SUMMARY
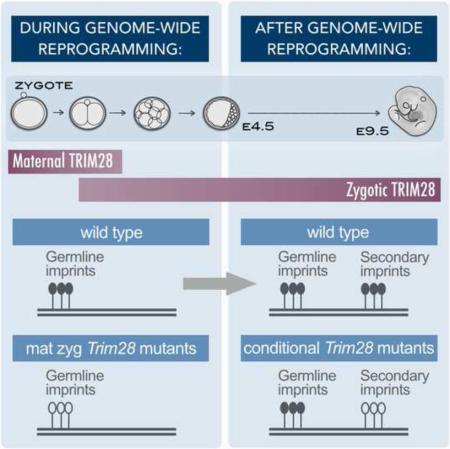
Genomic imprinting depends on the establishment and maintenance of DNA methylation at imprinting control regions. However, the mechanisms by which these heritable marks influence allele-specific expression are not fully understood. By analyzing maternal, zygotic, maternal-zygotic, and conditional Trim28 mutants, we found that the transcription factor TRIM28 controls genomic imprinting through distinct mechanisms at different developmental stages. During early genome-wide reprogramming, both maternal and zygotic TRIM28 are required to maintain methylation at germline imprints. However, in conditional Trim28 mutants, Gtl2 imprinted gene expression was lost despite normal methylation levels at the germline IG-DMR. These results provide evidence that TRIM28 controls imprinting after early embryonic reprogramming through a mechanism other than the maintenance of germline imprints. Additionally, our finding that secondary imprints were hypomethylated in TRIM28 mutants uncovers a requirement of TRIM28 after genome-wide reprogramming for interpreting germline imprints and regulating DNA methylation at imprinted gene promoters.
INTRODUCTION
Genomic imprinting is a process that regulates the allele-specific expression of certain genes depending on their maternal or paternal inheritance. To date, about a hundred genes have been described to be imprinted and defects in their expression have been associated with cancer and congenital disorders in humans (Lee and Bartolomei, 2013). Imprinted genes generally reside in gene clusters where their expression is controlled by regulatory sequences that are differentially methylated between the maternally and paternally-inherited chromosomes. Two types of these differentially methylated regions (DMRs), germline and secondary DMRs, have been identified in mammals (reviewed in Ferguson-Smith, 2011). DNA methylation at germline DMRs is established during gametogenesis and is later maintained in the zygote after fertilization (Ferguson-Smith, 2011). In contrast, allele-specific methylation at secondary DMRs is established only after fertilization (Bartolomei et al., 1993; Bhogal et al., 2004; Brandeis et al., 1993; Ferguson-Smith et al., 1993; Szabo and Mann, 1995; Tremblay et al., 1997; 1995) and is dependent on imprinting marks at germline DMRs (Lin et al., 2003; Lopes et al., 2003; Srivastava et al., 2003; 2000).
Once established, methylation at germline and secondary DMRs is maintained in somatic cells throughout embryonic development (reviewed in Messerschmidt et al., 2014). First, germline DMRs resist the genome-wide DNA demethylation event that reprograms the oocyte- and sperm-derived genomes shortly after fertilization (Kafri et al., 1993). During this process, germline DMRs are protected from enzymatic removal of DNA methylation that characterizes the early stages of genome-wide reprogramming. Additionally, both during and after genome-wide reprogramming, DNA methylation at DMRs is maintained to prevent the replication-dependent dilution of methyl marks as the zygote starts to divide (Shen et al., 2014). Studies in mice have identified a few molecular mechanisms that prevent loss of DNA methylation at imprinted DMRs. The developmental pluripotency-associated 3 gene (Dppa3, also known as stella or PGC7) encodes a DNA binding protein that is maternally required to protect imprinted loci from enzymatic demethylation during genome-wide reprogramming (Bian and Yu, 2014; Gu et al., 2011; Nakamura et al., 2007; 2012). Additionally, DNA methyltransferase 1 (DNMT1) is required both maternally and zygotically to prevent replication-dependent loss of methylation at DMRs (Hirasawa et al., 2008).
While many studies support a central role for DNA methylation in imprinting control, the mechanisms by which these heritable chromatin marks are interpreted to regulate allele-specific expression are still not entirely understood. The prevailing view is that differential methylation between the maternal and paternal germline DMRs influences the allele-specific recruitment of factors that function in cis to influence transcription. However, while a few proteins are known to bind specifically to either methylated DNA or unmethylated DMRs, some of these proteins are not required for imprinting (Filion et al., 2006; Hendrich et al., 2001; Monnier et al., 2013), and others only control imprinting at specific imprinted clusters (Balmer et al., 2002; Bell and Felsenfeld, 2000; Carr et al., 2007; Hark et al., 2000; Holmgren et al., 2001; Horike et al., 2005; Kanduri et al., 2000; Samaco et al., 2005; Szabo et al., 2004; Szabó et al., 2000). Therefore, the current data argues that rather than a universal mechanism to recognize methylated DNA, these epigenetic marks are interpreted in a locus-specific fashion to control transcription of nearby genes.
The transcriptional repressor TRIM28, also known as KAP1 and TIF1ß, is required for genomic imprinting (Lorthongpanich et al., 2013; Messerschmidt et al., 2012; Quenneville et al., 2011). TRIM28 has been described to bind to the methylated allele of all known imprinting control regions, a recruitment that is dependent on the KRüppel Associated Box (KRAB) domain zinc finger protein ZFP57 (Li et al., 2008; Quenneville et al., 2011). However, the molecular mechanisms by which TRIM28 controls imprinting are still unclear. TRIM28 interacts with a variety of effector proteins, including Heterochromatin protein 1 (Lechner et al., 2000; HP1; Nielsen et al., 1999; Ryan et al., 1999; Sripathy et al., 2006), histone deacetylases (Schultz et al., 2001), histone methyltransferases (Frietze et al., 2010; Ivanov et al., 2007; Schultz et al., 2002) and the DNA methyltransferases, DNMT1, DNMT3a and DNMT3b (Quenneville et al., 2011; Zuo et al., 2012). Whether one or more of these effectors is recruited by TRIM28 to control imprinting is not known. However, loss of function conditions for Zfp57 or Trim28 cause loss of DNA methylation and altered histone modifications at germline DMRs (Li et al., 2008; Lorthongpanich et al., 2013; Messerschmidt et al., 2012; Quenneville et al., 2011), indicating that these factors can, directly or indirectly, maintain epigenetic marks that preserve the imprinted status. Based on the facts that maternal depletion of TRIM28 causes loss of germline DNA methylation (Lorthongpanich et al., 2013; Messerschmidt et al., 2012) and that the zygote relies on maternally-deposited proteins during the early stages of genome-wide reprogramming, it has been proposed that TRIM28 functions by protecting imprinted loci from DNA demethylation during this early reprogramming event (Messerschmidt et al., 2012). However, despite the fact that TRIM28 binds to all known germline imprints, depletion of maternal TRIM28 is only known to disrupt imprinting with variable penetrance at some imprinted clusters (Lorthongpanich et al., 2013; Messerschmidt et al., 2012). Variable effects on imprinting were also observed in Zfp57 mutants, but the simultaneous loss of maternal and zygotic Zfp57 caused more drastic effects than either mutant condition alone (Li et al., 2008), suggesting that effective maintenance of germline imprints requires both maternal and zygotic ZFP57.
To address the requirements of maternal and zygotic TRIM28 for genomic imprinting at different embryonic stages, we analyzed imprinted gene expression and DMR methylation in maternal, zygotic, maternal-zygotic and conditional Trim28 mutants. Results from these studies showed that zygotic Trim28 is required to control imprinting at many imprinted loci, including imprinted clusters that were not previously identified in embryos depleted of maternal Trim28. Consistent with previous studies, our results support a role for maternal and zygotic TRIM28 in the maintenance of DNA methylation at germline DMRs during early embryonic reprogramming. Surprisingly, we also found that loss of TRIM28 at later embryonic stages disrupted allele-specific gene expression without affecting germline DMR methylation, providing evidence that TRIM28 controls imprinting through a molecular mechanism that is distinct from its role to preserve germline imprints during genome-wide reprogramming. Our analysis of conditional Trim28 mutants also revealed hypomethylation at the H19 and Gtl2 promoters. Together, our results provide insight into the in vivo requirements of TRIM28 and the mechanisms that govern allele-specific expression of imprinted genes at different stages of embryonic development.
RESULTS
Zygotic TRIM28 is required for proper allelic expression of many imprinted genes
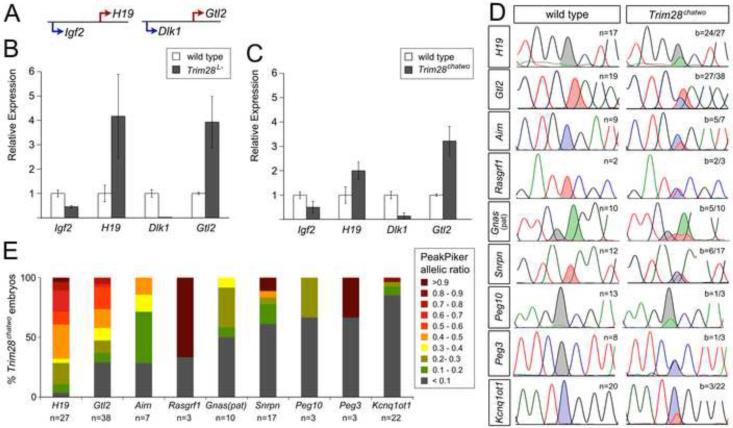
To determine whether zygotic TRIM28 is required for genomic imprinting, we evaluated imprinted gene expression in Trim28 null embryos (Trim28L-; Cammas et al., 2000), and in homozygote mutants for Trim28chatwo, a strong hypomorphic allele that causes developmental arrest at E8.5 and disrupts the protein stability and repressive activity of TRIM28 (Shibata et al., 2011). We first used quantitative RT-PCR (qRT-PCR) to test the levels of expression of imprinted genes in the Igf2-H19 and Dlk1-Gtl2 clusters (Figure 1A). This analysis revealed that the maternally-expressed genes, H19 and Gtl2, were upregulated in both Trim28L- and Trim28chatwo mutants, while the respective paternally-expressed genes from these clusters, Igf2 and Dlk1, were downregulated compared to wild type littermate controls (Figure 1B-C).
Figure 1. Imprinted gene expression in zygotic Trim28 mutants.
(A) Diagram of the Igf2-H19 and Dlk1-Gtl2 clusters, indicating maternally (red) and paternally (blue) expressed genes. (B-C) Expression of imprinted genes in the Igf2-H19 and Dlk1-Gtl2 clusters as determined by qRT-PCR in pools of 3-4 E7.5 Trim28L- (B) and E8.5 Trim28chatwo (C) embryos. Data shown is normalized to ß-actin and relative to wild type controls. Error bars represent the standard deviation from two biological replicates. (D) Selected Sanger sequencing traces of cDNAs for H19, Gtl2, Airn, Rasgrf1, Gnas (paternal isoform), Snrpn, Peg10, Peg3, and Kcnq1ot1 in individual E8.5 wild type and Trim28chatwo embryos containing allele-specific SNPs (shaded peaks). All imprinted genes were analyzed in embryonic tissues, except for Rasgrf1, which is only imprinted in E8.5 extraembryonic tissues (Dockery et al., 2009). b = embryos with biallelic expression over the total number of embryos analyzed. (E) Percent of Trim28chatwo embryos with biallelic expression of imprinted genes as analyzed by Sanger sequencing and quantified using PeakPicker. Wild type embryos showed PeakPicker allelic ratios between 0-0.1. Values higher than 0.1 were considered biallelic. A value of 1 corresponds to equal expression from both alleles. n = total number of embryos analyzed.
To resolve whether abnormal H19 and Gtl2 expression levels in zygotic Trim28 mutants were due to inappropriate biallelic expression, we sequenced cDNAs from embryos that contained single nucleotide polymorphisms (SNPs) distinguishing between the maternal and paternal alleles. While E8.5 wild type embryos expressed imprinted genes monoallelically, Trim28chatwo embryos showed biallelic expression of H19 and Gtl2, as well as Airn, Rasgfr1, Gnas (paternal isoform), Snrpn, Peg10, Peg3 and Kcnq1ot1 (Figure 1D). These results demonstrate that expression of Trim28 from the zygotic genome is required for allele-specific expression at many imprinted loci. Notably, our results show that loss of zygotic TRIM28 disrupts imprinted expression of Gtl2, which was previously only found to be marginally affected by loss of maternal TRIM28 (Messerschmidt et al., 2012). Thus, the analysis of Trim28chatwo mutants shows that TRIM28 has widespread requirements for controlling imprinting.
Loss of imprinting is fully penetrant in maternal-zygotic Trim28 mutants
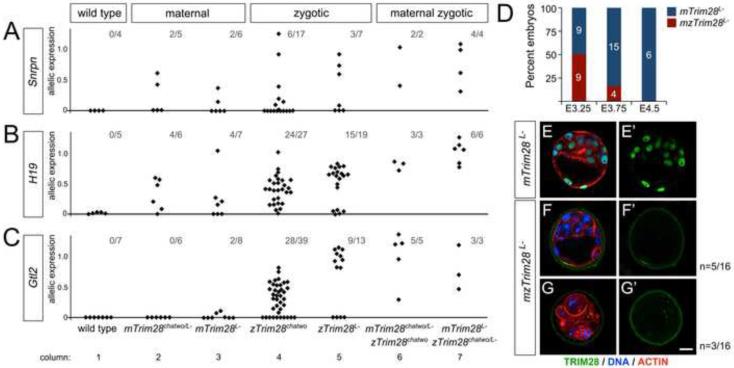
Our analysis of allele-specific imprinted gene expression in single embryos revealed that loss of imprinting was partially penetrant in zygotic Trim28chatwo mutants (Figure 1D-E; Figure 2A-C, column 4). This partial penetrance was not due to the hypomorphic nature of the chatwo mutation, since Trim28L- mutants also showed partially penetrant loss of imprinting (Figure 2A-C, column 5). We hypothesized that maternal TRIM28, which is present during the early development of zygotic Trim28 mutants, may account for the partial penetrance of imprinting defects in Trim28L- and Trim28chatwo embryos. To test this hypothesis, we generated embryos lacking both maternal and zygotic Trim28 (mzTrim28 mutants).
Figure 2. Analysis of maternal, zygotic, and maternal-zygotic Trim28 mutants.
Allelic expression in wild type, maternal (mTrim28chatwo/L- and mTrim28L-), zygotic (zTrim28chatwo and zTrim28L-), and hypomorphic maternal-zygotic (mTrim28L- - zTrim28chatwo/L- and mTrim28chatwo/L- - zTrim28chatwo) Trim28 mutants was analyzed at Snrpn (A), H19 (B), and Gtl2 (C) by Sanger sequencing and quantified with PeakPicker. Each diamond represents a single embryo. The fractional numbers indicate the number of mutants with biallelic expression over the total number of embryos analyzed. All embryos were analyzed at E8.5 except for zTrim28L- and hypomorphic maternal-zygotic mutants, which were analyzed at E7.5. Analysis of wild type samples at E7.5 and E8.5 showed similar results (not shown). (D) Percentage of mTrim28L- (blue) and mzTrim28L- (red) mutants found in dissections at E3.25 (n=18), E3.75 (n=19), and E4.5 (n=6). (E-G) Fluorescence staining of TRIM28 (green), DNA (DAPI, blue), and ACTIN (phalloidin, red) in mTrim28L- (E) and mzTrim28L- (F-G) blastocysts. TRIM28 localization (green channel) is shown separately in E’-G’. Scale bar = 20µm.
Maternal depletion of Trim28 was accomplished by using a conditional allele of Trim28 (Trim28L2; Cammas et al., 2000) in combination with the ZP3-Cre transgene, which expresses Cre-recombinase from the oocyte-specific Zona pellucida 3 promoter (de Vries et al., 2000). Mutants lacking both maternal and zygotic TRIM28 (mzTrim28L-) arrested before implantation at the mid-blastocyst stage (Figure 2D). Some of these mutants formed a blastocele cavity similar to that of wild type blastocysts, but showed morphological abnormalities as compared with littermate controls, including slightly larger cells in the inner cell mass (n=5/16, Figure 2F-F’). Other mzTrim28L- embryos failed to cavitate (n=11/16) and occasionally displayed fragmented nuclei characteristic of cell death (n=3/16, Figure 2G-G’). While the early lethality of these embryos prevented the analysis of imprinted gene expression in mzTrim28L- mutants, we found that embryos completely lacking maternal Trim28 and carrying the hypomorphic Trim28chatwo allele zygotically (mTrim28L- - zTrim28chatwo/L- embryos), or embryos carrying the Trim28chatwo allele maternally and zygotically (mTrim28chatwo/L- - zTrim28chatwo embryos), survived past implantation and had a morphology and developmental arrest similar to that of zygotic Trim28L- mutants in dissections at E7.5. These two allelic combinations, from here onwards referred to as hypomorphic mzTrim28 mutants, were used to analyze the effects of loss of maternal and zygotic Trim28 on imprinted gene expression.
In contrast to the partially penetrant biallelic expression of Snrpn, H19, and Gtl2 observed in maternal or zygotic Trim28 mutants (Figure 2A-C, columns 2-5), we found that simultaneous depletion of maternal and zygotic Trim28 in hypomorphic mzTrim28 embryos disrupted imprinting in all embryos analyzed (Figure 2A-C, columns 6-7; Snrpn n=6; H19 n=9; Gtl2 n=8). One possible explanation for the complete penetrance of imprinting defects in hypomorphic mzTrim28 mutants is that the amount of TRIM28 during the maternal to zygotic transition is critical for maintaining genomic imprinting during genome-wide reprogramming. In this scenario, loss of maternal TRIM28 could be partially compensated by zygotic TRIM28 and vice versa. However, it is also possible that the effects of lack of maternal and zygotic TRIM28 are additive, and represent separate requirements for TRIM28 during early embryonic reprogramming and during later stages of embryogenesis. To resolve the specific roles of TRIM28 at different developmental stages, we analyzed the effects of Trim28 depletion on DMR methylation in zygotic and conditional Trim28 mutants.
DNA methylation at germline DMRs is disrupted in zygotic Trim28 mutants
Because maternal TRIM28 has been previously implicated in the protection of germline DMRs from demethylation during early genome-wide reprogramming (Messerschmidt et al., 2012), we sought to evaluate whether zygotic TRIM28 is also required to maintain DNA methylation at germline DMRs. To this end, we used combined restriction-bisulfite analysis (COBRA), bisulfite sequencing, and quantitative pyrosequencing on Trim28L- and Trim28chatwo embryos.
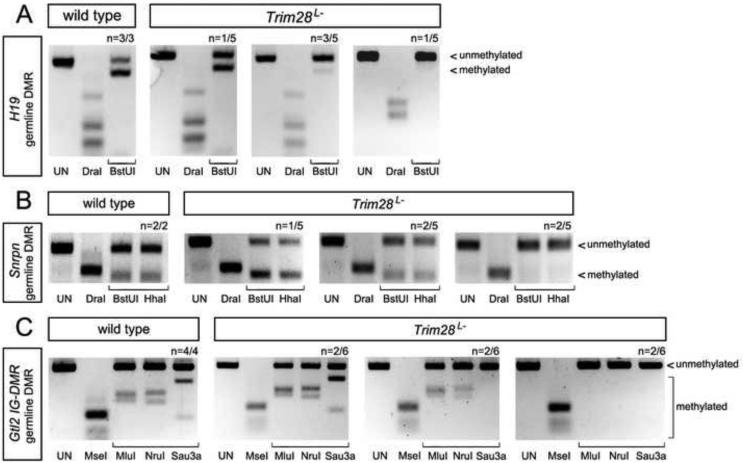
COBRA analysis showed loss of DNA methylation at the H19, Snrpn and Gtl2 germline DMRs in null Trim28L- mutants (Figure 3A-C) suggesting that, similar to maternal TRIM28, zygotic TRIM28 is also required to preserve DNA methylation at germline DMRs. Consistent with the variable penetrance of imprinting defects in Trim28L- mutants (Figure 2A-C), our COBRA experiments showed that hypomethylation at the H19, Snrpn and Gtl2 germline DMRs was also variable, with some Trim28L- embryos showing complete lack of methylation, while others only had partial or no effects (Figure 3A-C). To further analyze the relationship between loss of germline DNA methylation and imprinted gene expression, we used pyrosequencing to quantify the allele expression ratio and germline DNA methylation level in an extensive collection of individual Trim28chatwo mutants.
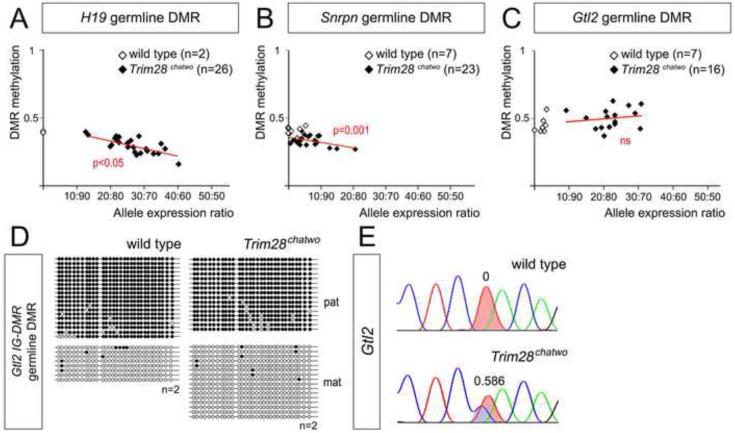
Figure 3. DNA methylation at the H19 and Gtl2 germline DMRs.
DNA methylation at the H19 (A), Snrpn (B) and Gtl2 (C) germline DMRs was detected by combined restriction-bisulfite analysis (COBRA) in single E7.5 wild type and Trim28L- embryos. n = number of embryos showing results similar to the one shown, relative to the total number of embryos analyzed. Restriction with DraI (A-B) and MseI (C) measured the efficiency of bisulfite conversion. All other restriction enzymes (lanes with brackets) only cut if the original sample was methylated. UN-undigested.
Consistent with the hypomorphic nature of the Trim28chatwo allele and with the variability of allele expression ratios in these mutants (Figure 2A-C), we found that Trim28chatwo embryos showed partial loss of methylation at the H19 and Snrpn germline DMRs (Figure 4A-B, Figure S1A-B, Figure S3). The extent of hypomethylation correlated with the ratio of H19 biallelic expression in a collection of twenty-six individual Trim28chatwo embryos (Figure 4A; p<0.05), supporting that abnormal Igf2-H19 imprinting in Trim28chatwo mutants was caused by loss of DNA methylation at the H19 germline DMR. Similarly, Snrpn allelic expression ratios also correlated with loss of methylation at the germline DMR (Figure 4B; p=0.001). In contrast, we found that the levels of Gtl2 germline DMR (IG-DMR) methylation in hypomorphic Trim28chatwo mutants were not significantly different from those of wild type littermates as analyzed with either COBRA (Figure S1C-D) or bisulfite sequencing (Figure 4C-D, Figure S3). Given that methylation of the IG-DMR was disrupted in null Trim28L- mutants (Figure 3C), the lack of effects in Trim28chatwo embryos indicates that this hypomorphic allele provides enough TRIM28 function to allow proper IG-DMR methylation maintenance. This result was nevertheless intriguing since the extent of IG-DMR methylation in Trim28chatwo embryos did not correlate with the ratio of biallelic Gtl2 expression observed in a collection of sixteen Trim28chatwo mutants (Figure 4C, p=0.56). Therefore, these results provide evidence that, upon loss of TRIM28 function, germline DMR methylation is not sufficient to dictate imprinted expression of Gtl2. Consequently, our analysis of hypomorphic Trim28chatwo embryos suggests that TRIM28 regulates imprinted expression at the Dlk1-Gtl2 imprinted cluster through a molecular mechanism that is distinct from its function to preserve DNA methylation at germline DMRs.
Figure 4. DNA methylation at germline DMRs.
DNA methylation at the H19 (A), Snrpn (B) and Gtl2 (C-D) germline DMRs was measured in single E8.5 wild type and Trim28chatwo embryos through pyrosequencing (A-C) and bisulfite sequencing (D). Plots in A-C represent the allelic expression ratio versus DMR methylation as measured by pyrosequencing. Figure S2 illustrates the relationship between allelic expression ratios quantified by PeakPicker and pyrosequencing. Red lines show the linear regression model for Trim28chatwo embryos. P-values (red) indicate the correlation between biallelic expression and DNA methylation. n = toal embryos analyzed. (D) Representative bisulfite sequencing results for wild type and Trim28chatwo embryos, additional results are shown in Figure S3. Filled circles, methylated DNA. Empty circles, unmethylated DNA. Maternal (mat) and paternal (pat) chromosomes were identified by allele-specific SNPs. (E) Sanger sequencing traces of cDNAs for Gtl2 in the embryos analyzed in D. Numbers indicate the PeakPicker allelic expression ratio.
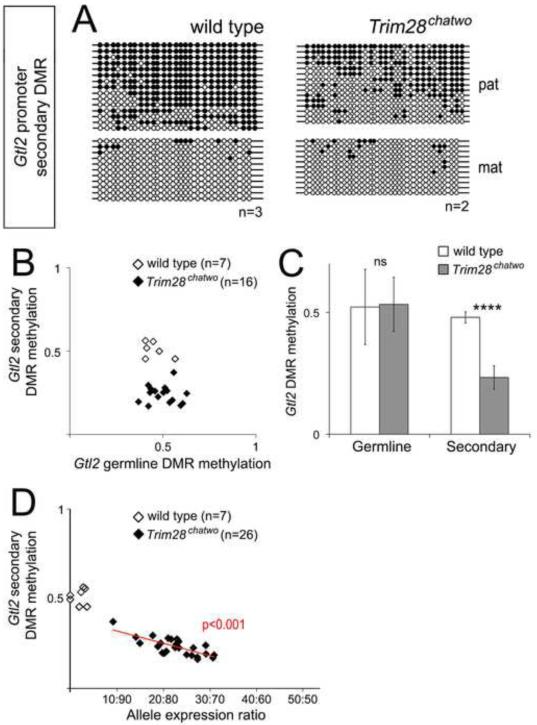
TRIM28 is required for DNA methylation at secondary DMRs
Because allele-specific methylation at secondary DMRs has been proposed to maintain the imprinted status at some imprinted clusters (reviewed in John and Lefebvre, 2011), we hypothesized that biallelic expression of Gtl2 in Trim28chatwo embryos could be due to disrupted methylation of the secondary DMR located at the Gtl2 promoter. To test this hypothesis, we used bisulfite sequencing and pyrosequencing to analyze DNA methylation at the Gtl2 secondary DMR in hypomorphic Trim28chatwo mutants. These experiments revealed that, despite normal levels of DNA methylation at the germline IG-DMR (Figure 4C-D; Figure 5B-C), Trim28chatwo embryos had decreased levels of DNA methylation at the Gtl2 secondary DMR as compared to wild type littermates (Figure 5A-C). While loss of DNA methylation at the Gtl2 secondary DMR was not complete in Trim28chatwo mutants, the extent of hypomethylation was highly correlated with the ratio of biallelic expression of Gtl2 (p<0.001, Figure 5D). Therefore, these results suggest that biallelic expression of Gtl2 in Trim28chatwo mutants could be due to loss of secondary DMR methylation. Additionally, the analysis of Trim28chatwo embryos demonstrates a requirement of TRIM28 for DNA methylation at the Gtl2 secondary DMR.
Figure 5. DNA methylation at the Gtl2 secondary DMR correlates with biallelic expression in Trim28chatwo mutants.
DNA methylation at the Gtl2 secondary DMR was measured in single E8.5 wild type and Trim28chatwo embryos by bisulfite sequencing (A) and pyrosequencing (B-D). (A) Representative bisulfite sequencing results for wild type and Trim28chatwo embryos, additional results are shown in Figure S3. Bisulfite sequencing in A and Figure S3 shows results from Trim28chatwo embryos that biallelically expressed Gtl2. Filled circles, methylated DNA. Empty circles, unmethylated DNA. Maternal (mat) and paternal (pat) chromosomes were identified by allele-specific SNPs. (B) DNA methylation at the germline IG-DMR versus the Gtl2 secondary DMR as measured by pyrosequencing in single wild type and Trim28chatwo embryos. (C) Average DMR methylation for all wild type and Trim28chatwo embryos analyzed. (D) Gtl2 allele expression ratios versus Gtl2 secondary DMR methylation as measured by pyrosequencing in single wild type and Trim28chatwo embryos. The red line in D shows the linear regression model for Trim28chatwo embryos. The P-value (red) indicates the correlation between biallelic expression and DNA methylation. The data represented in B-D includes the same E8.5 wild type and Trim28chatwo embryos as shown in Figure 4C. n = number of embryos analyzed. Error bars represent standard deviation. Statistical significance was measured using a paired student’s t-test, ns-not significant, ****p<0.0001.
TRIM28 has separate roles during and after genome-wide reprogramming
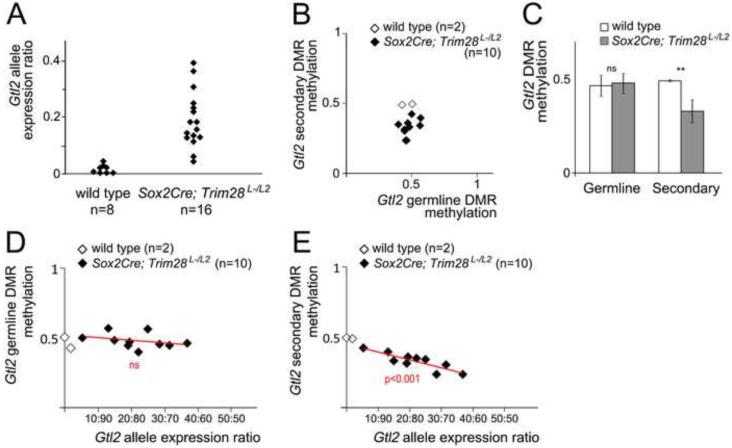
Since methylation at secondary DMRs is established after implantation (Brandeis et al., 1993; Ferguson-Smith et al., 1993; Sato et al., 2011), our finding that zygotic TRIM28 is required for DNA methylation at the Gtl2 secondary DMR supports the conclusion that zygotic TRIM28 controls imprinting beyond the stages of early embryonic reprogramming. To further explore the roles of TRIM28 after this genome-wide reprogramming event, we tested the effects of Trim28 loss of function using a conditional allele of Trim28 (Trim28L2) and the Sox2Cre transgenic line, which expresses Cre recombinase in all embryonic cells starting at implantation (Hayashi et al., 2002).
Analysis of imprinted gene expression using allele-specific SNPs revealed that Gtl2 was biallelically expressed in Sox2Cre;Trim28L-/L2 embryos (Figure 6A). Similar to our previous observations in Trim28chatwo mutants, we found that Sox2Cre;Trim28L-/L2 embryos had no significant differences in germline IG-DMR methylation (Figure 6B-C, p=0.75), but showed a consistent decrease in DNA methylation levels at the Gtl2 secondary DMR (Figure 6B-C, p<0.01). Most significantly, biallelic Glt2 expression in Sox2Cre;Trim28L-/L2 embryos did not correlate with the small variations in DNA methylation at the Gtl2 germline DMR (Figure 6D; p=0.32; n=10), but was highly correlated with loss of DNA methylation at the Gtl2 secondary DMR (Figure 6E; p<0.001; n=10). Therefore, together with our previous analysis of Trim28chatwo embryos, these results provide further support that the methylation at the Gtl2 promoter is linked to the allele-specific silencing of Gtl2. Furthermore, since TRIM28 is only effectively depleted in Sox2Cre;Trim28L-/L2 embryos after early embryonic genome-wide reprogramming (Shibata et al., 2011), our analysis of conditional Trim28 mutants provides conclusive evidence that zygotic TRIM28 has separate roles to control imprinting during and after early embryonic reprogramming.
Figure 6. Gtl2 DMR methylation and imprinted gene expression in Sox2Cre;Trim28L-/L2 embryos.
(A) PeakPicker allelic expression ratios for Gtl2 in single E8.5 wild type and Sox2Cre;Trim28L-/L2 embryonic tissues. (B) Germline IG-DMR methylation versus Gtl2 secondary DMR methylation in E8.5 wild type and Sox2Cre;Trim28L-/L2 embryonic tissues. (C) Average DMR methylation in wild type and Sox2Cre;Trim28L-/L2 embryos. Error bars represent standard deviation. Statistical significance was measured using a paired student’s t-test: ns-not significant, **p<0.01. (D-E) Gtl2 allele expression ratio versus Gtl2 germline DMR methylation (D) and Gtl2 secondary DMR methylation (E) in wild type and Sox2Cre;Trim28L-/L2 embryos. Red lines show the linear regression model for Sox2Cre;Trim28L-/L2 embryos. P-value indicates the correlation between biallelic expression and DNA methylation, ns-not significant. n = number of embryos analyzed.
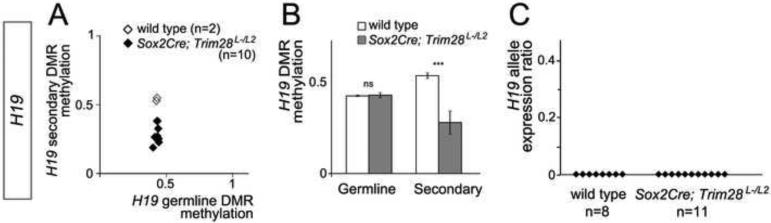
To determine whether TRIM28 also has separate roles during and after early embryonic reprogramming at other imprinted clusters, we analyzed DMR methylation and allele-specific expression of H19 and Snrpn in Sox2Cre;Trim28L-/L2 embryos. Similar to our findings at Gtl2, we found that there were no significant changes in the level of DNA methylation at the H19 and Snrpn germline DMRs in Sox2Cre;Trim28L-/L2 embryos (Figure 7A-B; Figure S4A). Consequently, our data argues against a general role of TRIM28 to prevent DMR methylation during the replication-dependent dilution of DNA methyl marks that takes place as cells undergo mitosis. Also similar to Gtl2, we found that Sox2Cre;Trim28L-/L2 embryos showed significant hypomethylation at the H19 secondary DMR (Figure 7A-B), demonstrating that the roles of TRIM28 to regulate the acquisition or maintenance of secondary DMR methylation are not exclusive to the Gtl2 cluster. Interestingly, neither H19 or Snrpn were biallelically expressed in Sox2Cre;Trim28L-/L2 mutants (Figure 7C and Figure S4B). Therefore, these results suggest that either the roles of TRIM28 after genome-wide reprogramming differ amongst imprinted clusters or that secondary DMR methylation has distinct roles for imprinting control at different imprinted loci. While more experiments will be required to uncover the mechanisms by which TRIM28 and secondary DMRs control imprinting after genome-wide reprogramming, our experiments with Trim28chatwo and conditional Trim28 mutants provide conclusive evidence that zygotic TRIM28 is not required to maintain germline DMR methylation beyond the stages of early genome-wide reprogramming at both maternally and paternally imprinted loci.
Figure 7. H19 DMR methylation and imprinted gene expression in Sox2Cre;Trim28L-/L2 embryos.
DNA methylation and allelic expression was measured at the H19 imprinted clusters by pyrosequencing (A-B) and Sanger sequencing (C). (A) Germline DMR methylation versus secondary DMR methylation in E8.5 wild type and Sox2Cre;Trim28L-/L2 embryonic tissues. The same data is shown in (B) as the average DNA methylation levels at germline and secondary DMRs. Error bars represent standard deviation. Statistical significance was measured using a paired student’s t-test: ns-not significant, ***p<0.001. (C) Allele expression ratios as quantified using PeakPicker.
DISCUSSION
Our study provides insights into how genomic imprinting is regulated during mammalian embryogenesis by uncovering distinct requirements for TRIM28 at different embryonic stages. First, we found that both maternal and zygotic TRIM28 are required to maintain DNA methylation at germline DMRs and that this function is exclusive to the first stages of embryonic development, when genome-wide reprogramming takes place. Furthermore, our experiments revealed that TRIM28 controls genomic imprinting at later stages of embryogenesis through a different mechanism that is independent of its role in maintaining DNA methylation at germline DMRs. The implications of these findings and the molecular mechanisms by which TRIM28 might regulate imprinting at these different stages of embryonic development are discussed below.
TRIM28 has widespread requirements for imprinting control
Based on the finding that TRIM28 binds all known imprinting control regions (Quenneville et al., 2011), TRIM28 has been lauded as a master regulator of genomic imprinting. However, loss of function studies in embryos lacking maternal TRIM28 showed abnormal imprinted gene expression only in a subset of imprinted clusters (Messerschmidt et al., 2012). We found that imprinted gene expression in zygotic Trim28 mutants was disrupted in all the maternally and paternally imprinted clusters we tested, including some loci that were not previously described to be disrupted by maternal Trim28 depletion, such as Airn, Rasgrf1, Gnas, Peg10, Peg3 and Kcnq1ot1. Therefore, the results described in this study provide genetic evidence that Trim28 has widespread requirements for genomic imprinting.
The amount of TRIM28 is critical for genomic imprinting
The maternal-to-zygotic transition in mouse embryos takes place at the 2-cell stage (reviewed in Li et al., 2013) and zygotic expression of TRIM28 is detectable as early as the 4-cell stage (Messerschmidt et al., 2012), when embryos are still undergoing genome-wide demethylation (Smith et al., 2012). Therefore, it is tempting to speculate that maternal and zygotic TRIM28 function redundantly to protect imprinted loci from demethylation during this early genome-wide reprogramming event. Consistent with this hypothesis, we found that either maternal or zygotic Trim28 mutants showed a partially penetrant loss of imprinting, but that simultaneous removal of both maternal and zygotic TRIM28 resulted in loss of imprinting in all the embryos analyzed.
The sensitivity of genomic imprinting to the amount of TRIM28 was also remarkable in zygotic Trim28 mutants. Specifically, we show that complete removal of zygotic TRIM28 in null Trim28L- mutants caused loss of germline DMR methylation at H19, Snrpn and Gtl2, but TRIM28 function in hypomorphic Trim28chatwo mutants was sufficient to maintain normal levels of DNA methylation at the Gtl2 germline IG-DMR. Because our previous observations support that the hypomorphic nature of the chatwo allele is largely due to a drastic decrease in TRIM28 protein levels (Shibata et al., 2011), we suspect that the different effects of the Trim28chatwo allele on imprinting at the H19, Snrpn and Gtl2 might be a reflection of the specific dose-sensitive requirements for TRIM28 at these imprinted clusters. Together, these results indicate that genomic imprinting is particularly sensitive to the amount of TRIM28.
TRIM28 maintains germline DMR methylation exclusively during genome-wide reprogramming
The ability of TRIM28 to interact with the maintenance DNA methyltransferase DNMT1 in embryonic stem cells, and the fact that loss of Zfp57 in ESCs leads to loss of germline DMR methylation (Quenneville et al., 2011; Zuo et al., 2012) have led to propose that TRIM28 maintains germline imprints throughout embryonic development (Messerschmidt, 2014). However, our finding that germline DNA methylation was not disrupted in conditional Sox2Cre;Trim28L-/L2 mutants at either the H19, Snrpn or Gtl2 germline DMRs provides genetic evidence that TRIM28 is not required for replication-dependent maintenance of germline imprints after genome-wide reprogramming in vivo. Therefore, our results support the conclusion that TRIM28 maintains DNA methylation at germline imprints exclusively during the early stages of embryonic development.
DNA demethylation during genome-wide reprogramming is accomplished through both active and passive mechanisms (Shen et al., 2014). Active demethylation takes place through enzymatic oxidation of methylated cytosine residues by the Ten-eleven translocation-3 (TET3) methylcytosine dioxigenase (Gu et al., 2011; Iqbal et al., 2011). Additionally, DNA methylation is passively lost through replication-dependent dilution of methylated cytosines, which is facilitated by the exclusion of the maintenance DNA methyltransferase DNMT1 from the nucleus during pre-implantation stages (Cardoso and Leonhardt, 1999; Doherty et al., 2002; Howell et al., 2001; Mertineit et al., 1998; Ratnam et al., 2002). Because TRIM28 has been shown to bind to the methylated allele of imprinting control regions (Quenneville et al., 2011), TRIM28 may interfere with active DNA demethylation by blocking the accessibility of TET3 to germline DMRs. However, it is also possible that TRIM28 interferes with passive mechanisms of DNA demethylation. In this respect, two studies have detected small amounts of DNMT1 in the nuclei of pre-implantation mouse embryos (Cirio et al., 2008; Kurihara et al., 2008). Given the ability of TRIM28 to interact with DNMT1 (Quenneville et al., 2011; Zuo et al., 2012), it is possible that TRIM28 might function by recruiting DNMT1 to germline DMRs during genome-wide reprogramming, ensuring that DNA methylation marks at these loci are perpetuated as the DNA replicates.
Separate roles for TRIM28 during and after early embryonic reprogramming
Perhaps the most striking result from our analysis of zygotic Trim28 mutants was the finding that imprinting at the Gtl2 cluster was disrupted in Trim28chatwo embryos despite normal levels of methylation at the IG-DMR. This result provides evidence for a role of TRIM28 in the regulation of genomic imprinting that is independent of DNA methylation maintenance at germline DMRs. Because this role of TRIM28 is also supported by our analysis of conditional Trim28 mutants, our data argues that the imprinting defects in Trim28chatwo embryos are not due to a neomorphic effect of the chatwo allele, but are rather caused by TRIM28 loss of function.
A role for TRIM28 interpreting germline imprints
While multiple studies support an essential role for germline DMRs in imprinting control (Lin et al., 2003; Thorvaldsen et al., 1998), there are still large gaps in our understanding of how differential methylation at these regulatory regions controls allele-specific expression of imprinted genes. One of the best characterized imprinting control regions is the H19 intergenic germline DMR (Ferguson-Smith, 2011). This imprinting control region is recognized in a methylation-specific manner by the chromatin insulator CCCTC-binding factor (CTCF), which is known to influence chromatin topology and favor the interaction of the H19 promoter with downstream enhancers (Bell and Felsenfeld, 2000; Engel et al., 2004; Hark et al., 2000; Murrell et al., 2004; Szabó et al., 2000). The finding that CTCF also binds to other imprinted clusters (Fitzpatrick et al., 2007; Yoon et al., 2005) has brought some support towards an “insulator model” of imprinting regulation (Wan and Bartolomei, 2008). However, CTCF does not bind to all imprinting control regions (Carr et al., 2007). Consequently, the mechanisms by which germline DMRs function in cis to control allele-specific expression are likely specific of each imprinted locus. In this respect, the mechanisms that operate at the Dlk1-Gtl2 germline IG-DMR to control imprinted gene expression are currently unknown (da Rocha et al., 2008). Chromatin immunoprecipitation experiments have failed to detect binding of CTCF, or methyl binding proteins MBD2 and MecP2, to the Gtl2 germline IG-DMR (Carr et al., 2007). However, TRIM28 can bind the Gtl2 germline IG-DMR in a methylation specific fashion (Quenneville et al., 2011). By identifying TRIM28 as a factor that controls allele-specific Gtl2 expression without disrupting germline DMR methylation, our studies provide genetic evidence supporting a role for TRIM28 in interpreting the epigenetic information inherited through the Gtl2 germline IG-DMR to ultimately influence imprinted gene expression.
TRIM28 can recruit several histone-modifying enzymes (Schultz et al., 2001; 2002) and alterations in histone modifications are known to disrupt imprinted gene expression (Carr et al., 2007; Mager et al., 2003). TRIM28 is also known to mediate long-range transcriptional repression through heterochromatin spreading (Groner et al., 2010; Quenneville et al., 2012) Therefore, it is tempting to speculate that TRIM28 might regulate imprinting after early embryonic reprogramming by binding to the methylated Gtl2 germline IG-DMR and spreading a repressive state through heterochromatin formation.
TRIM28 is required for secondary DMR methylation
DNA methylation at secondary DMRs has been proposed to control imprinting, but its role in regulating allele-specific expression is still controversial (reviewed in John and Lefebvre, 2011). Several studies support an instructive role of certain secondary DMRs for allele-specific expression. Secondary DMR methylation at the H19 and Gtl2 promoters has been shown to correlate with allele-specific silencing (Lin et al., 2003; Srivastava et al., 2000; Steshina et al., 2006; Thorvaldsen et al., 1998). Additionally, a study that conditionally deleted the paternal Igf2-H19 germline DMR late in embryogenesis suggested that, once established, secondary imprints can maintain the imprinted status in the absence of the germline DMR (Srivastava et al., 2000). Since methylation at the H19 and Gtl2 promoters has been found to depend on the allele-specific methylation at germline DMRs (Lin et al., 2003; Srivastava et al., 2000; Thorvaldsen et al., 1998), a model has been put forward that secondary DMRs perpetuate the imprinted status inherited from germline DMRs. However, monoallelic expression of H19 and Gtl2 is established before DNA methylation is acquired at their secondary DMR promoters (Sasaki et al., 1995; Sato et al., 2011), arguing that secondary DMR methylation is a consequence of the imprinted status, rather than an instructive mechanism for allele-specific expression.
The fact that decreased levels of secondary DMR methylation in Sox2Cre;Trim28L-/L2 embryos was highly correlated with biallelic expression of Gtl2 provides additional data supporting the relationship between secondary DMR methylation and allele-specific expression. However, in Sox2Cre;Trim28L-/L2 mutants, H19 was not biallelically expressed despite significant loss of methylation at the H19 promoter. These variable effects on different imprinted loci might be a reflection of the different mechanisms by which imprinting is regulated at specific clusters. For instance, it is possible that DNA methylation at the H19 secondary DMR is dispensable for H19 repression. Alternatively, it is possible that the roles of TRIM28 after genome-wide reprogramming differ at amongst imprinted loci. Although our experiments can not resolve whether methylation at secondary DMRs has an instructive role on imprinted gene expression or if is a secondary consequence of a previously established imprinted status, our results provide insight into the relationship between secondary DMR methylation and allele-specific expression of Gtl2 and H19. The lymphoid specific helicase LSH/HELLS is required for methylation of somatic imprints (Fan, 2005). However, LSH/HELLS seems to only be required at the Cdkn1c imprinted locus (Fan, 2005). Therefore, by identifying a requirement for TRIM28 in the regulation of DNA methylation at Gtl2 and H19 somatic imprints, our results provide an important contribution towards understanding the factors that control DNA methylation at secondary DMRs.
In conclusion, our analysis of maternal, zygotic and conditional Trim28 mutants not only provides additional insight about how TRIM28 maintains methylation at germline DMRs, but also uncovers a requirement of TRIM28 after genome-wide reprogramming for deciphering germline imprints and influencing secondary DMR methylation.
EXPERIMENTAL PROCEDURES
Mice
Trim28chatwo, Trim28L-, maternal Trim28 deletion (ZP3-Cre; Trim28L-/L2), and Sox2Cre; Trim28L-/L2 mutants (Cammas et al., 2000; Messerschmidt et al., 2012; Shibata et al., 2011). Trim28chatwo mutants were analyzed in a mixed CAST/Ei background, where mutants show developmental arrest at E8.5 (Shibata et al., 2011). To generate maternal Trim28chatwo/L- embryos, Zp3-Cre; Trim28chatwo/L2 females were mated to wild type males. For maternal-zygotic Trim28 mutants, Zp3-Cre;Trim28L-/L2 or Zp3-Cre;Trim28chatwo/L2 females were mated to Trim28L-/+ or Trim28chatwo/+ males. For genetic backgrounds, SNPs and primers used see Supplemental Table 1. Experiments involving mice were done according to standard operating procedures approved by Cornell’s Institutional Animal Care and Use Committee.
Embryo Collection
Embryos were dissected in phosphate buffered saline containing 4% bovine serum albumin. For post-implantation developmental stages (E6.5 and later), embryos were split into embryonic and extra-embryonic tissues that were processed separately for genotyping and imprinting analysis. In cases where allelic expression and DNA methylation were analyzed within the same embryo, the embryonic tissues were split and processed separately. At Rasgrf1, the embryonic tissues were used for genotyping and extra-embryonic tissues were used for analysis of imprinted gene expression. For pre-implantation developmental stages, embryos were flushed from the uterus and used directly for immunofluorescence.
Gene expression
Quantification of imprinted gene expression was tested by qRT-PCR on RNA samples extracted from independent pools of 3-4 E7.5 Trim28L- or E8.5 Trim28chatwo embryos and wild type littermates as previously described (Shibata et al., 2011). Allelic expression was detected by quantitative pyrosequencing and Sanger sequencing of RT-PCR products that amplified the SNP-containing region of the imprinted gene. Primer sequences and SNP positions are in Supplemental Table 1. Allele expression ratios were quantified with PeakPicker (Ge et al., 2005).
Immunofluorescence
Preimplantation embryos were fixed in 4% paraformaldehyde and used for staining with TRIM28 antibody (Santa Cruz, sc-33186), phalloidin, and 4’,6-diamidino-2-phenylindole (DAPI).
DNA methylation
DNA was bisulfite converted using the EZ DNA methylation-direct kit (Zymo, D5020). For bisulfite sequencing analysis, PCR products of bisulfite-converted DNA were cloned using the TOPO TA cloning kit (Invitrogen, K450001) and individual clones were analyzed by Sanger sequencing. The efficiency of bisulfite conversion was >99%. Pyrosequencing was done as previously described (Wang et al., 2014). For primer sequences see Supplemental Table 1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Timothy Bestor, Mathieu Boulard, Paul Soloway, Patrick Murphy and members of the laboratory for helpful discussions and comments on the manuscript; Joel Brown for help with graphical art; Florence Cammas, Shao-Cong Sun and Jim Hagarman for mice and reagents; and Cornell’s CARE staff for mice husbandry and care. This work was supported by NSF IOS-1020878 and NIH R01HD060581 to MJGG and NIH S10RR025502 to the Cornell Institute of Biotechnology. KAA was supported by NIH 5T32HD057854-04 training grant to Cornell University, Research and Career Training in Vertebrate Developmental Genomics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Balmer D, Arredondo J, Samaco RC, LaSalle JM. MECP2 mutations in Rett syndrome adversely affect lymphocyte growth, but do not affect imprinted gene expression in blood or brain. Hum. Genet. 2002;110:545–552. doi: 10.1007/s00439-002-0724-4. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bhogal B, Arnaudo A, Dymkowski A, Best A, Davis TL. Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics. 2004;84:961–970. doi: 10.1016/j.ygeno.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Bian C, Yu X. PGC7 suppresses TET3 for protecting DNA methylation. Nucleic Acids Research. 2014;42:2893–2905. doi: 10.1093/nar/gkt1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Kafri T, Ariel M, Chaillet JR, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. Embo J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dollé P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–2963. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J. Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr MS, Yevtodiyenko A, Schmidt CL, Schmidt JV. Allele-specific histone modifications regulate expression of the Dlk1–Gtl2 imprinted domain. Genomics. 2007;89:280–290. doi: 10.1016/j.ygeno.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev. Biol. 2008;8:9. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends in Genetics. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Dockery L, Gerfen J, Harview C, Rahn-Lee C, Horton R, Park Y, Davis TL. Differential methylation persists at the mouse Rasgrf1 DMR in tissues displaying monoallelic and biallelic expression. Epigenetics. 2009;4:241–247. doi: 10.4161/epi.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AS, Bartolomei MS, Schultz RM. Regulation of stage-specific nuclear translocation of Dnmt1o during preimplantation mouse development. Dev. Biol. 2002;242:255–266. doi: 10.1006/dbio.2001.0534. [DOI] [PubMed] [Google Scholar]
- Engel N, West AG, Felsenfeld G, Bartolomei MS. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat Genet. 2004;36:883–888. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- Fan T. Lsh controls silencing of the imprinted Cdkn1c gene. Development. 2005;132:635–644. doi: 10.1242/dev.01612. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC. Timeline: Genomic imprinting: the emergence of an epigenetic paradigm. Nature Publishing Group. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- Filion GJP, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez P-A. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Molecular and Cellular Biology. 2006;26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick GV, Pugacheva EM, Shin J-Y, Abdullaev Z, Yang Y, Khatod K, Lobanenkov VV, Higgins MJ. Allele-specific binding of CTCF to the multipartite imprinting control region KvDMR1. Molecular and Cellular Biology. 2007;27:2636–2647. doi: 10.1128/MCB.02036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frietze S, O'Geen H, Blahnik KR, Jin VX, Farnham PJ. ZNF274 Recruits the Histone Methyltransferase SETDB1 to the 3′ Ends of ZNF Genes. PLoS ONE. 2010;5:e15082. doi: 10.1371/journal.pone.0015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B, Gurd S, Gaudin T, Dore C, Lepage P, Harmsen E, Hudson TJ, Pastinen T. Survey of allelic expression using EST mining. Genome Research. 2005;15:1584–1591. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Dénervaud N, Bucher P, Trono D. KRAB–Zinc Finger Proteins and KAP1 Can Mediate Long-Range Transcriptional Repression through Heterochromatin Spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, Xie Z-G, Shi L, He X, Jin S-G, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 2002;119(Suppl 1):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren C, Kanduri C, Dell G, Ward A, Mukhopadhya R, Kanduri M, Lobanenkov V, Ohlsson R. CpG methylation regulates the Igf2/H19 insulator. Curr. Biol. 2001;11:1128–1130. doi: 10.1016/s0960-9822(01)00314-1. [DOI] [PubMed] [Google Scholar]
- Horike S-I, Cai S, Miyano M, Cheng J-F, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Iqbal K, Jin S-G, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.a. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain Required for Gene Silencing. Mol. Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John RM, Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81:270–280. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Kafri T, Gao X, Razin A. Mechanistic aspects of genome-wide demethylation in the preimplantation mouse embryo. Proc. Natl. Acad. Sci. U.S.a. 1993;90:10558–10562. doi: 10.1073/pnas.90.22.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kawamura Y, Uchijima Y, Amamo T, Kobayashi H, Asano T, Kurihara H. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev. Biol. 2008;313:335–346. doi: 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Molecular and Cellular Biology. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Li L, Lu X, Dean J. Molecular Aspects of Medicine. Molecular Aspects of Medicine. 2013;34:919–938. doi: 10.1016/j.mam.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A Maternal-Zygotic Effect Gene, Zfp57, Maintains Both Maternal and Paternal Imprints. Developmental Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-P, Youngson N, Takada S, Seitz H, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forné T, Murrell A, Constância M, Bartolomei M, Walter J, et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Human Molecular Genetics. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, Messerschmidt DM. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science. 2013;341:1110–1112. doi: 10.1126/science.1240617. [DOI] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP-M, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125:889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt D. Should I stay or should I go: Protection and maintenance of DNA methylation at imprinted genes. Epigenetics. 2014;7:969–975. doi: 10.4161/epi.21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proceedings of the National Academy of Sciences. 2013;110:20693–20698. doi: 10.1073/pnas.1310201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu Y-J, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. 2012:1–6. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. Embo J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In Embryonic Stem Cells, ZFP57/KAP1 Recognize a Methylated Hexanucleotide to Affect Chromatin and DNA Methylation of Imprinting Control Regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 System Contributes to the Early Embryonic Establishment of Site-Specific DNA Methylation Patterns Maintained during Development. CellReports. 2012;2:766–773. doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnam S, Mertineit C, Ding F, Howell CY, Clarke HJ, Bestor TH, Chaillet JR, Trasler JM. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev. Biol. 2002;245:304–314. doi: 10.1006/dbio.2002.0628. [DOI] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Molecular and Cellular Biology. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Human Molecular Genetics. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Ferguson-Smith AC, Shum AS, Barton SC, Surani MA. Temporal and spatial regulation of H19 imprinting in normal and uniparental mouse embryos. Development. 1995;121:4195–4202. doi: 10.1242/dev.121.12.4195. [DOI] [PubMed] [Google Scholar]
- Sato S, Yoshida W, Soejima H, Nakabayashi K, Hata K. Methylation dynamics of IG-DMR and Gtl2-DMR during murine embryonic and placental development. Genomics. 2011;98:120–127. doi: 10.1016/j.ygeno.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. Tet3 and DNA Replication Mediate Demethylation of Both the Maternal and Paternal Genomes in Mouse Zygotes. Stem Cell. 2014;15:459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Blauvelt KE, Liem KF, Garcia-Garcia MJ. TRIM28 is required by the mouse KRAB domain protein ZFP568 to control convergent extension and morphogenesis of extra-embryonic tissues. Development. 2011;138:5333–5343. doi: 10.1242/dev.072546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC. The KAP1 Corepressor Functions To Coordinate the Assembly of De Novo HP1-Demarcated Microenvironments of Heterochromatin Required for KRAB Zinc Finger Protein-Mediated Transcriptional Repression. Molecular and Cellular Biology. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Frolova E, Rottinghaus B, Boe SP, Grinberg A, Lee E, Love PE, Pfeifer K. Imprint Control Element-mediated Secondary Methylation Imprints at the Igf2/H19 Locus. Journal of Biological Chemistry. 2003;278:5977–5983. doi: 10.1074/jbc.M208437200. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Hsieh S, Grinberg A, Williams-Simons L, Huang SP, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- Steshina EY, Carr MS, Glick EA, Yevtodiyenko A, Appelbe OK, Schmidt JV. Loss of imprinting at the Dlk1-Gtl2 locus caused by insertional mutagenesis in the Gtl2 5' region. BMC Genet. 2006;7:44. doi: 10.1186/1471-2156-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo PE, Mann JR. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- Szabo PE, Tang SHE, Silva FJ, Tsark WMK, Mann JR. Role of CTCF Binding Sites in the Igf2/H19 Imprinting Control Region. Molecular and Cellular Biology. 2004;24:4791–4800. doi: 10.1128/MCB.24.11.4791-4800.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Duran KL, Bartolomei MS. A 5' 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Molecular and Cellular Biology. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- Wan LB, Bartolomei MS. Advances in Genetics. Elsevier; 2008. Chapter 7 Regulation of Imprinting in Clusters: Noncoding RNAs Versus Insulators; pp. 207–223. [DOI] [PubMed] [Google Scholar]
- Wang X, Douglas KC, VandeBerg JL, Clark AG, Samollow PB. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Research. 2014;24:70–83. doi: 10.1101/gr.161919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon B, Herman H, Hu B, Park YJ, Lindroth A, Bell A, West AG, Chang Y, Stablewski A, Piel JC, et al. Rasgrf1 imprinting is regulated by a CTCF-dependent methylation-sensitive enhancer blocker. Molecular and Cellular Biology. 2005;25:11184–11190. doi: 10.1128/MCB.25.24.11184-11190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. Zinc Finger Protein ZFP57 Requires Its Co-factor to Recruit DNA Methyltransferases and Maintains DNA Methylation Imprint in Embryonic Stem Cells via Its Transcriptional Repression Domain. Journal of Biological Chemistry. 2012;287:2107–2118. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.