Abstract
Until recently, dynein was the least understood of the cytoskeletal motors. However, a wealth of new structural, mechanistic, and cell biological data is shedding light on how this complicated minus-end–directed, microtubule-based motor works. Cytoplasmic dynein-1 performs a wide array of functions in most eukaryotes, both in interphase, in which it transports organelles, proteins, mRNAs, and viruses, and in mitosis and meiosis. Mutations in dynein or its regulators are linked to neurodevelopmental and neurodegenerative diseases. Here, we begin by providing a synthesis of recent data to describe the current model of dynein’s mechanochemical cycle. Next, we discuss regulators of dynein, with particular focus on those that directly interact with the motor to modulate its recruitment to microtubules, initiate cargo transport, or activate minus-end–directed motility.
Keywords: Lis1, Nudel/NudE, dynactin, microtubule, transport, cytoskeleton
INTRODUCTION
The spatial and temporal organization of intracellular components by the microtubule cytoskeleton is required for cell division, development, and neuronal function. Microtubules are dynamic, polar structures, with plus ends usually located near the cell periphery and minus ends typically embedded within internal microtubule organizing centers. Dyneins are molecular motors that move toward the minus ends of microtubules, whereas most kinesin motors move in the opposite direction.
Microtubule-based motors can be grouped based on the role they play during interphase, cell division, or ciliary function. In humans, cytoplasmic dynein-1 (originally described in Paschal et al. 1987, Paschal & Vallee 1987) and >15 kinesins are responsible for the transport of vesicles, organelles, proteins, and mRNAs during interphase (Vale 2003). This function is exploited by many viruses, which hijack these same motors to be transported through the host cell (Greber & Way 2006). In dividing cells, cytoplasmic dynein-1 and the mitotic kinesins organize the spindle and segregate chromosomes. Dyneins found in cilia are used to build the cilia and power their movement. Cytoplasmic dynein-2 and kinesin-2 are used for transport within cilia (intraflagellar transport), and multiple axonemal dyneins (14 in humans) power the movement of cilia (Yagi 2009). In this review, we focus on the structure, mechanism, and regulation of cytoplasmic dynein-1, which we refer to as dynein.
Dynein differs dramatically from kinesins and the actin-based myosins, the other cytoskeletal motors. Although kinesin and myosin are structurally related to G-proteins (Vale & Milligan 2000), the motor-containing subunit, or heavy chain, of dynein is a member of the AAA+ (ATPases Associated with various cellular Activities) superfamily (Neuwald et al. 1999). Dynein is also larger and more complicated in terms of its subunit composition than kinesins and myosins. Cytoplasmic dynein is a 1.4 MDa complex of at least eight polypeptides and requires multiple additional proteins and protein complexes for efficient motility. Furthermore, unlike myosins and kinesins, which are part of large gene families, dynein-1 is the only dynein isoform that functions in the cytoplasm of eukaryotic cells. Cytoplasmic dynein-1 is found in all eukaryotic genomes, with the exception of flowering plants, red algae, and Entamoeba, which lack dynein altogether (Wickstead & Gull 2007).
At the organismal level, dynein is essential in many species; for example, homozygous deletion of the dynein heavy chain causes embryonic lethality in mice (Harada et al. 1998). Single point mutations in dynein or its regulatory machinery result in various neurological diseases in humans (Lipka et al. 2013). Mutations in cytoplasmic dynein itself have been linked to peripheral neuropathy (Charcot-Marie-Tooth disease), spinal muscular atrophy, and malformations of cortical development (Harms et al. 2012, Poirier et al. 2013, Weedon et al. 2011). We will discuss additional disease mutations in dynein’s regulators in the following sections. Defects in microtubule-based axonal transport are also observed in other neurodegenerative diseases, including Alzheimer’s, Parkinson’s, and Huntington’s, suggesting that there may be additional links between dynein and neurological disease yet to be uncovered (Millecamps & Julien 2013).
As the only minus-end–directed, microtubule-based motor that can move over long distances in the cytoplasm, dynein performs diverse cellular functions. For example, dynein transports endosomes, peroxisomes, autophagosomes, lipid droplets and mitochondria, as well as many other membrane-bound compartments (Allan 2011). Dynein also transports mRNAs (Holt & Bullock 2009) and assembles and transports stress granules and processing bodies involved in RNA homeostasis (Loschi et al. 2009). Likewise, protein homeostasis requires dynein to transport protein aggregates (Egan et al. 2015, Johnston et al. 2002). Many viruses, including herpes simplex virus (HSV-1) and human immunodeficiency virus type 1 (HIV-1), recruit dynein to move toward the nucleus (Dodding & Way 2011). Finally, dynein has multiple roles during mitosis, including silencing the spindle assembly checkpoint (Wang et al. 2014).
Given dynein’s vast array of functions, it is not surprising that it is highly regulated. Lis1-Nudel/NudE and dynactin are two conserved protein complexes that regulate dynein-based motility. Adaptor and scaffolding proteins further modulate dynein activity (Fu & Holzbaur 2014a). Even in organisms such as the yeast Saccharomyces cerevisiae, in which dynein has only a single known function (spindle positioning), its activity is under tight control (Moore et al. 2009). Currently, key questions regarding dynein regulation include: How is cargo transport initiated? How is dynein recruited to a specific cargo at the right time and place? Given that dynein may exist in an inhibited state prior to cargo transport (Markus et al. 2009, McKenney et al. 2014, Schlager et al. 2014, Torisawa et al. 2014), how is dynein activated for motility? Are there multiple mechanisms of activation? Is dynein regulated to have different motile properties depending on the type of cargo it moves?
In this review, we first describe the subunit composition and structure of dynein, which will guide our discussion of dynein regulation. High-resolution structures of dynein have recently been obtained and highlight the structural complexity of this molecular motor (Bhabha et al. 2014; Kon et al. 2012; Schmidt et al. 2012, 2014). These structures, along with biochemical data, allow the first nearly complete view of dynein’s mechanochemical cycle—we present a synthesis here. Next, we discuss dynein regulation, with particular focus on Lis1-Nudel/NudE and dynactin, which directly interact with dynein to regulate its recruitment to microtubules, initiate cargo transport, or activate minus-end–directed motility.
CYTOPLASMIC DYNEIN
Structure of Cytoplasmic Dynein
Cytoplasmic dynein is a ~1.4 MDa complex composed of multiple polypeptide chains that are responsible for binding cargo and converting the energy from ATP hydrolysis into movement. The heavy chain is the largest polypeptide (~500 kDa) and comprises two functional moieties, the tail and motor (Figure 1a). The tail is required for dimerization and binding dynein’s other polypeptide chains (dimers of the intermediate chain, light intermediate chain, and up to three light chains), which are implicated in cargo binding (Figure 1a). The dynein intermediate chain (DIC) interacts with the dynein regulators dynactin and Nudel/NudE (McKenney et al. 2011, Nyarko et al. 2012, Schroer 2004, Wang et al. 2013). The motor is the minimal functional sequence required to generate movement along microtubules (Koonce & Samso 1996, Nishiura et al. 2004, Reck-Peterson et al. 2006) and interacts with the dynein regulators Lis1 (Huang et al. 2012, McKenney et al. 2010). Before discussing the details of dynein’s mechanochemical cycle, we describe the key structural and functional features of dynein’s motor.
Figure 1.
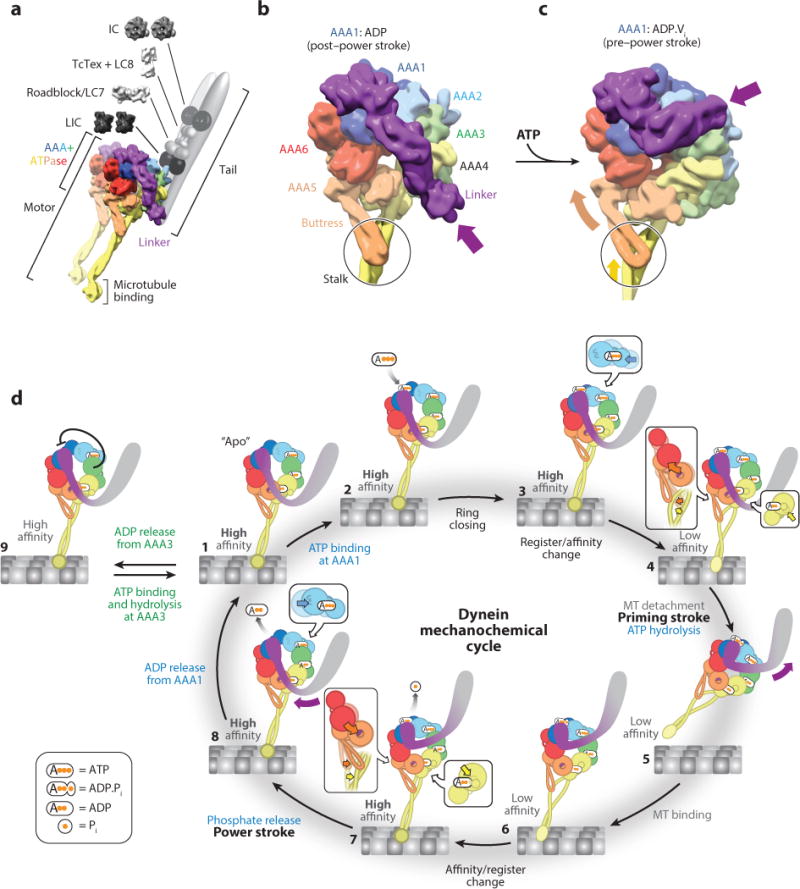
Dynein structure and mechanochemical cycle. (a) Structure of the cytoplasmic dynein complex. The motor domain contains the AAA+ (ATPases Associated with various cellular Activities) domains, the microtubule (MT)-binding domain (MTBD), and the linker. The N-terminal tail domain binds the intermediate (IC), light intermediate (LIC), and light chains (LC). We built the structure of the motor domain by combining entries 3VKG and 4RH7 from the Protein Data Bank (PDB) and displaying the model as a density at a resolution of 10 Å. (b) Structure of the dynein motor domain in the post–power stroke state, in which AAA1 is bound to ADP (PDB 3VKG). Dynein is colored according to the AAA+ domains. In this state, the linker adopts a straight position across the AAA ring; the purple arrow points to the N terminus of the linker, which is connected to the tail domain. The black circle highlights an interaction between the buttress (part of AAA5) and the coiled coil of the stalk (part of AAA4). We rendered the structure as a 10 Å density. (c) Structure of the dynein motor domain in the pre–power stroke state, in which AAA1 is bound to ADP.Vi, a transition state analog that mimics the posthydrolysis ADP.Pi state (PDB 4RH7). The bending of the linker domain, which brings its N terminus toward AAA2 (purple arrow), is accompanied by coordinate changes across the ring. These changes include an upward movement of AAA5 and AAA6 (orange arrow) that shifts the registry of the coiled coil in the stalk (yellow arrow inside the circle), lowering the affinity of the MTBD for the MT. We rendered the structure as a 10 Å density. (d) Overview of dynein’s mechanochemical cycle. A single dynein head is shown for clarity and is colored according to panel b. For each AAA+ module, the large and small domains are shown as large and small spheres, respectively. Steps 3, 4, 7, and 8 have insets highlighting how nucleotide binding affects the packing of the large and small domains together: Nucleotide binding and unbinding results in tight and loose packing, respectively.
The AAA+ ring
Dynein’s motor contains a ring of six concatenated AAA+ domains (termed AAA1–6; Figure 1b). AAA1 is the predominant site of ATP hydrolysis (Gibbons et al. 1987), and an active AAA1 domain is the minimal requirement for motility (Kon et al. 2004). Although AAA2 lacks the amino acids required for ATP hydrolysis, AAA3 and AAA4 both hydrolyze ATP in cytoplasmic dynein. Mutations that block nucleotide binding or hydrolysis in AAA3 impair dynein motility (Cho et al. 2008; DeWitt et al. 2015; Kon et al. 2004, 2005; Silvanovich et al. 2003), whereas the same mutations in AAA4 alter dynein’s motility to a lesser extent (Cho et al. 2008). Several lines of evidence suggest that dynein hydrolyzes only one ATP per step (DeWitt et al. 2015, Mallik et al. 2004, Reck-Peterson et al. 2006, Toba et al. 2006), implying that AAA3 and AAA4 do not hydrolyze an ATP molecule in each round of the mechanochemical cycle. Finally, AAA5 and AAA6 do not bind nucleotides; instead, they serve a structural role, transmitting conformational changes through the AAA+ ring (Schmidt et al. 2014).
The stalk, microtubule-binding domain, and buttress
Dynein’s microtubule-binding domain (MTBD) resides at the end of a 15-nm, coiled-coil stalk that emerges from AAA4 (Carter et al. 2011, Gee et al. 1997, Goodenough & Heuser 1984, Kon et al. 2011). The MTBD is a small α-helical domain that binds microtubules in the cleft between α- and β-tubulin (Carter et al. 2008, Redwine et al. 2012), a similar site to that bound by kinesin (Mizuno et al. 2004). Nucleotide binding at AAA1 and microtubule binding by the MTBD are coordinated allosterically through the stalk, where the coiled-coil helices can slide between different registers (Carter et al. 2008, Gibbons et al. 2005, Kon et al. 2009). To facilitate this allostery, AAA5 has an extension termed the buttress (or strut) (Carter et al. 2011, Kon et al. 2012), which interacts with and modulates register changes in the stalk (Schmidt et al. 2014).
The linker
The linker, composed of four α-helical segments and located N-terminal to dynein’s AAA+ ring, is required for motility (Reck-Peterson et al. 2006, Shima et al. 2006) and moves in response to the nucleotide state of AAA1 (Burgess et al. 2003). Its movement across the AAA ring from a bent, pre–power stroke conformation (Figure 1c) to a straight conformation (Figure 1b) constitutes dynein’s power stroke (see states 7 and 8 in Figure 1d) (Burgess et al. 2003; Kon et al. 2005; Roberts et al. 2009, 2012). ATP hydrolysis at AAA1 triggers conformational changes that result in the linker bending at a centrally located hinge in the middle of these segments (Figure 1c) (Kon et al. 2012, Schmidt et al. 2014).
The C-terminal domain
Following AAA6 is a C-terminal domain, which contacts AAA5, AAA6, and AAA1 on the opposite face of the ring relative to the linker (Carter et al. 2011, Kon et al. 2011, Roberts et al. 2009). In S. cerevisiae dynein, this domain is reduced in size to a short α-helix (Carter et al. 2011, Schmidt et al. 2012). Removal of the C-terminal domain from Dictyostelium discoideum dynein dramatically reduces the motility of individual dynein molecules in vitro (Numata et al. 2011), whereas a similar truncation in mammalian dynein results in a motor that can produce greater force (Nicholas et al. 2015).
Dynein’s Mechanochemical Cycle
Conformational changes that can do work and are linked to nucleotide hydrolysis are referred to as a protein’s mechanochemical cycle. Here, we present a synthesis of structural and biochemical studies that provide a nearly complete view of dynein’s mechanochemical cycle (Figure 1d, Supplemental Video 1; follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org). We limit our discussion of dynein’s mechanochemical cycle to one dynein motor domain (or head), but in reality cytoplasmic dynein exists as a dimer, and ATP hydrolysis occurs in both heads.
Overview of the mechanochemical cycle
We begin the mechanochemical cycle (Figure 1d, Supplemental Video 1) with dynein bound to the microtubule in the absence of ATP (the apo state, state 1). Binding of ATP to AAA1 (state 2) leads to closing of the ATP-binding pocket at AAA1 (state 3), lowering dynein’s affinity for the microtubule (state 4) and releasing the motor from its track (state 5). This is also when the linker moves to its bent, pre–power stroke conformation near AAA2 (state 5). Although ATP hydrolysis takes place during this part of the cycle (state 5), its precise timing and whether hydrolysis is coupled to the other changes taking place in the motor are not yet known. Dynein then rebinds the microtubule with a strong bias toward sites located in the direction of the microtubule minus end relative to its previous binding site (state 6). Subsequent phosphate release at AAA1 (state 7) induces the power stroke (states 7, 8), in which the linker moves from its pre–power stroke conformation to a straight, post–power stroke conformation near AAA4 and AAA5. This movement pulls the tail forward, along with the other head and attached cargo. Thus, by coupling a nucleotide hydrolysis cycle to conformational changes, dynein is capable of converting the energy from ATP hydrolysis into directional movement along its microtubule track. Below, we discuss each state of the cycle in more detail. The state numbers correspond to those listed in Figure 1d.
State 1
At the start of the cycle, AAA1 is nucleotide free and dynein is tightly bound to the microtubule. The nucleotide–binding site for AAA1 is in an open conformation, with AAA1 and AAA2 spaced far apart (compare the relative positions of AAA1 and AAA2 in states 1 and 3 in Figure 1d) (Carter et al. 2011, Roberts et al. 2012, Schmidt et al. 2012). At this point, AAA2 is bound to ATP and is expected to be so for the entire cycle (Schmidt et al. 2012). The nucleotide occupancy of AAA3 and AAA4 is less clear. It is likely that ATP is quickly hydrolyzed in both AAA3 and AAA4 (Kon et al. 2012) and that ADP remains bound to both sites for multiple cycles (DeWitt et al. 2015); we have chosen to represent this scenario in Figure 1d. The presence of nucleotide (ADP) at AAA3 and AAA4 results in a closely packed subdomain within the ring that can move as a rigid body (Schmidt et al. 2012, 2014). In this state of the AAA+ ring, the linker lies in the straight, post–power stroke conformation, docked at AAA5 (Bhabha et al. 2014, Schmidt et al. 2012).
States 2, 3, and 4
Upon ATP binding to AAA1 (state 2), changes within the dynein ring result in dynein releasing from the microtubule and the linker swinging to its pre–power stroke conformation. Binding of ATP at AAA1 closes the gap between AAA1 and AAA2 (Bhabha et al. 2014, Schmidt et al. 2014) (state 3, movement of AAA2 highlighted in inset); it is expected that this triggers conformational changes in the ring that cause the buttress, which interacts with the stalk, to alter the register of the coiled coil (Schmidt et al. 2014), in turn altering the conformation of the MTBD and weakening its interaction with the microtubule (Carter et al. 2008, Gibbons et al. 2005, Kon et al. 2009, Redwine et al. 2012) (state 4). Finally, the planar conformation adopted by the ring as a result of ATP binding at AAA1 leads to a steric clash between the linker and AAA4 (state 4), contributing to the linker adopting the pre–power stroke position (Schmidt et al. 2014) (state 5). Although state 4 has not yet been observed structurally, one would expect that release from the microtubule would precede the pre–power stroke movement of the linker, as the reverse order would be unproductive. Further work is necessary to understand the precise role played by ATP hydrolysis itself in the mechanochemical cycle.
States 5 and 6
The movement of the linker to the pre–power stroke position at AAA2 may play a role in biasing dynein further toward the minus end of the microtubule (state 5). The angle between the MTBD and dynein’s stalk (Carter et al. 2008, Mizuno et al. 2004) (see Figure 1a) may also play a role in setting this bias, but the source of the directionality in dynein’s movement is not yet fully understood. The MTBD of the newly freed, ADP.Pi-bound dynein head is now able to search the microtubule for a new binding site, loosely associating with the microtubule as it diffuses to a new site (state 6).
States 7 and 8
The interaction between the MTBD and a new binding site changes the MTBD’s conformation back to the high-affinity state (state 7) and is followed by a series of molecular events: (a) The stalk’s coiled coil returns to its high-affinity register, a change that is coupled to the ring via the buttress (state 7). (b) A movement in AAA4 removes the potential clash between it and a post–power stroke linker (state 7). (c) The buttress pulls AAA5 and the large domain of AAA6 away from AAA1 (state 7). (d) Pi is released from AAA1 (state 7). (e) The linker swings back toward its post–power stroke position, resulting in a conformation that is close to but distinct from the post–power stroke conformation and which has been captured in structures of both D. discoideum and S. cerevisiae dynein in the presence of ADP (Kon et al. 2012, Toropova et al. 2014). (f) The linker repositions further to its post–power stroke conformation, docking at a hydrophobic site in AAA5 (Schmidt et al. 2012) (state 8). (g) A gap opens between AAA1 and AAA2, which leads to ADP release and the resetting of the motor to begin a new cycle (state 1).
Slowed cycle
The entry of dynein into its mechanochemical cycle is allosterically regulated by the nucleotide state at AAA3. Mutations that block nucleotide hydrolysis at AAA3 impair dynein’s ability to release from microtubules when AAA1 binds ATP (DeWitt et al. 2015) and inhibit the linker from attaining the pre–power stroke conformation (Bhabha et al. 2014). The presence of ATP at AAA3 may lock dynein into a repressed state. The structure of the motor with an ATP analog (AMPPNP) bound to all four sites (AAA1–4) shows the linker in its post–power stroke position and no conformational changes in the buttress (Bhabha et al. 2014); this structure is more reminiscent of state 1 than states 4 or 5. These observations suggest that when dynein is actively stepping, AAA3 must be ADP bound, and experimental observations indicate that it remains ADP bound over the course of multiple steps (DeWitt et al. 2015).
Dynein processivity and stepping
Cytoplasmic dynein is a dimer in cells. Many cytoskeletal motors that transport cargo over long distances (e.g., kinesin-1 and myosin-V) are processive, capable of taking multiple steps along their track without releasing (Kull & Endow 2013). Yeast dynein is processive in vitro in the absence of regulators (Reck-Peterson et al. 2006) and because of its robust processivity has been used to characterize the dynein stepping mechanism (DeWitt et al. 2012, Gennerich et al. 2007, Qiu et al. 2012, Reck-Peterson et al. 2006). Although studies have shown that vertebrate dynein bound to beads is processive (for example, King & Schroer 2000, Mallik et al. 2004, Wang et al. 1995), direct observation of individual molecules in vitro has led to more variable results, with some studies reporting that dynein is processive and others reporting either weak or no processivity (reviewed in Jha & Surrey 2015). The processivity of dynein is enhanced by dynactin (Culver-Hanlon et al. 2006, Kardon et al. 2009, King & Schroer 2000, Ross et al. 2006, Tripathy et al. 2014); however, this effect is much more pronounced in the presence of a cargo adaptor for vertebrate dynein (McKenney et al. 2014, Schlager et al. 2014) (see below). Additionally, some cargo is moved by teams of dyneins, which are more processive than individual dynein molecules (reviewed in Mallik et al. 2013).
The average step size of dynein is ~8 nm, which corresponds well to the 8.3 nm spacing between tubulin dimers, although dynein can also take longer, backward, and sideways steps (DeWitt et al. 2012, Mallik et al. 2004, Qiu et al. 2012, Reck-Peterson et al. 2006, Toba et al. 2006). Other processive motors, such as kinesin-1 and myosin-V, step processively using gating mechanisms that coordinate the cycles of ATP hydrolysis between the two motor domains, so that one head releases only when the other head is bound to its track (Kull & Endow 2013). By contrast, purified yeast dynein exhibits processivity without strict coordination (DeWitt et al. 2012, Qiu et al. 2012).
HOW IS DYNEIN RECRUITED TO MICROTUBULE PLUS ENDS?
Dynein, which moves toward the minus ends of microtubules, often engages cargo from microtubule plus ends (for example, see Egan et al. 2012, Lee et al. 2003, Lenz et al. 2006, Lomakin et al. 2009, Sheeman et al. 2003, Vaughan et al. 2002, Zhang et al. 2010). Thus, an initial step in cargo transport in many cases is getting dynein to the microtubule plus end. Below, we discuss multiple mechanisms that are used to recruit dynein to microtubule plus ends, including transport by kinesin, recruitment from the cytoplasm by microtubule plus end–tracking proteins (+TIPs), and localization of dynein’s mRNA.
Transport by Kinesin
In budding yeast and filamentous fungi, genetics provided the first indication that dynein could be recruited to microtubule plus ends via a plus-end–directed kinesin (Carvalho et al. 2004, Zhang et al. 2003). In yeast, dynein recruitment to the plus ends of microtubules depends on a kinesin called Kip2, Lis1 (Pac1), and the microtubule-binding protein Clip170 (Bik1) (Carvalho et al. 2004, Lee et al. 2003, Sheeman et al. 2003). Recent in vitro reconstitution experiments using purified proteins on dynamic microtubules have led to a model for how these proteins transport dynein to microtubule plus ends (Roberts et al. 2014) (Figure 2a). In brief, dynein is brought to microtubule plus ends by Kip2, with Lis1 and Clip170 mediating the interaction between the two motors (Carvalho et al. 2004, Sheeman et al. 2003). Rather than being a passive passenger during this process, dynein can engage Kip2 in a tug-of-war, which Kip2 is able to win by recruiting the microtubule-binding proteins Clip170 and EB1 (Bim1) as processivity factors. There is also evidence that dynein is transported to microtubule plus ends in a kinesin-1–dependent manner in filamentous fungi (Egan et al. 2012; Lenz et al. 2006; Zhang et al. 2003, 2010) and neurons (Yamada et al. 2008, 2010). Direct interactions between mammalian dynein and kinesin subunits have been reported (Ligon et al. 2004), providing an alternative potential mechanism for dynein-kinesin interactions.
Figure 2.
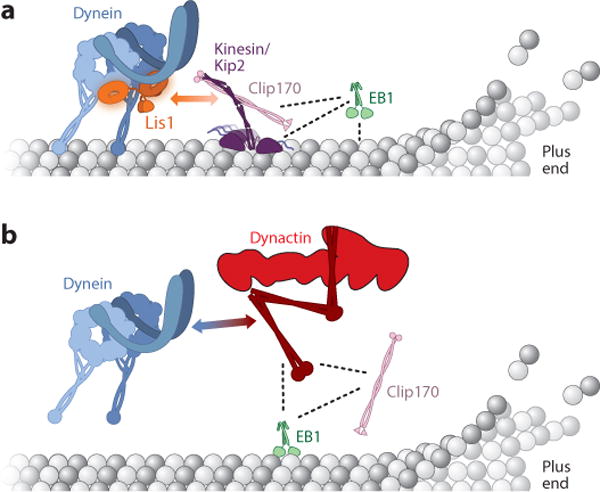
Models for the localization of dynein to the microtubule plus end. (a) Model for transport by kinesin in budding yeast (Roberts et al. 2014). Dynein is transported to the microtubule plus end directly by the kinesin Kip2. Lis1 and Clip170 mediate this interaction, and the plus end–tracking proteins (+TIPs) Clip170 and EB1 act as processivity factors for Kip2. The two-headed arrow indicates the Lis1-Clip170 interaction, whereas dotted lines indicate other direct interactions. Adapted from Roberts et al. 2014. (b) Model for the role of +TIPs in mammalian cells (Duellberg et al. 2014). Dynein is recruited to microtubule plus ends from the cytoplasm via microtubule +TIPs. Here, EB1 and Clip170 recruit the dynactin subunit p150 (the entire dynactin complex is shown in this figure) to microtubule plus ends, and p150 in turn recruits dynein. The two-headed arrow indicates the dynein-p150 interaction, whereas dotted lines indicate other direct interactions.
Recruitment via Microtubule Plus Tip Proteins from the Cytoplasm
Genetic studies in budding yeast suggest that a second pathway can target dynein to microtubule plus ends directly from the cytoplasm, in a process that requires Clip170 but not Kip2 (Caudron et al. 2008, Markus et al. 2009). In fission yeast, recruitment of dynein along the length of the microtubule from the cytoplasm has been visualized directly (Ananthanarayanan et al. 2013). In mammalian cells, dynein is recruited to microtubule plus ends by dynactin, Clip170, and EB1 (Lloyd et al. 2012, Lomakin et al. 2009, Moughamian et al. 2013, Moughamian & Holzbaur 2012, Splinter et al. 2012). Recent in vitro experiments have reconstituted this process, showing that EB1 and Clip170 can recruit the p150 subunit of dynactin to the plus ends of dynamic microtubules, from which it can recruit recombinant human dynein (Duellberg et al. 2014) (Figure 2b).
Plus End Localization of mRNA
The mechanisms discussed so far recruit dynein to microtubule plus ends in its protein form. Subcellular localization of the mRNA encoding dynein or its regulatory factors may be another way of ultimately targeting these proteins to microtubule plus ends. This mode of dynein localization could be particularly important in large, highly polarized cells such as neurons, in which transport of proteins from the cell body to destinations in axons could take days (Gumy et al. 2014). Evidence for this idea stems from the ability of the adenomatous polyposis coli (APC) protein to localize mRNA to the plus ends of microtubules in neurons (Preitner et al. 2014). These mRNAs include both the dynein light intermediate chain and dynein regulator Lis1, raising the possibility that they, as well as mRNAs encoding other dynein subunits and regulatory factors, are transported as messenger ribonucleoprotein particles (mRNPs) to microtubule plus ends, where they are locally translated.
HOW IS DYNEIN-BASED TRANSPORT INITIATED AND ACTIVATED?
As described in the previous section, several pathways deliver dynein to cargo loading sites, which are often at microtubule plus ends. After reaching microtubule plus ends or other sites of cargo interaction, dynein can be retained there, a step that appears to be important for initiating transport and/or loading cargo onto dynein. An emerging body of evidence suggests that dynein may exist in an inhibited or not-fully-activated state before transport commences (Markus et al. 2009, McKenney et al. 2014, Schlager et al. 2014, Torisawa et al. 2014). Thus, a final step in dynein-based transport is to activate dynein for motility. The dynein regulators Lis1, NudE/Nudel, and dynactin, as well as cargo adaptors, play important roles in these processes. Although these steps in cargo transport provide a useful framework for discussing dynein regulation, it remains to be determined whether these mechanisms are conserved in all organisms and cell types and whether there are alternative pathways for regulating dynein. Here, we discuss the roles that the dynein regulators Lis1, NudE/Nudel, and dynactin, as well as cargo adaptors, play in transport initiation and activation. We also present the most recent structural and mechanistic models for these regulatory factors.
Lis1 and Nudel/NudE
LIS1 was first described as the gene mutated in patients with type-1 lissencephaly, a neurodevelopmental disease characterized by a smooth cerebral surface, cognitive defects, and seizures (Reiner et al. 1993, Wynshaw-Boris 2007). Haploinsufficiency of LIS1 leads to lissencephaly (Saito et al. 2011), whereas overexpression of Lis1 results in microcephaly and neurodevelopmental delays (Lockrow et al. 2012). The cellular basis for these diseases is impaired neural migration, proliferation, and survival (Reiner & Sapir 2013). Lis1 interacts with the homologs Nudel and NudE, which also play key roles during neurodevelopment (Bradshaw et al. 2013). NudE knockout mice have reduced brain volume (Feng & Walsh 2004), whereas Nudel knockout mice are not viable (Sasaki et al. 2005).
Lis1 and Nudel/NudE are conserved dynein regulators that were first linked to the dynein pathway via genetic screens probing nuclear migration and positioning in filamentous fungi (Efimov & Morris 2000, Minke et al. 1999, Xiang et al. 1995). Shortly thereafter, genetic studies in Drosophila confirmed that this pathway is conserved in metazoans (Liu et al. 1999, 2000), and mammalian dynein, Lis1, and Nudel were shown to colocalize in vivo and physically interact in vitro (Faulkner et al. 2000, Sasaki et al. 2000, Smith et al. 2000).
Many dynein activities require Lis1 and Nudel/NudE. In HeLa cells, proper localization of Golgi complexes, endosomes, and lysosomes requires Lis1 (Lam et al. 2010), although other studies report little effect of Lis1 depletion in non-neuronal cells (Faulkner et al. 2000, Tai et al. 2002, Yi et al. 2011). In highly polarized cells, such as neurons and filamentous fungi, the motility of mitochondria, endosomes, peroxisomes, and lysosomes requires Lis1 (Egan et al. 2012, Lenz et al. 2006, Moughamian et al. 2013, Shao et al. 2013, Yi et al. 2011, Zhang et al. 2010). Lis1 is also required for positioning the nucleus and centrosome in organisms ranging from fungi to humans (Cockell et al. 2004, Lee et al. 2003, Sitaram et al. 2012, Tsai et al. 2007, Vallee & Tsai 2006, Xiang et al. 1995, Youn et al. 2009), localizing mRNA in Drosophila (Dix et al. 2013), and positioning and organizing the spindle pole in mitosis (Moon et al. 2014, Yingling et al. 2008). Nudel/NudE is also required for most of the same functions.
The molecular mechanism of Lis1 and Nudel/NudE
As a functional dimer, Lis1 is capable of interacting with both Nudel/NudE and dynein’s motor domain. Each Lis1 monomer contains an N-terminal dimerization domain, a coiled coil, a disordered loop, and a β-propeller domain containing seven WD repeats (Kim et al. 2004, Tarricone et al. 2004) (Figure 3a). Lis1’s C-terminal β-propeller domain interacts with AAA3 and AAA4 in dynein (Huang et al. 2012, Toropova et al. 2014), making Lis1 the only known dynein regulator to interact directly with the motor domain.
Figure 3.
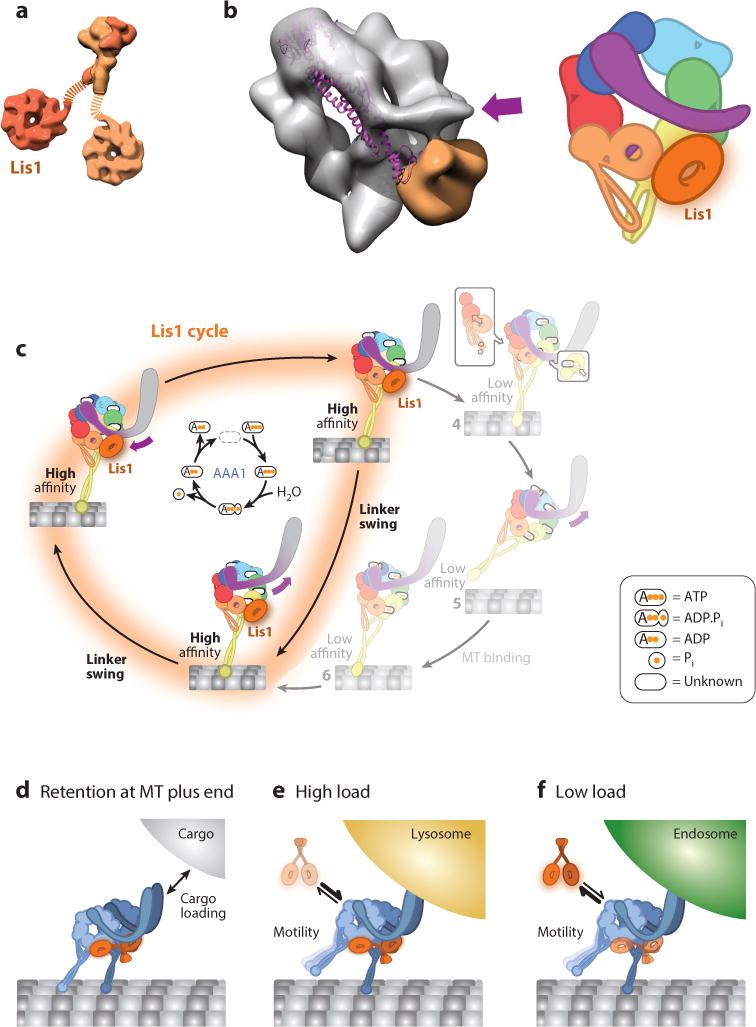
Structure and function of Lis1. (a) Structure of the Lis1 dimer. The N-terminal dimerization domain [Protein Data Bank (PDB) ID 1UUJ] is shown at the top and the C-terminal β-propeller domain (PDB 1VYH) at the bottom. The dotted lines represent the flexible linkers connecting the two domains. Lis1 is a homodimer; two different shades of orange distinguish the two monomers. (b, Left) Cryo-negative stain electron microscopy reconstruction of dynein-Lis1 (EMDB-6008); the dynein motor domain is shown as a semitransparent gray surface, and Lis1 is shown in orange. The purple arrow marks the N terminus of the linker in the dynein-Lis1 map. The linker domain from the crystal structure of dynein alone (PDB 3VKG) is shown as a purple ribbon diagram, aligned relative to the AAA+ (ATPases Associated with various cellular Activities) ring in the cryo-EM map. The post–power stroke position of the linker is sterically incompatible with the presence of Lis1. (Right) A representation of the dynein-Lis1 complex. Dynein subunits are colored according to the AAA+ domains, and Lis1 is shown as an orange ring interacting with AAA4 (yellow). The orange/purple circle in AAA5 indicates the normal docking site for the linker in the post–power stroke conformation. Since the resolution of the dynein-Lis1 map (21 Å) does not allow for the precise positioning of the AAA+ domains, we reduced the level of detail relative to Figure 1. (c) Model of dynein’s mechanochemical cycle in the presence of Lis1. By binding at AAA4, Lis1 blocks the linker from reaching its normal AAA5 docking site, preventing microtubule release. The ATP hydrolysis cycle at AAA1 (center) continues when Lis1 is bound to dynein. The linker is capable of moving between its normal pre–power stroke position at AAA2 and the new Lis1-induced position; we call this movement linker swing to distinguish it from the canonical power stroke. (d–f) Models for cellular functions of Lis1. Lis1 assists in retaining dynein at microtubule (MT) plus ends, facilitating cargo loading (d). Following loading, Lis1 may also be required for the transport of high-load cargo (e) but not low-load cargo (f).
Nudel and NudE are large, dimeric, coiled coil–containing proteins. The N terminus of Nudel/NudE binds the DIC, whereas the C-terminal region binds Lis1 (Wang & Zheng 2011, Zylkiewicz et al. 2011). Nudel/NudE likely tethers Lis1 to the dynein motor domain. Overexpression of Lis1 can partially rescue a NudE deletion phenotype in yeast and filamentous fungi (Efimov 2003, Li et al. 2005). The tethering role of Nudel/NudE is further supported by in vitro studies: high concentrations of Lis1 can overcome spindle pole defects in Xenopus laevis extracts depleted of Nudel (Wang et al. 2013), and in S. cerevisiae, Nudel enhances Lis1-mediated effects on dynein motility (Huang et al. 2012). Nudel/NudE’s tethering role could be regulated by phosphorylation; for example, phosphorylation by Cdk5 enhances the interaction between Nudel/NudE and Lis1 (Hebbar et al. 2008, Pandey & Smith 2011). Nudel/NudE may also have activities other than tethering—mammalian Nudel/NudE decreases dynein’s microtubule-binding properties in the absence of Lis1 (McKenney et al. 2010, Yamada et al. 2008).
Biochemical experiments performed with both yeast and mammalian proteins suggest that Lis1 increases dynein’s affinity for microtubules (Huang et al. 2012, McKenney et al. 2010, Toropova et al. 2014, Yamada et al. 2008). Yeast Lis1 causes dynein to remain attached to microtubules in the presence of ATP, a condition that would normally release dynein from microtubules (Huang et al. 2012, Toropova et al. 2014). Lis1 also slows dynein’s velocity in both single-molecule motility assays and ensemble microtubule gliding assays (Huang et al. 2012, Yamada et al. 2008). Finally, in a bead-based optical trap assay, mammalian Lis1 and NudE allow dynein to achieve a prolonged force-producing state (McKenney et al. 2010).
Interestingly, Lis1 does not increase dynein’s microtubule-binding affinity by preventing ATP hydrolysis (Huang et al. 2012, McKenney et al. 2010, Toropova et al. 2014, Yamada et al. 2008). This has led to the idea that Lis1 acts like a clutch, uncoupling the cycles of nucleotide hydrolysis and microtubule binding and release (Huang et al. 2012). A recent study suggests that the clutch is engaged when Lis1 binds at AAA4 in dynein, interrupting the allosteric pathway between AAA1 and the stalk/MTBD (Figure 3b) (Toropova et al. 2014). Lis1 appears to achieve this, at least in part, by blocking dynein’s linker from docking at AAA5 (Figure 1d, state 8; Figure 3c) (Toropova et al. 2014). Further work is necessary to understand why dynein continues to hydrolyze ATP when bound to Lis1 or if additional layers of regulation are required to block ATP hydrolysis.
The cellular mechanism of Lis1 and Nudel/NudE
How does Lis1’s ability to increase dynein’s affinity for microtubules influence dynein activity in cells? Although much remains to be done to answer this question, current evidence suggests three possible, not mutually exclusive, cellular mechanisms for Lis1: (a) Lis1 participates in recruiting dynein to the microtubule plus end, (b) Lis1 helps initiate transport, and (c) Lis1 helps dynein withstand load when moving large cargos (Figure 3d–f). We discussed Lis1’s role in recruiting dynein to microtubule plus ends above. Here, we discuss how Lis1’s biochemical properties may be important for its other two proposed cellular functions.
Transport initiation
Lis1 may play a role in transport initiation by recruiting dynein to binding partners (receptors) on transported cargo. For example, in yeast cells, in addition to playing a role in localizing dynein to microtubule plus ends (Lee et al. 2003, Roberts et al. 2014, Sheeman et al. 2003), Lis1 appears to be important for maintaining dynein in a conformation that can be offloaded to its receptor (Num1) on the cell cortex (Markus & Lee 2011). In filamentous fungi, in which endosomes are a dynein cargo, Lis1 is required for the initiation but not the maintenance of endosome motility (Egan et al. 2012, Lenz et al. 2006), as the absence of Lis1 reduces endosome flux and Lis1 is not observed on moving endosomes (Egan et al. 2012). Similarly, Lis1 is required to recruit dynein to mRNA localization complexes in Drosophila (Dix et al. 2013) and membranes in HeLa cells (Lam et al. 2010, Splinter et al. 2012). In mouse dorsal root ganglion (DRG) neurons, Lis1 knockdown reduces retrograde flux of lysosomes (Moughamian et al. 2013), suggesting that Lis1 could play an analogous role in transport initiation in neurons.
Transport of high-load cargo
The idea that Lis1 may allow dynein to move high-load cargo (Figure 3e,f) is supported by in vitro optical trapping experiments showing that mammalian Lis1 and NudE allow dynein to achieve a prolonged force-producing state (McKenney et al. 2010). Furthermore, in rat neurons, inhibition of Lis1 via injection of a function-blocking antibody does not affect transport of smaller cargo but severely impairs transport of larger (≥1 μm) membranous cargo (Yi et al. 2011). By contrast, in mouse DRG neurons, Lis1’s effects on organelle transport appear to be size independent (Moughamian et al. 2013).
Dynactin
Dynactin, a dynein regulator, was originally discovered as a factor capable of activating dynein-mediated vesicle motility in vitro (Gill et al. 1991, Schroer & Sheetz 1991). Since its discovery, dynactin has been implicated in almost all functions performed by dynein (Schroer 2004). The activities in which dynactin is involved include recruiting dynein to kinetochores (Bader & Vaughan 2010, Yeh et al. 2013), initiating transport by recruiting dynein to microtubule plus ends (Lloyd et al. 2012, Moughamian et al. 2013, Moughamian & Holzbaur 2012, Splinter et al. 2012, Zhang et al. 2003), and moving with dynein cargo complexes (Deacon et al. 2003, Egan et al. 2012, Lenz et al. 2006, Maday et al. 2012, Moughamian et al. 2013), where it most likely acts as a processivity factor (see below). The central role played by dynactin during dynein transport is underscored by genetic disorders that are linked to point mutations in the microtubule-binding domain of p150, the largest subunit in dynactin (Farrer et al. 2009, Lipka et al. 2013, Puls et al. 2003, Vilarino-Guell et al. 2009). These point mutations result in motor neuron disease and Perry syndrome, which are both adult-onset neurodegenerative diseases (Farrer et al. 2009, Puls et al. 2003). The molecular basis for these diseases may lie in the aberrant recruitment of dynactin to microtubule plus ends or changes in microtubule dynamics (Lazarus et al. 2013, Moughamian & Holzbaur 2012).
Structure of dynactin
Dynactin is a large 1.2 MDa multisubunit protein complex consisting of 23 distinct polypeptides in vertebrates. The overall architecture of vertebrate dynactin was originally determined using biochemistry and low-resolution electron microscopy (EM) (Eckley et al. 1999; Imai et al. 2006, 2014; Schafer et al. 1994; Schroer 2004). Recent work using cryo-electron microscopy (cryo-EM) has provided the first near-atomic resolution models of parts of the dynactin complex (Chowdhury et al. 2015, Urnavicius et al. 2015). Dynactin is composed of a long, thin, segmented arm and a shorter helical filament (Figure 4a). The arm, called p150, consists of a homodimer that forms three parallel coiled-coil segments (CC1a, CC1b, and CC2), with a microtubule-binding CAP-Gly (cytoskeleton-associated protein glycine-rich) domain at the N terminus of each monomer. It is linked to the filament at its other end through a shoulder complex containing the dynamitin/p50 and p24 subunits (Figure 4a). The helical filament, ~40 nm in length, is built mainly from actin-related protein 1 (Arp1).
Figure 4.
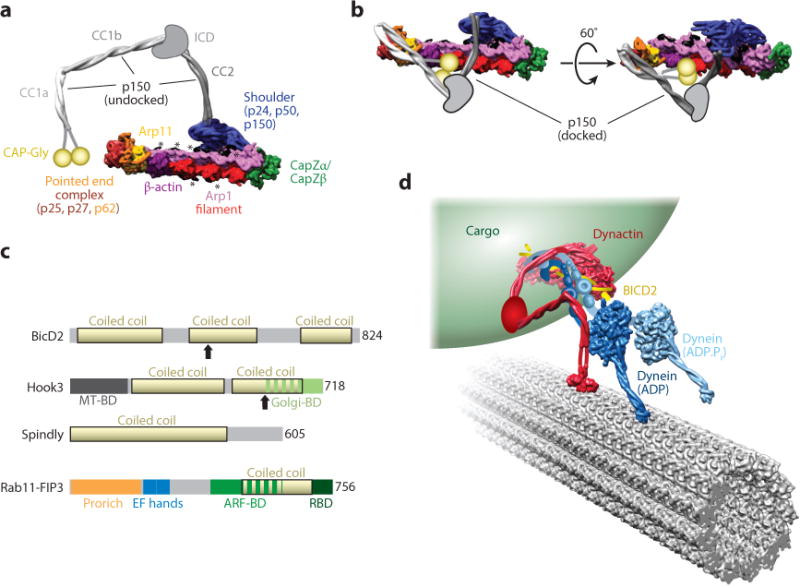
Structure and function of dynactin and cargo adaptors. (a) Overview of the structure of the dynactin complex. In this panel, p150 is shown in the undocked conformation, in which CC1a, CC1b, and CC2 extend away from the filament. The structure was generated from PDB 5AFT and was rendered as a 10 Å–resolution molecular surface. The positions of the p150 coiled coils were modified from the docked conformation in the cryo-EM map (shown in panel b). The ICD connecting CC2 and CC1b and the CAP-Gly domains, which are not part of the atomic model, are shown in cartoon form. Peptides emanating from the shoulder, most likely from p50, that are proposed to be involved in setting the length of the Arp1 filament are shown in black and highlighted with asterisks. (b) Cryo-EM structure of dynactin in the docked conformation, in which p150 folds back on itself and docks onto the Arp1 filament. The structure shown is a 10 Å–resolution molecular surface generated from PDB 5AFT. As in panel a, the ICD and CAP-Gly domains are shown in cartoon form. (c) Domain organization of cargo adaptors that activate dynein motility. Coiled-coil domains for all adaptors are shown in yellow. Arrows indicate the sites of truncation for N-terminal fragments of BICD2 and Hook3 that stimulate dynein activity. (d) Model of dynein-dynactin-BICD2 bound to an organelle (e.g., a Rab6 vesicle) and moving along a microtubule. The model was drawn to account for known interactions within the complex. The N terminus of dynein’s tail (blue) binds to a complex between dynactin (red) and BICD2 (yellow). This binding results in the undocked conformation of the p150 subunit of dynactin. Its three coiled coils (red) extend away from the Arp1 filament such that CC1b can interact with dynein’s intermediate chain at the base of the tail. This model was generated using PDBs 3VKG and 4RH7 for dynein, 5AFT for dynactin, and 3TQ7 for the CAP-Gly domain. Dynein’s tail, its associated chains, dynactin’s ICD, and the connection between CC1a and the CAP-Gly domain in p150 are shown in schematic form. Abbreviations: ARF-BD, Arf GTPase binding domain; CAP-Gly, cytoskeleton-associated protein glycine-rich; cryo-EM, cryo-electron microscopy; EF hand, calmodulin-binding EF-hand motif; Golgi-BD, Golgi-binding domain; ICD, intercoiled domain; MTBD, microtubule-binding domain; PDB, Protein Data Bank; Pro rich, proline rich; RBD, Rab GTPase–binding domain.
A recent cryo-EM structure (Urnavicius et al. 2015) has revealed the mechanisms responsible for controlling the length of the Arp1 filament. Extended peptides originating from the shoulder complex coat the Arp1 filament. These peptides, which appear to originate from the p50 subunit, interact exclusively with the Arp1 components of the filament, suggesting that they may be important for setting its length. The dynactin filament contains eight Arp1 subunits, five in one protofilament and three in the other (Chowdhury et al. 2015, Urnavicius et al. 2015). The fourth position in the shorter Arp1 protofilament is occupied by β-actin (Urnavicius et al. 2015), which in turn recruits Arp11, a polymerization-incompetent Arp that prevents further filament growth at that end (the pointed end). Additional subunits bind to Arp11, but they do not appear to be involved in length control. These additional subunits—p25, p27, and p62—are absent from yeast and are involved in tethering dynein-dynactin to cargo, such as endosomes and the nuclear envelope (Yeh et al. 2012, Zhang et al. 2011). At the other end of the Arp1 filament (the barbed end), capping occurs via a heterodimer of the proteins CapZα and CapZβ (Figure 4a).
Dynein activation by dynactin and cargo adaptors
Although dynactin has been shown to increase dynein processivity on its own (Ayloo et al. 2014, Culver-Hanlon et al. 2006, Kardon et al. 2009, King & Schroer 2000, Ross et al. 2006, Tripathy et al. 2014), two recent studies have revealed that the combined addition of dynactin and a cargo adaptor [Bicaudal-D2 (BICD2), Hook3, Spindly, or Rab11-FIP3; Figure 4c] converts mammalian dynein from a diffusive motor into a highly processive one in vitro (McKenney et al. 2014, Schlager et al. 2014).
How do dynactin and a cargo adaptor activate dynein’s motility? Structural studies of a dynein-dynactin-BICD2 (DDB) complex have shown that dynactin and BICD2 interact with the distal end (relative to the motor domain) of dynein’s tail domain (Chowdhury et al. 2015, McKenney et al. 2014, Schlager et al. 2014, Urnavicius et al. 2015). Recent cryo-EM structures of dynactin alone and dynactin in complex with dynein’s tail and BICD2 have provided additional insights into the activating functions of BICD2 and dynactin (Urnavicius et al. 2015). First, p150 (CC1a) and BICD2 (its N-terminal coiled coils, called CC1 and CC2) have overlapping binding sites on the pointed-end complex in both structures. Although it is not yet clear whether the docked conformation of p150 observed in the cryo-EM structure (Figure 4b) occurs in vivo, the mutual exclusivity of p150 CC1a and BICD2 CC1 binding suggests that binding of BICD2 may release p150, leading to its undocked conformation (Figure 4a,b). Given that previous biochemical studies have shown that p150 is likely to contact the DIC via CC1b (Karki & Holzbaur 1995, King et al. 2003, Siglin et al. 2013, Vaughan & Vallee 1995) (Figure 4a,b), it is possible that the BICD2-dependent release of p150 CC1a is required for this interaction to take place. Second, a terminal fragment of dynein’s tail (distal to the motor domain) binds dynactin’s filament through an interface that includes both dynactin and BICD2 (Urnavicius et al. 2015), explaining why the adaptor is required for a functional interaction between dynein and dynactin. Finally, the structure of the dynactin-BICD2–tail complex suggests that the dynactin filament induces asymmetry in the dimer of dynein tails. Although both tail fragments show an overall translational symmetry that matches that of the filament (i.e., they are arranged side by side following the repeating pattern of the Arp1 subunits), the two fragments do not bind the filament in identical ways (Urnavicius et al. 2015). One of them has unique interactions with BICD2 and an Arp1 subunit that make its path on the filament unique. This asymmetry in the paths taken by the two tails on the filament may activate dynein, possibly by relieving an internally symmetric, autoinhibited state (Torisawa et al. 2014).
How does the entire DDB complex interact with microtubules? A recent EM study has provided key insights into the architecture of the transport-competent ternary DDB complex and its interaction with its track (Chowdhury et al. 2015). The tail-dynactin-BICD2 portion of the DDB complex is directed away from the motor domains, with the cargo-binding pointed end of dynactin stretching back (i.e., toward the plus end of the microtubule; Figure 4d). The dynein motor domains, which are located at the front of the DDB complex (i.e., facing the minus end of the microtubule), show what would be expected to be a walking-competent arrangement: the motor domains bind to the microtubule side by side, with their stalks and microtubule-binding domains pointing toward the minus end of the microtubule (Figure 4d). Finally, an analysis of dimers of the dynein holoenzyme has revealed a dramatic kink in the tail domain, in a region that connects the motor domain to the portion of the tail that interacts with dynactin and BICD2 (Chowdhury et al. 2015). This structural feature may uncouple, to some extent, the motions in the motor domain that result from dynein’s mechanochemical cycle from those that would take place in the tail-dynactin-BICD2 portion of the DDB complex during cargo transport.
Cargo Adaptors
Proteins that link dynein and kinesin motors to their cargo are referred to as cargo adaptors or scaffolding proteins (recently reviewed by Fu & Holzbaur 2014b). Interestingly, many of the known cargo adaptors (e.g., BICD2, Hook3, Spindly, FIP3, HAP1, RILP, Snapin, and TRAK1 and 2; Figure 4c) contain long coiled coils predicted to be >20 nm. Although these adaptors do not appear to share any sequence homology, conserved motifs in the extended coiled coils may allow them to bind dynactin through the same interaction surface seen for BICD2. Although only four cargo adaptors are verified activators of dynein motility in vitro (BICD2, Hook3, Spindly, and Rab11-FIP3) (McKenney et al. 2014, Schlager et al. 2014), these adaptors provide a useful model for understanding how motor activity can be regulated in a cargo-specific manner. Below we discuss the roles of BICD2, Hook proteins, Spindly, and Rab11-FIP3 in regulating dynein-based motility.
BICD2
As the best-characterized cargo adaptor, BICD2’s role in dynein-mediated transport has been dissected at both the biochemical and cell biological levels. BICD2 was originally discovered in Drosophila, where mutations in BicD resulted in two-tailed (bicaudal) embryos (Mohler & Wieschaus 1986). In humans, mutations in BICD2 are associated with dominant congenital spinal muscular atrophy (Neveling et al. 2013, Oates et al. 2013, Peeters et al. 2013), similar to some mutations reported in the dynein heavy chain (Harms et al. 2012). At the cellular level, BicD plays a role in dynein-mediated RNA localization (Bianco et al. 2010, Bullock et al. 2006), Rab6 secretory vesicle transport (Matanis et al. 2002, Splinter et al. 2012), and nuclear positioning (Splinter et al. 2010).
BICD2’s coiled-coil domains provide the specificity to bind both cargo and dynein-dynactin. These coiled-coil domains are found at the N terminus (CC1), the middle region (CC2), and the C terminus (CC3) of BICD2. In addition to maintaining BICD2 as a parallel homodimer, the coiled-coil domains are capable of intradimer interactions, whereby CC3 can fold back onto CC1 to block access to the dynein-dynactin binding sites (Hoogenraad et al. 2001). The CC1-CC3 interaction appears to be inhibitory, as truncation of CC3 drives the robust formation of a DDB complex (Splinter et al. 2012). When autoinhibition is released, CC1-CC2 binds dynein-dynactin and CC3 binds intracellular cargo.
The CC3 domain of BICD2 has several known cargos. These include the GTP-bound conformation of Rab6/Rab6a, a GTPase that regulates the recruitment of kinesin and dynein to the Golgi complex, endoplasmic reticulum, and cytoplasm-derived vesicles (Coutelis & Ephrussi 2007; Hoogenraad et al. 2001, 2003; Januschke et al. 2007; Matanis et al. 2002); the clathrin heavy chain, to facilitate endocytic processes (Li et al. 2010); and the RNA-binding proteins Egalitarian and FMRP (Fragile X mental retardation protein), which allow mRNA to be transported by dynein (Bianco et al. 2010, Liu et al. 2013).
Hook
The hook family of proteins was originally discovered through a mutation in the Drosophila Hook protein that resulted in defects in the endocytic pathway (Kramer & Phistry 1996). The Drosophila protein has three mammalian homologs (Hook1, Hook2, and Hook3) that have distinct functions. Hook1 associates with membranes that are derived from clathrin-independent endocytosis and subsequently sorted into endosomes (Maldonado-Baez et al. 2013). Hook2 associates with centrosomes and mediates primary cilium formation (Baron Gaillard et al. 2011; Szebenyi et al. 2007a,b). Hook3 is associated with the Golgi complex (Walenta et al. 2001). Filamentous fungi have a single Hook protein that is responsible for endosome trafficking (Bielska et al. 2014, Zhang et al. 2014). Thus, the Hook proteins represent a diverse group of cargo adaptors that localize to distinct subsets of intracellular targets.
The Hook proteins share a common architecture: a microtubule-binding domain, a long coiled-coil region, and a C-terminal cargo-interaction domain (Figure 4c). The N-terminal microtubule-binding domain shares a similar architecture to the CLIP-170 family of microtubule-binding proteins (Simpson et al. 2005). This domain may tether Hook-bound cargo to microtubules (Walenta et al. 2001). Adjacent to the microtubule-binding domain is a long coiled-coil region that is responsible for dimerizing the Hook family of proteins and likely interacts with the Arp1 filament in the activated dynein-dynactin-Hook3 complex (McKenney et al. 2014, Urnavicius et al. 2015). Like the C terminus of BICD2, the C termini of Hook proteins contain a cargo interacting domain. This domain, which diverges among the Hook proteins, likely specifies different cargo interactions.
Spindly
Spindly (also called Ccdc99) plays a central role in silencing the spindle assembly checkpoint (Griffis et al. 2007), a mitotic checkpoint that persists until all spindle microtubules become attached to kinetochores (Barisic & Geley 2011). Spindly incorporates into the kinetochore via the Rod/ZW10/Zwilch (RZZ) complex and is required to recruit dynein to kinetochores (Chan et al. 2009, Gassmann et al. 2008, Griffis et al. 2007). Once microtubules are attached, the Mad1/Mad2 checkpoint proteins, RZZ, and Spindly are transported away from the kinetochore (Barisic & Geley 2011). Like BICD2 and Hook, Spindly is primarily a coiled-coil protein (Figure 4c). The Spindly motif, located at the beginning of one of the coiled coils, is necessary for dynein/dynactin recruitment to kinetochores in HeLa cells (Gassmann et al. 2010). Because full length Spindly is able to immunoprecipitate dynein/dynactin and activate its processivity in vitro (McKenney et al. 2014), a key question for the future is how dynein is activated for motility in vivo only after kinetochore biorientation is achieved.
Rab11-FIP3
Rab11 family members are involved in numerous endosomal-recycling pathways (Kelly et al. 2012). Mammals have five Rab11 family–interacting proteins, or FIPs, that are effectors of Rab11 GTPases (Baetz & Goldenring 2013). One of these, Rab11-FIP3, recruits dynein onto Rab11-positive membranes through an interaction with the dynein light intermediate chain (Horgan et al. 2010). Like the other dynein adaptor proteins, Rab11-FIP3 also contains a long coiled-coil region at its C terminus, which could interact with dynein-dynactin in a manner similar to BICD2 (McKenney et al. 2014, Urnavicius et al. 2015) (Figure 4c). Rab11-FIP3 also contains an N-terminal proline-rich domain and two EF-hand motifs (Horgan et al. 2010), suggesting the potential for interactions with SH3 domains and calcium regulation.
CONCLUSIONS AND FUTURE DIRECTIONS
A wealth of new structural and biochemical data provides a detailed view of dynein’s mechanochemical cycle and the regulation of cytoplasmic dynein by the conserved Lis1-Nudel/NudE and dynactin complexes. Likewise, many roles for dynein and its regulators at the organismal and cellular level have been described. A major goal going forward will be to link mechanistic understanding of dynein and its regulation to its organismal and cellular functions. Mutations in the genes encoding dynein, the dynactin complex, Lis1, Nudel/NudE, and dynein’s cargo adaptors cause a range of neurodegenerative diseases. Why are specific types of neurons sensitive to mutations in dynein and its regulators? Why do mutations at different sites in the same gene lead to very different neurological diseases?
Another important future direction will be to determine how cytoplasmic dynein, whose heavy chain is encoded by only one gene, is regulated to perform dozens, if not hundreds, of functions, ranging from the transport of small vesicles to pulling the entire mitotic spindle. Although there have been important advances in identifying dynein cargo adaptors (Maday et al. 2014), for many cargos, the mechanism of dynein recruitment and how recruitment is regulated remains unknown. Finally, we are only beginning to understand how dynein’s mechanochemical properties are modulated to perform functions that require varying degrees of force and processivity.
Supplementary Material
Acknowledgments
We apologize to any colleagues whose work we failed to cite owing to space and reference number constraints. We thank Andrew Carter, Anthony Roberts, Xin Xiang, Mark McClintock, Martin Egan, Bret Redwine, and John Salogiannis for critical reading of the manuscript. MD is supported by a Jane Coffin Childs fellowship, MC by a Damon Runyon Cancer Research fellowship, and SRP and AL by the NIH (R01GM107214 to SRP and AL and R01GM100947 to SRP).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39:1169–78. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan V, Schattat M, Vogel SK, Krull A, Pavin N, Tolic-Norrelykke IM. Dynein motion switches from diffusive to directed upon cortical anchoring. Cell. 2013;153:1526–36. doi: 10.1016/j.cell.2013.05.020. [DOI] [PubMed] [Google Scholar]
- Ayloo S, Lazarus JE, Dodda A, Tokito M, Ostap EM, Holzbaur EL. Dynactin functions as both a dynamic tether and brake during dynein-driven motility. Nat Commun. 2014;5:4807. doi: 10.1038/ncomms5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader JR, Vaughan KT. Dynein at the kinetochore: timing, interactions, and functions. Semin Cell Dev Biol. 2010;21:269–75. doi: 10.1016/j.semcdb.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz NW, Goldenring JR. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol Biol Cell. 2013;24:643–58. doi: 10.1091/mbc.E12-09-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic M, Geley S. Spindly switch controls anaphase: Spindly and RZZ functions in chromosome attachment and mitotic checkpoint control. Cell Cycle. 2011;10:449–56. doi: 10.4161/cc.10.3.14759. [DOI] [PubMed] [Google Scholar]
- Baron Gaillard CL, Pallesi-Pocachard E, Massey-Harroche D, Richard F, Arsanto JP, et al. Hook2 is involved in the morphogenesis of the primary cilium. Mol Biol Cell. 2011;22:4549–62. doi: 10.1091/mbc.E11-05-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabha G, Cheng HC, Zhang N, Moeller A, Liao M, et al. Allosteric communication in the Dynein motor domain. Cell. 2014;159:857–68. doi: 10.1016/j.cell.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A, Dienstbier M, Salter HK, Gatto G, Bullock SL. Bicaudal-D regulates fragile X mental retardation protein levels, motility, and function during neuronal morphogenesis. Curr Biol. 2010;20:1487–92. doi: 10.1016/j.cub.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielska E, Schuster M, Roger Y, Berepiki A, Soanes DM, et al. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J Cell Biol. 2014;204:989–1007. doi: 10.1083/jcb.201309022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw NJ, Hennah W, Soares DC. NDE1 and NDEL1: twin neurodevelopmental proteins with similar ‘nature’ but different ‘nurture’. Biomol Concepts. 2013;4:447–64. doi: 10.1515/bmc-2013-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16:1447–52. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421:715–8. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–65. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, et al. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–5. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–29. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Caudron F, Andrieux A, Job D, Boscheron C. A new role for kinesin-directed transport of Bik1p (CLIP-170) in Saccharomyces cerevisiae. J Cell Sci. 2008;121:1506–13. doi: 10.1242/jcs.023374. [DOI] [PubMed] [Google Scholar]
- Chan YW, Fava LL, Uldschmid A, Schmitz MH, Gerlich DW, et al. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J Cell Biol. 2009;185:859–74. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283:25839–45. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Ketcham SA, Schroer TA, Lander GC. Structural organization of the dynein-dynactin complex bound to microtubules. Nat Struct Mol Biol. 2015;22:345–47. doi: 10.1038/nsmb.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell MM, Baumer K, Gonczy P. lis-1 is required for dynein-dependent cell division processes in C. elegans embryos. J Cell Sci. 2004;117:4571–82. doi: 10.1242/jcs.01344. [DOI] [PubMed] [Google Scholar]
- Coutelis JB, Ephrussi A. Rab6 mediates membrane organization and determinant localization during Drosophila oogenesis. Development. 2007;134:1419–30. doi: 10.1242/dev.02821. [DOI] [PubMed] [Google Scholar]
- Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol. 2006;8:264–70. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- Deacon SW, Serpinskaya AS, Vaughan PS, Lopez Fanarraga M, Vernos I, et al. Dynactin is required for bidirectional organelle transport. J Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335:221–25. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA, Cypranowska CA, Cleary FB, Belyy V, Yildiz A. The AAA3 domain of cytoplasmic dynein acts as a switch to facilitate microtubule release. Nat Struct Mol Biol. 2015;22:73–80. doi: 10.1038/nsmb.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix CI, Soundararajan HC, Dzhindzhev NS, Begum F, Suter B, et al. Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J Cell Biol. 2013;202:479–94. doi: 10.1083/jcb.201211052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–39. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duellberg C, Trokter M, Jha R, Sen I, Steinmetz MO, Surrey T. Reconstitution of a hierarchical +TIP interaction network controlling microtubule end tracking of dynein. Nat Cell Biol. 2014;16:804–11. doi: 10.1038/ncb2999. [DOI] [PubMed] [Google Scholar]
- Eckley DM, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the Arp1 minifilament pointed end. J Cell Biol. 1999;147:307–20. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP. Roles of NUDE and NUDF proteins of Aspergillus nidulans: insights from intracellular localization and overexpression effects. Mol Biol Cell. 2003;14:871–88. doi: 10.1091/mbc.E02-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–8. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, McClintock MA, Hollyer IHL, Elliot HL, Reck-Peterson SL. Cytoplasmic dynein is required for the spatial organization of protein aggregates in filamentous fungi. Cell Rep. 2015;11:201–9. doi: 10.1016/j.celrep.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012;197:971–82. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer MJ, Hulihan MM, Kachergus JM, Dachsel JC, Stoessl AJ, et al. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41:163–5. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, et al. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–91. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–93. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Fu MM, Holzbaur EL. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014a;24:564–74. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu MM, Holzbaur EL. MAPK8IP1/JIP1 regulates the trafficking of autophagosomes in neurons. Autophagy. 2014b;10:2079–81. doi: 10.4161/auto.34451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Essex A, Hu JS, Maddox PS, Motegi F, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–99. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Holland AJ, Varma D, Wan X, Civril F, et al. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–71. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390:636–9. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–65. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Garbarino JE, Tan CE, Reck-Peterson SL, Vale RD, Carter AP. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J Biol Chem. 2005;280:23960–5. doi: 10.1074/jbc.M501636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Lee-Eiford A, Mocz G, Phillipson CA, Tang WJ, Gibbons BH. Photosensitized cleavage of dynein heavy chains. Cleavage at the “V1 site” by irradiation at 365 nm in the presence of ATP and vanadate. J Biol Chem. 1987;262:2780–6. [PubMed] [Google Scholar]
- Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1991;115:1639–50. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Heuser J. Structural comparison of purified dynein proteins with in situ dynein arms. J Mol Biol. 1984;180:1083–118. doi: 10.1016/0022-2836(84)90272-9. [DOI] [PubMed] [Google Scholar]
- Greber UF, Way M. A superhighway to virus infection. Cell. 2006;124:741–54. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177:1005–15. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Katrukha EA, Kapitein LC, Hoogenraad CC. New insights into mRNA trafficking in axons. Dev Neurobiol. 2014;74:233–44. doi: 10.1002/dneu.22121. [DOI] [PubMed] [Google Scholar]
- Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141:51–9. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Ori-McKenney KM, Scoto M, Tuck EP, Bell S, et al. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology. 2012;78:1714–20. doi: 10.1212/WNL.0b013e3182556c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol. 2008;182:1063–71. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–6. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, et al. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 2001;20:4041–54. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad CC, Wulf P, Schiefermeier N, Stepanova T, Galjart N, et al. Bicaudal D induces selective dynein-mediated microtubule minus end–directed transport. EMBO J. 2003;22:6004–15. doi: 10.1093/emboj/cdg592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan CP, Hanscom SR, Jolly RS, Futter CE, McCaffrey MW. Rab11-FIP3 binds dynein light intermediate chain 2 and its overexpression fragments the Golgi complex. Biochem Biophys Res Commun. 2010;394:387–92. doi: 10.1016/j.bbrc.2010.03.028. [DOI] [PubMed] [Google Scholar]
- Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150:975–86. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Narita A, Schroer TA, Maeda Y. Two-dimensional averaged images of the dynactin complex revealed by single particle analysis. J Mol Biol. 2006;359:833–9. doi: 10.1016/j.jmb.2006.03.071. [DOI] [PubMed] [Google Scholar]
- Imai H, Narita A, Maeda Y, Schroer TA. Dynactin 3D structure: implications for assembly and dynein binding. J Mol Biol. 2014;426:3262–71. doi: 10.1016/j.jmb.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J, Nicolas E, Compagnon J, Formstecher E, Goud B, Guichet A. Rab6 and the secretory pathway affect oocyte polarity in Drosophila. Development. 2007;134:3419–25. doi: 10.1242/dev.008078. [DOI] [PubMed] [Google Scholar]
- Jha R, Surrey T. Regulation of processive motion and microtubule localization of cytoplasmic dynein. Biochem Soc Trans. 2015;43:48–57. doi: 10.1042/BST20140252. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Illing ME, Kopito RR. Cytoplasmic dynein/dynactin mediates the assembly of aggresomes. Cell Motil Cytoskeleton. 2002;53:26–38. doi: 10.1002/cm.10057. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. PNAS. 2009;106:5669–74. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–11. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- Kelly EE, Horgan CP, McCaffrey MW. Rab11 proteins in health and disease. Biochem Soc Trans. 2012;40:1360–7. doi: 10.1042/BST20120157. [DOI] [PubMed] [Google Scholar]
- Kim MH, Cooper DR, Oleksy A, Devedjiev Y, Derewenda U, et al. The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure. 2004;12:987–98. doi: 10.1016/j.str.2004.03.024. [DOI] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–97. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- Kon T, Imamula K, Roberts AJ, Ohkura R, Knight PJ, et al. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat Struct Mol Biol. 2009;16:325–33. doi: 10.1038/nsmb.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle–dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12:513–19. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43:11266–74. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, et al. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484:345–50. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol. 2011;18:638–42. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Samso M. Overexpression of cytoplasmic dynein’s globular head causes a collapse of the interphase microtubule network in Dictyostelium. Mol Biol Cell. 1996;7:935–48. doi: 10.1091/mbc.7.6.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer H, Phistry M. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmem-brane ligand into multivesicular bodies. J Cell Biol. 1996;133:1205–15. doi: 10.1083/jcb.133.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci. 2013;126:9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C, Vergnolle MA, Thorpe L, Woodman PG, Allan VJ. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J Cell Sci. 2010;123:202–12. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- Lazarus JE, Moughamian AJ, Tokito MK, Holzbaur EL. Dynactin subunit p150Glued is a neuron-specific anti-catastrophe factor. PLOS Biol. 2013;11:e1001611. doi: 10.1371/journal.pbio.1001611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–64. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–86. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee WL, Cooper JA. NudEL targets dynein to microtubule ends through LIS1. Nat Cell Biol. 2005;7:686–90. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kuromi H, Briggs L, Green DB, Rocha JJ, et al. Bicaudal-D binds clathrin heavy chain to promote its transport and augments synaptic vesicle recycling. EMBO J. 2010;29:992–1006. doi: 10.1038/emboj.2009.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, Tokito M, Finklestein JM, Grossman FE, Holzbaur EL. A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J Biol Chem. 2004;279:19201–8. doi: 10.1074/jbc.M313472200. [DOI] [PubMed] [Google Scholar]
- Lipka J, Kuijpers M, Jaworski J, Hoogenraad CC. Mutations in cytoplasmic dynein and its regulators cause malformations of cortical development and neurodegenerative diseases. Biochem Soc Trans. 2013;41:1605–12. doi: 10.1042/BST20130188. [DOI] [PubMed] [Google Scholar]
- Liu Y, Salter HK, Holding AN, Johnson CM, Stephens E, et al. Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev. 2013;27:1233–46. doi: 10.1101/gad.212381.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Steward R, Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat Cell Biol. 2000;2:776–83. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xie T, Steward R. Lis1, the Drosophila homolog of a human lissencephaly disease gene, is required for germline cell division and oocyte differentiation. Development. 1999;126:4477–88. doi: 10.1242/dev.126.20.4477. [DOI] [PubMed] [Google Scholar]
- Lloyd TE, Machamer J, O’Hara K, Kim JH, Collins SE, et al. The p150Glued CAP-Gly domain regulates initiation of retrograde transport at synaptic termini. Neuron. 2012;74:344–60. doi: 10.1016/j.neuron.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockrow JP, Holden KR, Dwivedi A, Matheus MG, Lyons MJ. LIS1 duplication: expanding the phenotype. J Child Neurol. 2012;27:791–95. doi: 10.1177/0883073811425972. [DOI] [PubMed] [Google Scholar]
- Lomakin AJ, Semenova I, Zaliapin I, Kraikivski P, Nadezhdina E, et al. CLIP-170–dependent capture of membrane organelles by microtubules initiates minus-end directed transport. Dev Cell. 2009;17:323–33. doi: 10.1016/j.devcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–82. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur EL. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–17. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Baez L, Cole NB, Kramer H, Donaldson JG. Microtubule-dependent endosomal sorting of clathrin-independent cargo by Hook1. J Cell Biol. 2013;201:233–47. doi: 10.1083/jcb.201208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–52. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- Mallik R, Rai AK, Barak P, Rai A, Kunwar A. Teamwork in microtubule motors. Trends Cell Biol. 2013;23:575–82. doi: 10.1016/j.tcb.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Markus SM, Lee WL. Microtubule-dependent path to the cell cortex for cytoplasmic dynein in mitotic spindle orientation. Bioarchitecture. 2011;1:209–15. doi: 10.4161/bioa.18103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Punch JJ, Lee WL. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–92. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–41. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–14. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J Biol Chem. 2011;286:39615–22. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14:161–76. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- Minke PF, Lee IH, Plamann M. Microscopic analysis of Neurospora ropy mutants defective in nuclear distribution. Fungal Genet Biol. 1999;28:55–67. doi: 10.1006/fgbi.1999.1160. [DOI] [PubMed] [Google Scholar]
- Mizuno N, Toba S, Edamatsu M, Watai-Nishii J, Hirokawa N, et al. Dynein and kinesin share an overlapping microtubule-binding site. EMBO J. 2004;23:2459–67. doi: 10.1038/sj.emboj.7600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler J, Wieschaus EF. Dominant maternal-effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics. 1986;112:803–22. doi: 10.1093/genetics/112.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HM, Youn YH, Pemble H, Yingling J, Wittmann T, Wynshaw-Boris A. LIS1 controls mitosis and mitotic spindle organization via the LIS1-NDEL1-dynein complex. Hum Mol Genet. 2014;23:449–66. doi: 10.1093/hmg/ddt436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton. 2009;66:546–55. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moughamian AJ, Holzbaur EL. Dynactin is required for transport initiation from the distal axon. Neuron. 2012;74:331–43. doi: 10.1016/j.neuron.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moughamian AJ, Osborn GE, Lazarus JE, Maday S, Holzbaur EL. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J Neurosci. 2013;33:13190–203. doi: 10.1523/JNEUROSCI.0935-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Neveling K, Martinez-Carrera LA, Holker I, Heister A, Verrips A, et al. Mutations in BICD2, which encodes a golgin and important motor adaptor, cause congenital autosomal-dominant spinal muscular atrophy. Am J Hum Genet. 2013;92:946–54. doi: 10.1016/j.ajhg.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas MP, Hook P, Brenner S, Wynne CL, Vallee RB, Gennerich A. Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat Commun. 2015;6:6206. doi: 10.1038/ncomms7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura M, Kon T, Shiroguchi K, Ohkura R, Shima T, et al. A single-headed recombinant fragment of Dictyostelium cytoplasmic dynein can drive the robust sliding of microtubules. J Biol Chem. 2004;279:22799–802. doi: 10.1074/jbc.M313362200. [DOI] [PubMed] [Google Scholar]
- Numata N, Shima T, Ohkura R, Kon T, Sutoh K. C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett. 2011;585:1185–90. doi: 10.1016/j.febslet.2011.03.036. [DOI] [PubMed] [Google Scholar]
- Nyarko A, Song Y, Barbar E. Intrinsic disorder in dynein intermediate chain modulates its interactions with NudE and dynactin. J Biol Chem. 2012;287:24884–93. doi: 10.1074/jbc.M112.376038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates EC, Rossor AM, Hafezparast M, Gonzalez M, Speziani F, et al. Mutations in BICD2 cause dominant congenital spinal muscular atrophy and hereditary spastic paraplegia. Am J Hum Genet. 2013;92:965–73. doi: 10.1016/j.ajhg.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey JP, Smith DS. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J Neurosci. 2011;31:17207–19. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Shpetner HS, Vallee RB. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol. 1987;105:1273–82. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330:181–83. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- Peeters K, Litvinenko I, Asselbergh B, Almeida-Souza L, Chamova T, et al. Molecular defects in the motor adaptor BICD2 cause proximal spinal muscular atrophy with autosomal-dominant inheritance. Am J Hum Genet. 2013;92:955–64. doi: 10.1016/j.ajhg.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet. 2013;45:639–47. doi: 10.1038/ng.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Quan J, Nowakowski DW, Hancock ML, Shi J, et al. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell. 2014;158:368–82. doi: 10.1016/j.cell.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–6. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Qiu W, Derr ND, Goodman BS, Villa E, Wu D, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19:193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–48. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwine WB, Hernandez-Lopez R, Zou S, Huang J, Reck-Peterson SL, Leschziner AE. Structural basis for microtubule binding and release by dynein. Science. 2012;337:1532–6. doi: 10.1126/science.1224151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, et al. Isolation of a Miller-Dieker lissencephaly gene containing G protein β–subunit-like repeats. Nature. 1993;364:717–21. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Reiner O, Sapir T. LIS1 functions in normal development and disease. Curr Opin Neurobiol. 2013;23:951–56. doi: 10.1016/j.conb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Goodman BS, Reck-Peterson SL. Reconstitution of dynein transport to the microtubule plus end by kinesin. eLife. 2014;3:e02641. doi: 10.7554/eLife.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Malkova B, Walker ML, Sakakibara H, Numata N, et al. ATP-driven remodeling of the linker domain in the dynein motor. Structure. 2012;20:1670–80. doi: 10.1016/j.str.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136:485–95. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–70. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- Saito T, Hanai S, Takashima S, Nakagawa E, Okazaki S, et al. Neocortical layer formation of human developing brains and lissencephalies: consideration of layer-specific marker expression. Cerebral Cortex. 2011;21:588–96. doi: 10.1093/cercor/bhq125. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Mori D, Toyo-oka K, Chen A, Garrett-Beal L, et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol Cell Biol. 2005;25:7812–27. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, et al. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–96. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–12. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager MA, Hoang HT, Urnavicius L, Bullock SL, Carter AP. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–68. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat Struct Mol Biol. 2012;19:492–97. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Zalyte R, Urnavicius L, Carter AP. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature. 2014;518:435–38. doi: 10.1038/nature14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–79. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–18. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao CY, Zhu J, Xie YJ, Wang Z, Wang YN, et al. Distinct functions of nuclear distribution proteins LIS1, Ndel1 and NudCL in regulating axonal mitochondrial transport. Traffic. 2013;14:785–97. doi: 10.1111/tra.12070. [DOI] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, et al. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–72. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- Shima T, Kon T, Imamula K, Ohkura R, Sutoh K. Two modes of microtubule sliding driven by cytoplasmic dynein. PNAS. 2006;103:17736–40. doi: 10.1073/pnas.0606794103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siglin AE, Sun S, Moore JK, Tan S, Poenie M, et al. Dynein and dynactin leverage their bivalent character to form a high-affinity interaction. PLOS ONE. 2013;8:e59453. doi: 10.1371/journal.pone.0059453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvanovich A, Li MG, Serr M, Mische S, Hays TS. The third P-loop domain in cytoplasmic dynein heavy chain is essential for dynein motor function and ATP-sensitive microtubule binding. Mol Biol Cell. 2003;14:1355–65. doi: 10.1091/mbc.E02-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Martin S, Evans TM, Kerr M, James DE, et al. A novel hook-related protein family and the characterization of hook-related protein 1. Traffic. 2005;6:442–58. doi: 10.1111/j.1600-0854.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Sitaram P, Anderson MA, Jodoin JN, Lee E, Lee LA. Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis. Development. 2012;139:2945–54. doi: 10.1242/dev.077511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, et al. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–75. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Splinter D, Razafsky DS, Schlager MA, Serra-Marques A, Grigoriev I, et al. BICD2, dynactin, and LIS1 cooperate in regulating dynein recruitment to cellular structures. Mol Biol Cell. 2012;23:4226–41. doi: 10.1091/mbc.E12-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLOS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Hall B, Yu R, Hashim AI, Kramer H. Hook2 localizes to the centrosome, binds directly to centriolin/CEP110 and contributes to centrosomal function. Traffic. 2007a;8:32–46. doi: 10.1111/j.1600-0854.2006.00511.x. [DOI] [PubMed] [Google Scholar]
- Szebenyi G, Wigley WC, Hall B, Didier A, Yu M, et al. Hook2 contributes to aggresome formation. BMC Cell Biol. 2007b;8:19. doi: 10.1186/1471-2121-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–68. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarricone C, Perrina F, Monzani S, Massimiliano L, Kim MH, et al. Coupling PAF signaling to dynein regulation: structure of LIS1 in complex with PAF-acetylhydrolase. Neuron. 2004;44:809–21. doi: 10.1016/j.neuron.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Overlapping hand-overhand mechanism of single molecular motility of cytoplasmic dynein. PNAS. 2006;103:5741–45. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisawa T, Ichikawa M, Furuta A, Saito K, Oiwa K, et al. Autoinhibition and cooperative activation mechanisms of cytoplasmic dynein. Nat Cell Biol. 2014;16:1118–24. doi: 10.1038/ncb3048. [DOI] [PubMed] [Google Scholar]
- Toropova K, Zou S, Roberts AJ, Redwine WB, Goodman BS, et al. Lis1 regulates dynein by sterically blocking its mechanochemical cycle. eLife. 2014;3:e03372. doi: 10.7554/eLife.03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathy SK, Weil SJ, Chen C, Anand P, Vallee RB, Gross SP. Autoregulatory mechanism for dynactin control of processive and diffusive dynein transport. Nat Cell Biol. 2014;16:1192–201. doi: 10.1038/ncb3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–79. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, et al. The structure of the dynactin complex and its interaction with dynein. Science. 2015;347:1441–46. doi: 10.1126/science.aaa4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–80. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA. The way things move: looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Vallee RB, Tsai JW. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006;20:1384–93. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–16. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150Glued to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–19. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarino-Guell C, Wider C, Soto-Ortolaza AI, Cobb SA, Kachergus JM, et al. Characterization of DCTN1 genetic variability in neurodegeneration. Neurology. 2009;72:2024–28. doi: 10.1212/WNL.0b013e3181a92c4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta JH, Didier AJ, Liu X, Kramer H. The Golgi-associated Hook3 protein is a member of a novel family of microtubule-binding proteins. J Cell Biol. 2001;152:923–34. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ketcham SA, Schon A, Goodman B, Wang Y, et al. Nudel/NudE and Lis1 promote dynein and dynactin interaction in the context of spindle morphogenesis. Mol Biol Cell. 2013;24:3522–33. doi: 10.1091/mbc.E13-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng Y. Identification of a novel dynein binding domain in Nudel essential for spindle pole organization in Xenopus egg extract. J Biol Chem. 2011;286:587–93. doi: 10.1074/jbc.M110.181578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jin F, Higgins R, McKnight K. The current view for the silencing of the spindle assembly checkpoint. Cell Cycle. 2014;13:1694–701. doi: 10.4161/cc.29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Khan S, Sheetz MP. Single cytoplasmic dynein molecule movements: characterization and comparison with kinesin. Biophys J. 1995;69:2011–23. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Hastings R, Caswell R, Xie W, Paszkiewicz K, et al. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet. 2011;89:308–12. doi: 10.1016/j.ajhg.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. Dyneins across eukaryotes: a comparative genomic analysis. Traffic. 2007;8:1708–21. doi: 10.1111/j.1600-0854.2007.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and LIS1: insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T. Bioinformatic approaches to dynein heavy chain classification. Methods Cell Biol. 2009;92:1–9. doi: 10.1016/S0091-679X(08)92001-X. [DOI] [PubMed] [Google Scholar]
- Yamada M, Hirotsune S, Wynshaw-Boris A. The essential role of LIS1, NDEL1 and Aurora-A in polarity formation and microtubule organization during neurogensis. Cell Adh Migr. 2010;4:180–84. doi: 10.4161/cam.4.2.10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, et al. LIS1 and NDEL1 coordinate the plus-end–directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–83. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TY, Kowalska AK, Scipioni BR, Cheong FK, Zheng M, et al. Dynactin helps target Polo-like kinase 1 to kinetochores via its left-handed β-helical p27 subunit. EMBO J. 2013;32:1023–35. doi: 10.1038/emboj.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin’s pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–37. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP, Vallee RB. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, et al. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–86. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn YH, Pramparo T, Hirotsune S, Wynshaw-Boris A. Distinct dose-dependent cortical neuronal migration and neurite extension defects in Lis1 and Ndel1 mutant mice. J Neurosci. 2009;29:15520–30. doi: 10.1523/JNEUROSCI.4630-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Fischer R, Xiang X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell. 2003;14:1479–88. doi: 10.1091/mbc.E02-08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Qiu R, Arst HN, Jr, Penalva MA, Xiang X. HookA is a novel dynein–early endosome linker critical for cargo movement in vivo. J Cell Biol. 2014;204:1009–26. doi: 10.1083/jcb.201308009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Peñalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein–early endosome interaction. J Cell Biol. 2011;193:1245–55. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhuang L, Lee Y, Abenza JF, Penalva MA, Xiang X. The microtubule plus-end localization of Aspergillus dynein is important for dynein–early-endosome interaction but not for dynein ATPase activation. J Cell Sci. 2010;123:3596–604. doi: 10.1242/jcs.075259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylkiewicz E, Kijanska M, Choi WC, Derewenda U, Derewenda ZS, Stukenberg PT. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J Cell Biol. 2011;192:433–45. doi: 10.1083/jcb.201011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


