaA | GuaX |
|---|---|---|---|---|---|---|
|
|
||||||
| Nucleotide, nM | ||||||
| cAMP | 0.8 ± 0.3 | 1.4 ± 0.5 ×105 | 11.7 ± 4.9 | 78.6 ± 16.9 | 2.3 ± 0.3 ×102 | 4.0 ± 0.9 ×102 |
| cGMP | 1.3 ± 0.7 | 49.5 ± 14.1 | 5.2 ± 2.8 ×103 | 3.8 ± 0.3 ×104 | 28.0 ± 5.2 | 1.0 ± 0.3 ×105 |
CONCLUSIONS
The triple, R123N T127C S128T, mutant of the cAMP receptor protein, CRPG, binds DNA in vitro equally well in the presence of either cAMP or cGMP. In E. coli, CRPG activates lacZ gene expression in response to both cyclic nucleotides but shows preference to cGMP. The E. coli CRP/CRPG reporter system developed here presents a reliable tool for distinguishing between adenylate and guanylate cyclase activities of nucleotide cyclases without the need of enzyme purification. We expect this system to facilitate (i) characterization of cGMP-dependent signal transduction in bacteria and (ii) engineering of guanylate cyclases with desired properties.
Acknowledgments
We are grateful to Drs. Carl Bauer, Rob Ryan and Shusei Sato for plasmid and genomic DNA and to Dr. Kurt Miller for manuscript editing. This study was supported by National Institutes of Health grants 5R21CA167862 (to MG) and 1R15AI101919-01 (to HY).
Footnotes
The authors declare no conflict of interest.
References
- 1.Linder JU cGMP production in bacteria. Mol Cell Biochem. 2010;334:215–219. doi: 10.1007/s11010-009-0321-0. [DOI] [PubMed] [Google Scholar]
- 2.Gomelsky M. cAMP, c-di-GMP, c-di-AMP and now cGMP: Bacteria use them all! Mol Microbiol. 2011;79:562–565. doi: 10.1111/j.1365-2958.2010.07514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marden JN, Dong Q, Roychowdhury S, Berleman JE, Bauer CE. Cyclic GMP controls Rhodospirillum centenum cyst development. Mol Microbiol. 2011;79:600–615. doi: 10.1111/j.1365-2958.2010.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An SQ, Chin KH, Febrer M, McCarthy Y, Yang JG, Liu CL, Swarbreck D, Rogers J, Maxwell Dow J, Chou SH, Ryan RP. A cyclic GMP-dependent signalling pathway regulates bacterial phytopathogenesis. EMBO J. 2013;32:2430–2438. doi: 10.1038/emboj.2013.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zubay G, Schwartz D, Beckwith J. Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc Natl Acad Sci USA. 1970;66:104–110. doi: 10.1073/pnas.66.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fic E, Bonarek P, Gorecki A, Kedracka-Krok S, Mikolajczak J, Polit A, Tworzydlo M, Dziedzicka-Wasylewska M, Wasylewski Z. cAMP receptor protein from Escherichia coli as a model of signal transduction in proteins--a review. J Mol Microbiol Biotechnol. 2009;17:1–11. doi: 10.1159/000178014. [DOI] [PubMed] [Google Scholar]
- 7.Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem. 2010;285:41501–41508. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryu MH, Kang IH, Nelson MD, Jensen TM, Lyuksyutova AI, Siltberg-Liberles J, Raizen DM, Gomelsky M. Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci USA. 2014;111:10167–72. doi: 10.1073/pnas.1324301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes i Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youn H, Kerby RL, Conrad M, Roberts GP. Study of highly constitutively active mutants suggests how cAMP activates cAMP receptor protein. J Biol Chem. 2006;281:1119–1127. doi: 10.1074/jbc.M509421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 12.Youn H, Koh J, Roberts GP. Two-state allosteric modeling suggests protein equilibrium as an integral component for cyclic AMP (cAMP) specificity in the cAMP receptor protein of Escherichia coli. J Bacteriol. 2008;190:4532–4540. doi: 10.1128/JB.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serate J, Roberts GP, Berg O, Youn H. Ligand responses of Vfr, the virulence factor regulator from Pseudomonas aeruginosa. J Bacteriol. 2011;193:4859–4868. doi: 10.1128/JB.00352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterkofsky A, Gazdar C. Glucose and metabolism of adenosine 3’-5’-cyclic monophosphate in Escherichia coli. Proc Natl Acad Sci USA. 1971;68:2794–2798. doi: 10.1073/pnas.68.11.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Youn H, Kerby RL, Koh J, Roberts GP. A C-helix residue, Arg-123, has important roles in both the active and inactive forms of the cAMP receptor protein. J Biol Chem. 2007;282:3632–3639. doi: 10.1074/jbc.M606602200. [DOI] [PubMed] [Google Scholar]
- 16.Lee EJ, Glasgow J, Leu SF, Belduz AO, Harman JG. Mutagenesis of the cyclic AMP receptor protein of Escherichia coli: targeting positions 83, 127 and 128 of the cyclic nucleotide binding pocket. Nucleic Acids Res. 1994;22:2894–2901. doi: 10.1093/nar/22.15.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin SH, Lee JC. Communications between the high-affinity cyclic nucleotide binding sites in Escherichia coli cyclic AMP receptor protein: effect of single site mutations. Biochemistry. 2002;41:11857–11867. doi: 10.1021/bi026099z. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi M, Blazy B, Baudras A. Ligand-modulated binding of a gene regulatory protein to DNA: quantitative analysis of cyclic-AMP induced binding of CRP from Escherichia coli to nonspecific and specific DNA targets. J Mol Biol. 1989;207:783–796. doi: 10.1016/0022-2836(89)90244-1. [DOI] [PubMed] [Google Scholar]
- 19.Seok SH, Im H, Won HS, Seo MD, Lee YS, Yoon HJ, Cha MJ, Park JY, Lee BJ. Structures of inactive CRP species reveal the atomic details of the allosteric transition that discriminates cyclic nucleotide second messengers. Acta Cryst. 2014;D70:1726–1742. doi: 10.1107/S139900471400724X. [DOI] [PubMed] [Google Scholar]
- 20.Passner JM, Schultz SC, Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution. J Mol Biol. 2000;304:847–859. doi: 10.1006/jmbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- 21.Popovych N, Tzeng SR, Tonelli M, Ebright RH, Kalodimos CG. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci USA. 2009;106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomelsky M, Galperin MY. Bacterial second messengers, cGMP and c-di-GMP, in a quest for regulatory dominance. EMBO J. 2013;32:2421–2423. doi: 10.1038/emboj.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yathindra N, Sunderalingam M. Conformations of cyclic 3’,5’-nucleotides. Effect of the base on the Syn-Anti conformer distribution. Biochem Biophys Res Commun. 1974;56:119–126. doi: 10.1016/s0006-291x(74)80323-2. [DOI] [PubMed] [Google Scholar]
- 24.Kanacher T, Schultz A, Linder JU, Schultz JE. A GAF-domain-regulated adenylyl cyclase from Anabaena is a self-activating cAMP switch. EMBO J. 2002;21:3672–3680. doi: 10.1093/emboj/cdf375. [DOI] [PMC free article] [PubMed] [Google Scholar]


