Abstract
A key challenge to the clinical implementation of decellularized scaffold-based tissue engineering lies in understanding the process of removing cells and immunogenic material from a donor tissue/organ while maintaining the biochemical and biophysical properties of the scaffold that will promote growth of newly seeded cells. Current criteria for evaluating whole organ decellularization are primarily based on nucleic acids, as they are easy to quantify and have been directly correlated to adverse host responses. However, numerous proteins cause immunogenic responses and thus should be measured directly to further understand and quantify the efficacy of decellularization. In addition, there has been increasing appreciation for the role of the various protein components of the extracellular matrix (ECM) in directing cell growth and regulating organ function. We performed in-depth proteomic analysis on four types of biological scaffolds and identified a large number of both remnant cellular and ECM proteins. Measurements of individual protein abundances during the decellularization process revealed significant removal of numerous cellular proteins, but preservation of most structural matrix proteins. The observation that decellularized scaffolds still contain many cellular proteins, although at decreased abundance, indicates that elimination of DNA does not assure adequate removal of all cellular material. Thus, proteomic analysis provides crucial characterization of the decellularization process to create biological scaffolds for future tissue/organ replacement therapies.
Keywords: mass spectrometry, proteomics, matrisome, Matrigel, decellularized rat liver, decellularized human lung
Introduction
A promising tissue engineering approach for functional organ replacement involves using synthetic or biological scaffolds, within which seeded cells, potentially from the transplant recipient, grow to form new tissues. In contrast to synthetic scaffolds, biological scaffolds are composed of naturally occurring extracellular matrix (ECM) and retain much of the correct anatomical structure, native ECM architecture, and biomechanical properties, as well as many of the cell adhesion ligands found in living tissues. These beneficial characteristics also modulate the immunological response, and thus biological scaffolds have been widely used in various cell culture and tissue reconstruction applications. These scaffolds range from simple purified proteins, to extracted protein mixtures, to very complex decellularized whole organs. A key challenge to achieving the ultimate therapeutic goal – creation of a functional organ – lies in understanding the process of removing cells and immunogenic material from a donor organ while maintaining the biochemical and biophysical properties of the scaffold that will promote growth of newly seeded cells. In the present study, we employ mass spectrometry-based proteomics to extract crucial information regarding the ECM proteins of the scaffold as well as remnant cellular proteins.
Three types of biological scaffolds of increasing complexity – collagen, Matrigel, and decellularized organs – were studied in this work. Purified collagen is often employed in gel form as a very simple biological matrix, providing structure but limited molecular heterogeneity. Type I collagen gel has long been used in studies of cell proliferation and differentiation to sustain continuous cell growth [1, 2], or to promote expression of cell-specific morphology and function [3, 4]. It has also been used for the in vitro reconstruction of various tissues such as thyroid follicles [5], cornea [6], and urinary bladder mucosa [7]. Another commonly used biological matrix is Matrigel, a mixture of basement membrane proteins extracted from the Engelbreth-Holm-Swarm mouse tumor. It is often used to mimic the ECM in cancer research and in stem cell studies due to its ability to maintain stem cells in an undifferentiated state. Matrigel, either alone [8-10] or in combination with collagen gel [11-13], has been shown to recapitulate certain tissue-like structures. The most sophisticated biological scaffolds are decellularized whole tissues or organs. Such decellularized scaffolds retain the organ’s macroscopic three-dimensional architecture and organ-specific cues that can support the growth of cells to reconstruct the original organ type [14]. After seeding these allogeneic or xenogeneic acellular scaffolds with organ-matched cell populations, researchers have created partially functional replacements for key organs including heart [15], liver [16], lung [17-19], and kidney [20]. These preclinical studies demonstrate the potential of this strategy to transform human medicine. A common characteristic of these naturally occurring scaffolds is their ability to support cell growth and even tissue regeneration. Although not yet well understood, it is very likely that both tissue structure and composition contribute to this capability. Thus, a deeper knowledge of the composition of these scaffolds is expected to have high value in elucidating the mechanisms that drive their functional capacity.
It is crucial to evaluate the effectiveness of the decellularization process in terms of removing antigenic cellular materials while also preserving the composition and biomechanical properties of the ECM. The composition of biological scaffolds varies depending on their source, as well as the processes employed to decellularize them. Current criteria for evaluating whole organ decellularization are based primarily on nucleic acids, as they are easy to quantify and have been directly correlated to adverse host responses [21]. However, other cellular materials, such as proteins, are likely culprits in eliciting immunogenic responses [22-24], and therefore additional analyses, such as proteomics, will greatly enhance understanding of the decellularization process and provide improved molecular readout to guide engineering efforts.
ECM proteomics is attracting increased attention due to the crucial role of the ECM in cell development and important physiological and pathological processes [25-27]. ECM proteins have been notoriously difficult to analyze because of their covalent crosslinking and numerous modifications [25, 28]. Researchers have developed various protein extraction strategies and have performed extensive fractionation at the peptide and/or protein level, in order to obtain deeper coverage of ECM proteins [25, 29-35]. When evaluating scaffolds for tissue engineering applications, however, the identification of remnant cellular proteins is of equal importance to the analysis of ECM proteins. After comparing different sample preparation strategies, filter-aided sample preparation (FASP) [36] stood out as the one that provided in-depth coverage for both cellular and ECM proteins, making it an appealing choice for evaluating decellularized scaffolds. A further benefit of the FASP method is that it efficiently removes the sodium dodecyl sulfate (SDS) routinely employed for tissue/organ decellularization, as SDS is detrimental to MS-based proteomics. This concomitant benefit of FASP is especially important in evaluating tissues that are sampled during the perfusion process (such as the partially-decellularized rat livers presented in this work) or ones that have not gone through extensive washing after SDS perfusion.
We employed the FASP method of protein extraction and digestion to analyze four types of biological scaffolds: rat-tail type I collagen, growth factor reduced (GFR)-Matrigel, decellularized rat livers, and decellularized human lungs. We identified both remnant cellular proteins, which may be unfavorable upon transplantation, and a set of ECM proteins that are potentially crucial determinants of constructive tissue remodeling. Furthermore, we characterized the early- versus late-stage of the decellularization process in the rat liver model by measuring protein abundances for naïve, partially-decellularized, and “fully” decellularized samples. Our study reports, for the first time, an in-depth examination of the proteome of partially functional decellularized organs and a quantitative tracking of specific protein losses during the decellularization process. The findings advance understanding of biological scaffolds and underscore the need for incorporating proteomic techniques into tissue engineering research.
Materials and Methods
Tissue Acquisition and Decellularization
All animal care, handling and surgical procedures were performed in accordance with the guidelines set by the Institutional Animal Care and Use Committee (IACUC) at Massachusetts General Hospital. Livers were harvested from female Lewis rats (Charles River Laboratories, Wilmington, MA) weighing 175–200 g, gently flushed with and stored in 0.9% NaCl at −80 °C until utilized for decellularization. Organ decellularization was performed as previously described [37]. Briefly, the liver was thawed at room temperature and washed by portal vein perfusion with PBS overnight at a flow rate of 1.0 mL/min to clear residual blood from the organ. Then, the liver was perfused with 0.01% (w/v) SDS (Sigma-Aldrich, St. Louis, MO) for increasing durations of 5, 10, 15 and 20 min with 60 min PBS washes in between. After the last PBS wash, the liver was perfused with 0.01% SDS for 24 h followed by perfusion with 0.1% (w/v) SDS for 24 h, and 0.2% (w/v) SDS for 2 h. Next, the liver was perfused with 0.1% (w/v) Triton X-100 (Sigma-Aldrich, St. Louis, MO) for 30 min to remove the residual components and rinsed with PBS for at least 2 h. Tissue biopsies (50–100 mg) were taken at three separate time points during this process: 1) immediately after harvest (naïve), 2) after the 24 h wash with 0.01% SDS (partially decellularized), and 3) at the end of the final PBS wash (decellularized). These liver tissues were flash frozen for DNA, total collagen (see Supplemental Notes) and proteomic analyses.
Human lungs were obtained from the International Institute for the Advancement of Medicine (IIAM); they were recovered under sterile conditions within 60 min of cessation of cardiovascular circulation, but were otherwise unsuitable for transplantation. Perfusion decellularization was performed according to previously published methodology [38, 39].
DNA Measurement
Genomic DNA from rat liver samples (naïve, partially decellularized and decellularized) was extracted using the PureLink® Genomic DNA Kit (Life Technologies, Grand Island, NY). The DNA concentration in the extracts was measured using the PicoGreen® Assay Kit (Life Technologies, Grand Island, NY).
Protein Extraction and Digestion
For rat-tail type I collagen and GFR-Matrigel (both from BD Bioscience, San Jose, CA), aliquots of approximately 200 μg of protein were added to 150 μL of SDT solution—4% SDS, 0.1 M Tris-HCl (pH 7.6) and 0.1 M dithiothreitol (DTT) (all reagents from Sigma-Aldrich, St. Louis, MO). For decellularized or naïve tissues, aliquots of approximately 15 mg of tissue were washed with ice-cold PBS solution (Invitrogen, Grand Island, NY) and ground with disposable pellet pestles (Kimble Chase Kontes, Vineland, NJ) for 1 min in 1.5-mL tubes, followed by addition of 150 μL SDT solution. Samples were then heated at 95 °C for 7 min. and sonicated on ice with a probe sonicator (Misonix XL2015, Misonix microtip PN/418, Farmingdale, NY)—alternating 20 seconds on and 20 seconds off for 6 min, followed by centrifugation at 22 °C for 5 min. at 16,100 g.
The FASP protocol was used for SDS removal and on-filter digestion [36, 40]. Briefly, a 30 μL aliquot of the sample supernatant was mixed with 200 μL of 8M urea/0.1 M Tris buffer pH 8.0 in a 30K MW Vivacon 500 filter (Sartorius, Bohemia, NY). Note that even small proteins (<10 kDa) are retained in this 30K filter because they are denatured/unfolded by the SDT solution [41]. The sample was washed, alkylated with iodoacetamide, washed further, then digested with trypsin (Promega, Madison, WI; protein:enzyme ratio of 50:1) overnight at 37 °C, and the digested peptides were collected by centrifugation. After the digestion was quenched with 10% trifluoroacetic acid (TFA) to a final concentration of 0.5% TFA, samples were desalted using Sep-Pak C18 1 cc Vac Cartridges (Waters, Milford, MA). The cartridge was wetted with 2×200 μL acetonitrile (ACN) and equilibrated with 150 μL H2O. Tryptic peptides were loaded onto the sorbent, and then washed with 2×100 μL 0.1% TFA and 50 μL 0.1% formic acid (FA). Finally, peptides were eluted with 2×100 μL of 75% ACN, 0.1% FA. The eluent was dried down using a Savant SpeedVac (Thermo Scientific, Waltham, MA) and reconstituted in 5% ACN, 2% FA.
High-pH Fractionation
As noted in the Results section, certain digested peptide samples were fractionated prior to LC-MS. In such cases, Sep-Pak desalting was skipped, and the quenched digests were subjected to high-pH fractionation on an HPLC system (Shimadzu, Columbia, MD) using a C18 Gemini 3μ, 110Å, 3.0x150mm column (Phenomenex, Torrance, CA). Mobile phase A was aqueous 20 mM ammonium formate and mobile phase B was 20 mM ammonium formate in 70% ACN; the gradient of 0–100% mobile phase B occurred over 20 min. The HPLC flow rate was 1 mL/min and the eluent was collected into 6 fractions, each of which was evaporated to dryness in the SpeedVac and reconstituted in 5% ACN, 2% FA.
Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
Approximately 0.6 μg protein digest, estimated by BCA protein assay, was injected into a Waters nanoAcquity HPLC coupled to an ESI ion-trap/Orbitrap mass spectrometer (LTQ Orbitrap Velos, Thermo Scientific, Waltham, MA). Peptides were separated on a 100 μm inner diameter column packed with 20 cm of 3 μm MAGIC aqC18 beads (Bruker-Michrom, Auburn, CA), and eluted at 0.3 μL/min in 0.1% FA with a gradient of increasing ACN over 2.5 h. As noted in the Results section, certain samples were separated on a column packed with 20 cm of 1.7 μm BEH C18 particles (Waters, Milford, MA), and a heater cartridge was used to keep the capillary column at 60 °C. A full-mass scan (300-1500 m/z) was performed in the Orbitrap at a resolution of 60,000. The ten most intense peaks were selected for fragmentation by higher-energy collisional dissociation (HCD) at 42% collision energy, then analyzed with a resolution of 7500 and an isolation width of 2.5 m/z. Dynamic exclusion was enabled with a repeat count of 1 over 30 s and an exclusion duration of 120 s. All MS raw files may be downloaded from the PeptideAtlas data repository [42] by the following link: http://www.peptideatlas.org/PASS/PASS00557.
Proteomic Data Analysis
The acquired raw files were analyzed by MaxQuant [43, 44] (version 1.4.1.2). The resulting peak lists were searched with Andromeda [45] against UniProt canonical protein databases for the appropriate organism (Rattus norvegicus: 7853 reviewed sequences downloaded on May 4, 2013 for rat-tail collagen search, and 33,607 sequences (reviewed plus unreviewed) downloaded on May 29, 2014 for rat liver searches; Mus musculus: 16,642 reviewed sequences downloaded on December 16, 2013; Homo sapiens: 20,278 reviewed sequences downloaded on December 5, 2013), each supplemented with 247 common contaminants (Supplemental Table S1). Precursor and fragment ion mass tolerances were set to 4.5 ppm and 20 ppm, respectively. Static cysteine carbamidomethylation (+57.0215 Da) and up to 5 variable methionine and proline oxidations (+15.9949 Da) were specified. The modification parameters differed somewhat for the collagen gel search: oxidation of lysine (+15.9949 Da) and glycosylation of lysine (monosaccharide, +178.0473 Da, disaccharide, +340.0995 Da) were also included as variable modifications, and up to 7 variable modifications were allowed. A false discovery rate of 1% at both the peptide and the protein level was allowed. Up to two missed cleavages were allowed and a minimum of two unique peptides per protein was required. Protein groups containing matches to proteins from the reversed database or contaminants were discarded. Intensity-based absolute quantification (iBAQ) and label-free quantification (LFQ) algorithms embedded in the MaxQuant software package were employed. Only unique and razor peptides were used for quantification and a minimum count of two was required. We multiplied the LFQ intensity of the part-decell and the naïve sample by correction factors of 10.3 and 28.4, respectively, based on tissue wet and dry weight information (see Supplemental Notes for details of the calculation). Perseus software (version 1.5.0.15) was used for downstream statistical analyses. Proteins were filtered by requiring at least two valid values in at least one sample condition. The corrected intensities were log2 transformed and missing values were replaced using data imputation by employing a width of 0.3 and a downshift of 1.2.
Western Blot Analysis
Protein from rat liver tissue was extracted with 150 μL SDT solution containing an aliquot of 1.5 μL Calbiochem Protease Inhibitor Cocktail Set I (Merck Millipore, Billerica, MA). The protein extract was incubated at 95 °C and sonicated as described above. A NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE) was used to estimate protein concentration. Reducing SDS-PAGE was performed using a pre-cast 4-20% acrylamide gel (Bio-Rad, Hercules, CA) with 142 μg, 52 μg, and 5 μg total protein load for the naïve, part-decell, and decell samples, respectively; these relative amounts were based on the correction factors reported above. Following transfer, polyvinylidene fluoride membranes were blotted using the following primary antibodies: rabbit anti-fibronectin (1:1500); mouse anti-GAPDH (1:20,000) (both from Sigma Aldrich); goat anti-serpina3k (1:200; Santa Cruz Biotechnology, Dallas, TX); and rabbit anti-filamin A (1:1000; Cell Signaling Technology, Beverly, MA). Blots were detected using the Clarity western ECL substrate kit (Bio-Rad) with relevant horseradish peroxidase (HRP)-conjugated anti-mouse (1:20,000), anti-rabbit (1:20,000) (both from Bio-Rad), and anti-goat (1:5,000) (Santa Cruz Biotechnology) IgG secondary antibodies, according to the manufacturer’s instructions. An ImageQuant LAS 4000 mini system (GE Healthcare, Pittsburgh, PA) was used to produce digital images of the chemiluminescent membranes.
Statistical Analysis
Gene Ontology (GO) enrichment analyses were performed using BiNGO 2.44 (hypergeometric model and Benjamini-Hochberg (BH) correction) [46]. Two-sample t-tests with BH correction were performed to quantitatively compare the LFQ values of the naïve and decellularized rat liver samples.
Results
We performed proteomic analysis on four types of biological scaffolds. The first two types, collagen I gel and GFR-Matrigel, are relatively less complex than decellularized organs and thus were employed to test the proteomics workflow. This workflow, in brief, was as follows: samples were extracted with an SDS/DTT/Tris buffer at 95 °C, sonicated, subjected to FASP to remove detergent and digest the proteins into peptides, and finally analyzed by LC-MS/MS. After success with these simpler scaffolds, we quantitatively compared the proteins found in decellularized, partially-decellularized, and naïve rat livers. The last type of biological scaffold studied, decellularized human lungs, demonstrated the applicability of the method to a different tissue type.
Rat-Tail Type I Collagen
Type I collagen (collagen I) is one of the primary constituents of the ECM. An important analytical consideration is that collagen I is comprised of a large fraction of proline residues (~18%), which are frequently posttranslationally modified by prolyl 4-hydroxylase to generate hydroxyproline residues that increase the stability of the collagen triple helix [47]. It is thus crucial to include proline hydroxylation (oxidation +15.9949 Da) as a variable modification during proteomic database searching. In addition, hydroxylation of lysine and glycosylation (mono- and disaccharide) of hydroxylysine were included as variable modifications, as they are known to occur on collagen I.
Proteomic analysis of rat-tail type I collagen yielded numerous unique peptides and high sequence coverage (~75%) for collagen I alpha-1 and alpha-2 chains, as shown in Table 1. The ~25% of undetected protein sequences for these collagen chains were mostly in the signal and propeptide regions. These results are superior to those reported in previous collagen proteomic studies [25, 48], indicating that the FASP method is suitable for proteomics of collagenous samples.
Table 1.
Proteomic results for rat-tail type I collagen (n=3 biological replicates; mean±standard deviation; see Supplemental Table S2 for the full protein list).
| Protein description | Unique peptides |
Sequence coverage (%) |
iBAQ (%) |
|---|---|---|---|
| Collagen I alpha-2 chain | 89±8 | 73.4±3.6 | 29.8±11.2 |
| Collagen I alpha-1 chain | 91±1 | 76.5±1.2 | 22.2±9.4 |
| Serum albumin | 28±3 | 48.8±2.1 | 15.7±2.6 |
| Fibromodulin | 7±1 | 20.9±4.5 | 8.5±1.7 |
| Decorin | 9±1 | 23.7±4.3 | 7.8±1.9 |
| 71 other protein groups | / | / | 16.0±4.7 |
Numerous other proteins were identified in this rat-tail type I collagen sample, and their relative abundances were measured by intensity-based absolute quantification (iBAQ) using the MaxQuant software package [49]. iBAQ values are normalized by the number of theoretical peptides for each protein, which allows for comparison of different proteins within a single sample [50]. The most abundant proteins apart from collagen I include serum albumin, fibromodulin, and decorin (Table 1). Note that the iBAQ% values for the most abundant proteins (e.g. collagen I) are an underestimate, as our mass spectrometry settings facilitate the observation of lower-abundance proteins. Although tendons have limited blood supply, some plasma proteins were relatively abundant among the 74 co-extracted proteins, including serum albumin, hemoglobin, and serotransferrin, which is reportedly able to stimulate cell proliferation [51]. Several small leucine-rich proteoglycans were present, including fibromodulin, decorin, prolargin, and biglycan, which associates with fibrillar collagens to help regulate collagen fibrillogenesis [52]. Procollagen C-endopeptidase enhancer, known to be highly expressed in rat-tail tendon [53], was also co-extracted. Additional identified proteins included vimentin, which associates with and stabilizes collagen I mRNAs, thus contributing to high levels of collagen expression [54]; fibronectin and thrombospondin-4, two glycoproteins that contribute to cell proliferation, migration and adhesion [55, 56]; and complement component C9, a component of the membrane attack complex that plays a key role in the innate and adaptive immune response by forming pores in the plasma membrane of target cells [57]. Various muscle proteins and proteases were also identified. Besides collagen I, other fibril-forming collagens were present, including collagens type II, III, V, and XI, as well as collagen chaperone protein SERPINH1 at low abundance.
Growth Factor Reduced (GFR) Matrigel
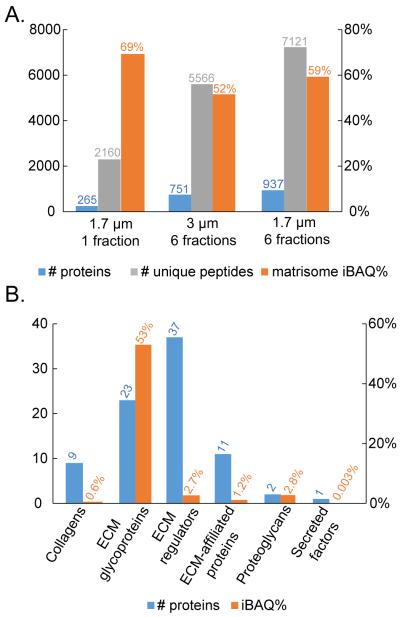
Given the increased proteomic complexity of the GFR-Matrigel sample, as compared to the type I collagen sample, we chose to examine the utility of enhanced separation conditions. We optionally employed high-pH reverse-phase HPLC for offline fractionation of the peptides and also compared 1.7 versus 3.0 μm particle sizes for the low-pH online reverse-phase HPLC-MS analysis. The proteomic results from this comparison are given in Figure 1A. Fractionation led to a greater than 3-fold increase in the number of proteins and unique peptides. This deep proteomic coverage may provide understanding of less abundant but important components of biological scaffolds, but it comes at a cost of 6X increased instrument time. The smaller 1.7 μm packing particles also increased the number of protein identifications (by about 25%). With both the fractionation step and the smaller particle size column, a total of 937 proteins were identified among the three replicates (Supplemental Table S3).
Figure 1.
Proteomic results for GFR-Matrigel samples. A) Numbers of proteins and unique peptides identified, along with the percentage of iBAQ intensity (iBAQ%) arising from matrisome proteins. B) Under the condition of 1.7 μm column and 6 peptide fractions, the number of proteins and the percentage of iBAQ intensity for each matrisome subcategory. n=3 biological replicates.
The importance of matrix structure and function to biological scaffolds led us to employ a recently developed categorization approach offered by Naba et al. called the “matrisome”, which encompasses ECM and ECM-associated proteins [58]. The current mouse matrisome inventory consists of 1122 protein entries (note that Matrigel is derived from mouse). The percentage of iBAQ intensity attributed to matrisome proteins is shown in Figure 1A for each separation condition. The decrease in the percentage of matrisome protein intensity after fractionation may reflect improved detection of low-abundance cellular (i.e. non-matrisome) proteins.
Among the 937 proteins identified in GFR-Matrigel, the most abundant ones (based on iBAQ values) were laminin, nidogen, vimentin, basement membrane-specific heparan sulfate proteoglycan core protein, and fibrinogen. Numerous cellular proteins were observed, which is in agreement with previous report [31]. A total of 83 proteins were assigned to the matrisome category and their qualitative and quantitative distribution among subcategories is depicted in Figure 1B. The GFR-Matrigel matrisome was composed primarily of abundant ECM glycoproteins, which possess domains and motifs that promote cell adhesion and form interactions involving ECM assembly [59]. There also existed a large number of low-abundance ECM regulators, which are important for matrix modification and remodeling.
Naïve, Partially-decellularized and Decellularized Rat Livers
With encouraging proteomic results from collagen and Matrigel, we next applied the proteomic approach to analysis of decellularized rat livers. Three biological replicates for each of three conditions (naïve, part-decell, and decell) were analyzed. For this study, peptides were not pre-fractionated off-line, but they were on-line separated using the more efficient chromatographic condition (1.7 μm particle size at 60 °C) prior to MS/MS analysis.
Proteomic results obtained for the three sample conditions are shown in Figure 2. As expected, fewer distinct proteins were detected as decellularization progressed, dropping from 1019 to 854 and then 517 protein identifications in the naïve, part-decell and decell conditions (Supplemental Table S4), respectively. For a closer look at ECM composition, we again employed the matrisome/non-matrisome categorization, but in this case we first needed to generate a list of rat matrisome proteins, since curated matrisome inventories currently only exist for mouse and human. It is not immediately possible to create a completely accurate rat matrisome database by the methodology previously used for mouse and human because the rat genome sequence is less complete than that of the other two species. Therefore, as a reasonable approximation, we generated a matrisome protein list for rat by combining those previously curated for human (1062 entries) and mouse (1122 entries) [58]. The resulting 1284 gene symbols (900 were shared) were used to categorize matrisome proteins in our rat liver proteomic results. The number of identified matrisome proteins was comparable for the naïve livers and the part-decell livers (32 and 33 proteins, respectively), but increased substantially to 58 proteins for the decell livers. Nonetheless, the “fully” decellularized rat liver proteome was still dominated by non-matrisome or cellular proteins, as shown in Figure 2.
Figure 2.
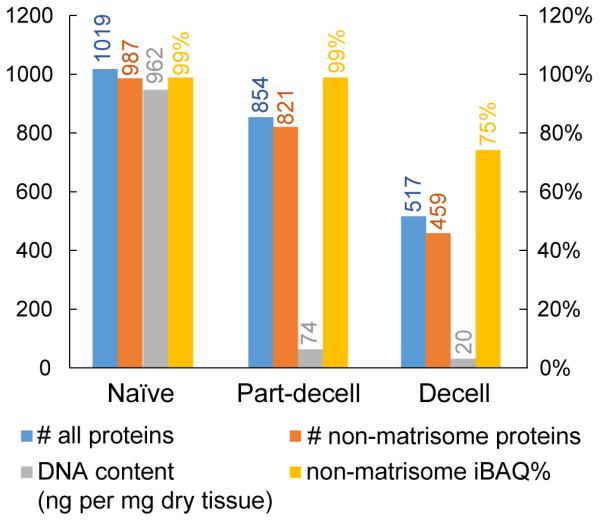
Proteomic results and DNA content for the naïve, partially-decellularized (part-decell), and decellularized (decell) rat livers.
DNA quantification, a classic assay for evaluating decellularization effectiveness, was also performed for each sample condition, and the results are given in Figure 2. The DNA content dropped dramatically in the first stage of decellularization, from 962 to 74 ng per mg of tissue (dry weight), and then decreased further to 20 ng/mg for the decell samples. This value for the decell liver satisfies one of the traditional decellularization criteria—less than 50 ng DNA per mg of dry tissue weight [21]. These DNA results contrast drastically with the proteomic results where the non-matrisome iBAQ% remained at 99% for the part-decell and 75% for the decell samples, indicating that the majority of the protein content remaining after decellularization is not related to the ECM.
A total of 58 matrisome proteins (Supplemental Table S4) were identified in at least two biological replicates of the decellularized livers: the most abundant was collagen VI, followed by transglutaminase 2, biglycan, fibrinogen, and fibronectin. Collagen VI is a network-forming collagen that anchors interstitial structures such as blood vessels and collagen fibers into surrounding connective tissues [60]. It has been demonstrated to promote mesenchymal cell proliferation and is also a potent inhibitor of apoptosis [61], making it an important component of biological scaffolds used for tissue reconstruction. The other 14 collagen forms identified included: fibrillar collagens I, II, III, and V, which contribute tensile strength; network-forming collagen IV and multiplexin collagen XVIII, which anchor and stabilize other basement membrane components; and fibril-associated collagens XII, XIV, and XVI, which regulate fibrillar collagen growth [62]. Several ECM glycoproteins were observed, including fibrinogen, fibronectin, dermatopontin, laminin, vitronectin, and elastin. A number of proteoglycans, ECM-affiliated proteins, ECM regulators, and secreted factors were also identified. These matrisome proteins together drive the enrichment of Gene Ontology (GO) terms that are favorable for subsequent cell seeding and tissue regeneration, such as the terms: protein complex assembly, liver development, extracellular matrix organization, and cell-substrate adhesion (corrected p-values = 4.12×10−8, 1.01×10−5, 1.72×10−4, and 4.43×10−3, respectively).
Conversely, the 459 identified cellular proteins included those that are unfavorable, such as the previously reported xenoantigens serum albumin [23], hemoglobin alpha-1 chain [22], and peroxiredoxin 1 [63-65], as well as tyrosine-protein kinase Lyn, a protein with an inflammatory response function [66]. The detection of a large number of remnant cellular proteins indicates that the decellularized rat livers are not as free of cellular material as suggested by the DNA assay. The presence of these proteins, along with the possibility of other intracellular molecules, may provide clues to explain why some decellularized scaffolds contribute to tissue/organ rejection upon in vivo transplantation.
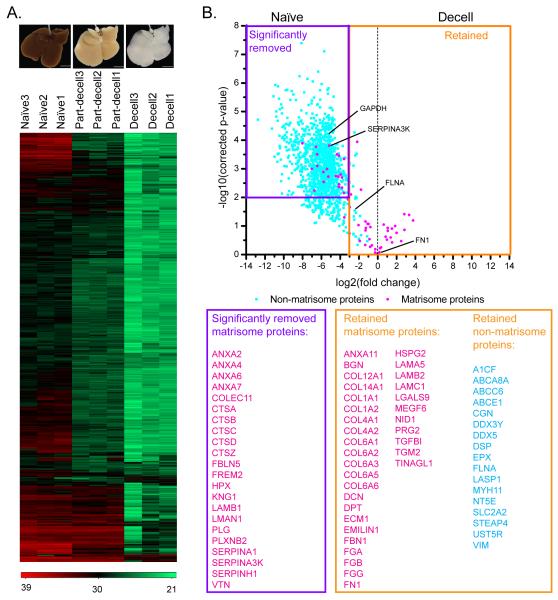
To quantitatively characterize differential protein removal and preservation during the decellularization process, we performed label-free quantification based on the LFQ intensity generated by the MaxQuant software. The heatmap in Figure 3A shows the abundance change of each protein in each biological sample. The majority of proteins decreased in abundance as decellularization progressed, but a number did not (rows at the bottom of the heatmap). Figure 3B includes a volcano plot showing fold change in protein abundance (decell versus naïve) plotted against statistical p-value. The purple rectangle denotes cutoff criteria for protein underrepresentation (fold change < 1/8; p-value < 0.01) in the decellularized sample, i.e., proteins that were significantly removed after decellularization. The orange rectangle denotes proteins that were retained after decellularization (fold change > 1/8 compared to naïve). A number of ECM regulators and ECM-affiliated proteins were significantly depleted, such as cathepsins B, D, and Z, serpins A1, A3K, and H1, and annexins 2, 4, 6, and 7. However, the “core-matrisome,” which is critical to overall scaffold structure and facilitates cell attachment and growth, was well preserved as indicated by retention of: collagens I, IV, VI, XII, and XIV; ECM glycoproteins including fibrinogen, dermatopontin, and fibronectin; as well as proteoglycans including biglycan and decorin. Seventeen non-matrisome, or cellular, proteins were retained in the decellularized samples, including: filamin A, a large (280 kDa) protein that binds to numerous proteins such as transmembrane receptors and signaling molecules [67]; LASP1, an actin-binding protein that may play a role in cytoskeletal organization [68]; vimentin, an intermediate filament protein that anchors the organelles in the cytosol and regulates focal contact [69]; as well as a few other membrane and ATP-binding-related proteins.
Figure 3.
Quantitative proteomic results for rat livers. (A) Images of naïve, part-decell, and decell livers (scale bar: 10 mm), along with a heatmap showing changes in protein abundances for these three conditions. Numbers in the color code are log-transformed-corrected LFQ intensity. (B) Volcano plot showing quantitative comparison of the naïve versus the decell rat livers. The purple rectangle denotes cutoff criteria for significant removal of a protein in the decell sample compared to the naïve sample (fold change < 1/8; Benjamini Hochberg-adjusted p-value < 0.01). The orange rectangle encompasses the proteins that are considered retained in the decell sample (fold change > 1/8). Four proteins that were chosen for further validation are highlighted in the volcano plot. n=3 biological replicates.
We validated these proteomic results using western blots for four different proteins, one from each major category (“matrisome” versus “non-matrisome” and “retained” versus “removed”). The western blots are shown in Figure 4, and in each case they confirm our quantitative proteomic data: (i) fibronectin (FN) – a matrisome protein – was retained; serine protease inhibitor A3K (SERPINA3K) – a matrisome protein – was removed; filamin alpha (FLNA) – a non-matrisome protein – was retained; and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) – a non-matrisome protein – was removed.
Figure 4.
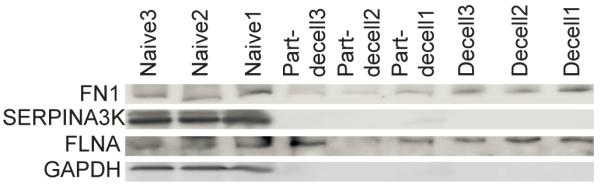
Western blots of four proteins – FN1, SERPINA3K, FLNA, GAPDH – in the naïve, partially-decellularized (part-decell), and decellularized (decell) rat livers. n=3 biological replicates.
Naïve and Decellularized Human Lungs
Another type of decellularized organ scaffold, human lung, was analyzed and compared to its naïve counterpart. Peptides from these samples were off-line separated into 6 fractions using high-pH HPLC, and then each fraction was analyzed by LC-MS/MS (3 μm particle-size, room temperature). The total and matrisome protein numbers as well as the iBAQ% arising from the non-matrisome proteins are shown in Figure 5. The number of protein identifications dramatically decreased from 2147 to 384 after decellularization. Matrisome proteins were well retained during decellularization, comprising 152 of the 384 proteins and 59% of the total iBAQ intensity. These results demonstrate the applicability of our proteomic workflow to different tissue types, as well as its capacity for achieving in-depth proteome coverage for both cellular and matrisome proteins.
Figure 5.
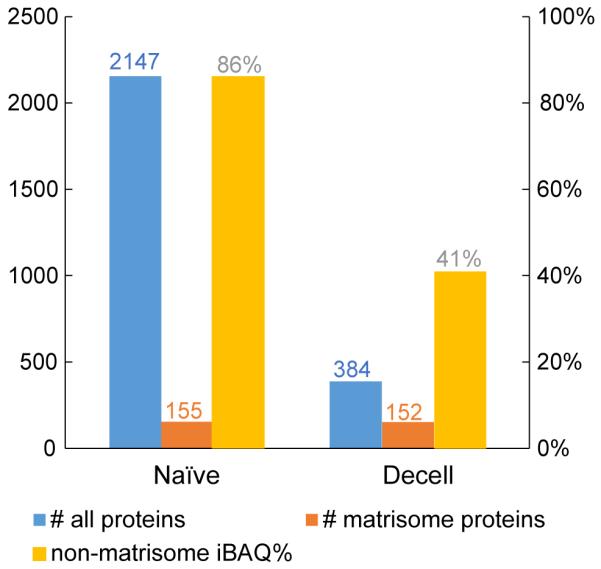
Proteomic results for naïve and decellularized (decell) human lung. Both samples were subjected to peptide pre-fractionation (6 fractions).
Discussion
Besides identifying the myriad of ECM components that play important roles in facilitating the engraftment, survival, and long-term function of reseeded cells, a comprehensive profile of the remnant cellular components in biological scaffolds is also imperative in revealing specific molecules involved in the host immune response after transplantation. The few published reports of proteome composition of decellularized biological scaffolds focus primarily on ECM proteins [38, 70, 71]. In this work, we have shown that protein extraction with SDS/DTT/Tris buffer followed by FASP cleanup and digestion enables identification and quantification of numerous low-abundance cellular proteins that represent the residual component of all the cell-related complexity, along with a variety of ECM proteins that are crucial in tissue reconstruction. The quantitative proteomic results are reassuring in that most of the remnant cellular proteins have significantly decreased in abundance, whereas the structural matrix proteins are well preserved after decellularization.
Collagen I and Matrigel are used extensively in biological research. We identified 76 proteins in “purified” collagen I, and many of the observed minor components may play a role in supporting cell growth or differentiation when collagen I is employed as a scaffold. GFR-Matrigel is a variant of Matrigel that has reduced levels of bioreactive growth factors such as fibroblast growth factor, epidermal growth factor, insulin-like growth factor 1, and transforming growth factor-beta [31]. The absence of these growth factors in our proteomic results confirms the quality of this “growth factor reduced” matrix, but hepatoma-derived growth factor and insulin-like growth factor 2 mRNA-binding protein 1 were still identified, although at very low abundance. The identification of nearly 1000 additional cellular and matrisome proteins, beyond the classically reported basement membrane-associated proteins laminin, nidogen, fibronectin and collagen IV, suggests caution in interpreting the functional consequences of cell culture or tissue regeneration experiments using Matrigel.
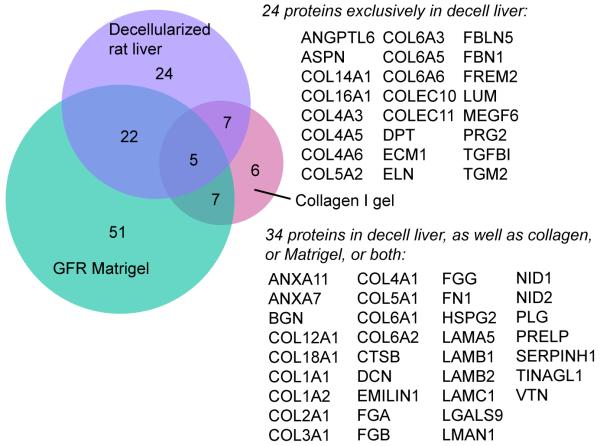
Collagen gel, Matrigel and decellularized liver have all been used as biological scaffolds for hepatocyte culture. Decellularized liver matrix is superior to the other two matrices in terms of supporting hepatocyte attachment and long-term survival [72-75]. To potentially explain these differences, we compared and contrasted their matrisome proteome composition; the Venn diagram is shown in Figure 6. A total of 34 matrisome proteins in the decellularized liver were also identified in either collagen or Matrigel, or both. This group represents a key set of proteins that may be essential for fundamental cell attachment, growth, and differentiation of hepatocytes. There were 24 matrisome proteins exclusively identified in the decellularized liver but absent in rat-tail collagen and Matrigel; these represent the uniqueness of the decellularized rat liver compared to collagen gel and Matrigel. Of note, COL4A1 and COL4A2 were shared by the three sample types, whereas COL4A3, A5, and A6 were unique to the decellularized rat livers. This observation is consistent with the fact that the “major” A1 and A2 chains of collagen IV are present ubiquitously in basement membranes, whereas the “minor” A3 through A6 chains are more spatially and temporally restricted [60]. The 24 unique components in decellularized livers could be critical determinants for directing constructive tissue remodeling, especially when combined with the original organ architecture and preserved microvascular network, thereby making them superior scaffolds for hepatocyte culture and liver regeneration.
Figure 6.
Venn diagram with the number of matrisome protein identifications for rat-tail type I collagen, GFR-Matrigel, and decellularized rat liver. Note that the GFR-Matrigel samples were subjected to peptide fractionation (6 fractions), while the collagen gel and rat liver samples were not, which could account for the relatively large number of proteins only detected in the GFR-Matrigel sample.
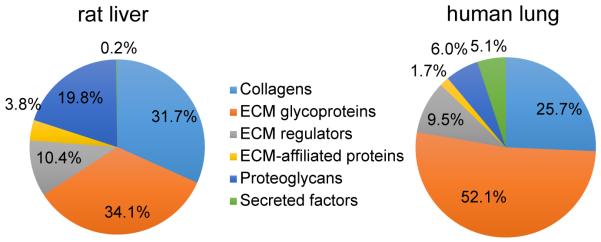
The decellularized ECM scaffold composition varies depending upon the tissue/organ source and the decellularization protocol. This observation is illustrated in Figure 7, where we show the iBAQ% attributed to each matrisome subcategory for the decellularized rat liver and human lung samples. Compared with decellularized human lung, decellularized rat liver has higher collagen and proteoglycan content, but proportionally lower abundance of ECM glycoproteins and secreted factors. These distinct matrisome compositions, together with differences in ECM architecture, help explain the fact that decellularized liver tissue may be the preferred ECM substrate for hepatocytes [73], while ECM from the lung may be preferred for respiratory epithelial cells [76]. We note that comparison of matrisome composition in different tissue types may be affected by different protein extraction efficiencies. However, in the present case, both the liver and lung scaffolds were nearly completely solubilized (visual evidence) and thus extraction efficiencies were likely similar. We have also analyzed cricoid cartilage and noticed that tissue solubilization was visually incomplete, and therefore quantitative comparisons to other tissues would not be appropriate. The proteomic results from cricoid cartilage included identification of >600 total proteins and >100 matrisome proteins with a single MS run (data not shown), thereby demonstrating that in-depth proteomic analysis is possible even for dense structural musculoskeletal tissues.
Figure 7.
Matrisome compositions by sub-category for decellularized rat liver and decellularized human lung.
Proteomic analysis revealed that the rat liver prior to decellularization has a higher fraction of cellular protein than the human lung – iBAQ values of 99% for the naïve liver and 86% for the naïve lung (Figures 2 & 5). This difference in baseline cellularity may partially explain why the decellularized lung sample appears more decellularized than the liver, when evaluated at the proteomic level – iBAQ values of 75% non-matrisome for the liver and only 41% for the lung. As mentioned above, these iBAQ values probably overestimate the cellular protein abundance, as our proteomics method is somewhat biased toward them, especially in tissues that possess compact ECM structures that are difficult to solubilize completely with sonication in SDS/DTT/Tris solution. Nonetheless, the relative comparison of iBAQ values (e.g. higher cellular percentage for liver than lung) should be valid. Notably, Hill et al. recently reported a method that involves protein fractionation and stable isotope labeled peptides to allow unbiased protein characterization and quantification [71]. Our present work employs a simple sample preparation method to reveal numerous ECM proteins that can be used to evaluate ECM preservation and loss, along with numerous low-abundance cellular proteins, which could have significant clinical consequences.
During creation of a decellularized scaffold, it is crucial to preserve the organ-specific ECM “zip codes” that direct or support site-appropriate cell attachment and differentiation [21]. A total of 22 matrisome proteins were significantly removed during either the first stage (naïve to part-decell), the second stage (part-decell to decell), or both stages of rat liver decellularization. Only three of these are “core matrisome” proteins: fibulin-5, laminin subunit beta-1, and vitronectin. The Space of Disse, lying between sinusoidal endothelial cells (SEC) and hepatocytes, has a unique matrix that is essential for maintaining the differentiation of these two types of cells [77]. The Space of Disse primarily consists of abundant fibronectin and collagen VI; common basement membrane proteins such as collagen IV and laminin; as well as fibrillar collagens I, III, and V [78]. These proteins were preserved during the decellularization process in the overall liver matrix, and thus it is reasonable to conclude that they were also preserved in the Space of Disse. Although it is unknown to what extent the loss of three core matrisome proteins and 19 other ECM regulators and ECM-affiliated proteins will affect subsequent cell seeding and proliferation, it is encouraging that the majority of matrisome proteins are preserved in this complex matrix, especially those associated with the biologically important Space of Disse.
In this work, we showed that cellular proteins are more difficult to remove than DNA and still constitute a large fraction of the decellularized scaffold proteome. The rat liver data presented in Figure 2 and Supplemental Figure S1 (two volcano plots: naïve vs. part-decell; part-decell vs. decell) reveal that more cellular proteins were removed during the latter stage of the decellularization process than during the earlier stage. This is likely due to the fact that the SDS concentration was higher (0.1-0.2%) during the latter stage (part-decell to decell), as compared to only 0.01% initially (naïve to part-decell). Although the decellularized rat livers are still dominated by remnant cellular proteins in terms of protein numbers (confirming that elimination of DNA does not assure adequate removal of all cellular material), quantitative comparison shows that most of these cellular proteins have significantly decreased abundance after decellularization. The majority of the 17 well-retained cellular proteins were large plasma membrane or intermediate filament proteins. These results suggest that although vascular perfusion decellularization is a powerful technique for preserving key structural components, disrupting cell membranes, and removing nucleic acids, it remains a challenge to remove all of the large anchoring and adhesion complexes [79]. The association between the relative abundance of remnant cellular proteins and subsequent in vivo host response requires further systematic investigation that combines proteomic analysis with transplantation studies. It is also possible that reseeded cells will facilitate degradation of these cellular remnants, and research is underway to study the turnover of these proteins.
Biological scaffold-based tissue engineering is an important approach within the sphere of regenerative medicine. A crucial step is to acquire scaffolds that provide microenvironments hospitable to reseeded cells and conducive to the growth of three-dimensional structured tissue. Our study provides the first insight into the complexity of the proteome of biological scaffolds and sheds new light on the quantitative protein changes that occur as decellularization progresses. We believe proteomics will be an extremely useful tool in facilitating understanding of the connection between the scaffold composition and future clinical transplantation outcomes.
Supplementary Material
Acknowledgments
This work was supported by the following grants: R01 DC004428 (to NVW), R01 DC010777 and R01 DC010777-S1 (to NVW and BLF) from the National Institute on Deafness and Other Communication Disorders; R01 DK084053 (to MLY) and R00 DK088962 (to BEU) from the National Institute of Diabetes and Digestive and Kidney Diseases.
References
- [1].Yang J, Elias JJ, Petrakis NL, Wellings SR, Nandi S. Effects of Hormones and Growth-Factors on Human Mammary Epithelial-Cells in Collagen Gel Culture. Cancer Res. 1981;41:1021–7. [PubMed] [Google Scholar]
- [2].Rocha V, Ringo DL, Read DB. Casein Production during Differentiation of Mammary Epithelial-Cells in Collagen Gel Culture. Exp Cell Res. 1985;159:201–10. doi: 10.1016/s0014-4827(85)80049-5. [DOI] [PubMed] [Google Scholar]
- [3].Sirica AE, Richards W, Tsukada Y, Sattler CA, Pitot HC. Fetal Phenotypic Expression by Adult Rat Hepatocytes on Collagen Gel-Nylon Meshes. Proc Natl Acad Sci U S A. 1979;76:283–7. doi: 10.1073/pnas.76.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Balasubramanian P, Prabhakaran MP, Sireesha M, Ramakrishna S. Collagen in Human Tissues: Structure, Function, and Biomedical Implications from a Tissue Engineering Perspective. In: Abe A, Kausch HH, Moller M, Pasch H, editors. Polymer Composites - Polyolefin Fractionation - Polymeric Peptidomimetics – Collagens. 2013. pp. 173–206. [Google Scholar]
- [5].Toda S, Sugihara H. Reconstruction of Thyroid-Follicles from Isolated Porcine Follicle Cells in 3-Dimensional Collagen Gel Culture. Endocrinology. 1990;126:2027–34. doi: 10.1210/endo-126-4-2027. [DOI] [PubMed] [Google Scholar]
- [6].Minami Y, Sugihara H, Oono S. Reconstruction of Cornea in 3-Dimensional Collagen Gel Matrix Culture. Invest Ophthalmol Vis Sci. 1993;34:2316–24. [PubMed] [Google Scholar]
- [7].Fujiyama C, Masaki Z, Sugihara H. Reconstruction of the Urinary-Bladder Mucosa in 3-Dimensional Collagen Gel Culture: Fibroblast-Extracellular Matrix Interactions on the Differentiation of Transitional Epithelial Cells. J Urol. 1995;153:2060–7. [PubMed] [Google Scholar]
- [8].Ito KI, Ryuto M, Ushiro S, Ono M, Sugenoya A, Kuraoka A, et al. Expression of Tissue-Type Plasminogen Activator and Its Inhibitor Couples with Development of Capillary Network by Human Microvascular Endothelial Cells on Matrigel. J Cell Physio. 1995;162:213–24. doi: 10.1002/jcp.1041620207. [DOI] [PubMed] [Google Scholar]
- [9].Braun H, Buhnemann C, Neumann J, Reymann KG. Preparation of a tissue-like cortical primary culture from embryonic rats using Matrigel and serum free Start V Medium. J Neurosci Methods. 2006;157:32–8. doi: 10.1016/j.jneumeth.2006.03.019. [DOI] [PubMed] [Google Scholar]
- [10].Eldred JR, Rogers K, Boughner D. Tissue Engineering the Aortic Valve Spongiosa Using Matrigel-Cell-Scaffold-Composites (MCSCs) FASEB J. 2008;22:903–6. [Google Scholar]
- [11].Lu SH, Wang HB, Liu H, Wang HP, Lin QX, Li DX, et al. Reconstruction of Engineered Uterine Tissues Containing Smooth Muscle Layer in Collagen/Matrigel Scaffold In Vitro. Tissue Eng Part A. 2009;15:1611–8. doi: 10.1089/ten.tea.2008.0187. [DOI] [PubMed] [Google Scholar]
- [12].Lu SH, Lin QX, Liu YN, Gao Q, Hao T, Wang Y, et al. Self-assembly of renal cells into engineered renal tissues in collagen/Matrigel scaffold in vitro. J Tissue Eng Regen Med. 2012;6:786–92. doi: 10.1002/term.484. [DOI] [PubMed] [Google Scholar]
- [13].Zhou J, Shu Y, Lu SH, Li JJ, Sun HY, Tang RY, et al. The Spatiotemporal Development of Intercalated Disk in Three-Dimensional Engineered Heart Tissues Based on Collagen/Matrigel Matrix. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0081420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13:27–53. doi: 10.1146/annurev-bioeng-071910-124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med. 2008;14:213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- [16].Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16:814–20. doi: 10.1038/nm.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–41. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Price AP, England KA, Matson AM, Blazar BR, Panoskaltsis-Mortari A. Development of a Decellularized Lung Bioreactor System for Bioengineering the Lung: The Matrix Reloaded. Tissue Eng Part A. 2010;16:2581–91. doi: 10.1089/ten.tea.2009.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–33. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- [20].Song JJ, Guyette JP, Gilpin SE, Gonzalez G, Vacanti JP, Ott HC. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat Med. 2013;19:646–51. doi: 10.1038/nm.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–43. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Griffiths LG, Choe LH, Reardon KF, Dow SW, Orton EC. Immunoproteomic identification of bovine pericardium xenoantigens. Biomaterials. 2008;29:3514–20. doi: 10.1016/j.biomaterials.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Beretta B, Conti A, Fiocchi A, Gaiaschi A, Galli CL, Giuffrida MG, et al. Antigenic determinants of bovine serum albumin. Int Arch Allergy Immunol. 2001;126:188–95. doi: 10.1159/000049513. [DOI] [PubMed] [Google Scholar]
- [24].Benjamin DC, Berzofsky JA, East IJ, Gurd FRN, Hannum C, Leach SJ, et al. The Antigenic Structure of Proteins - a Reappraisal. Annu Rev Immunol. 1984;2:67–101. doi: 10.1146/annurev.iy.02.040184.000435. [DOI] [PubMed] [Google Scholar]
- [25].Hansen KC, Kiemele L, Maller O, O’Brien J, Shankar A, Fornetti J, et al. An in-solution ultrasonication-assisted digestion method for improved extracellular matrix proteome coverage. Mol Cell Proteomics. 2009;8:1648–57. doi: 10.1074/mcp.M900039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Angel PM, Nusinow D, Brown CB, Violette K, Barnett JV, Zhang B, et al. Networked-based Characterization of Extracellular Matrix Proteins from Adult Mouse Pulmonary and Aortic Valves. J Proteome Res. 2011;10:812–23. doi: 10.1021/pr1009806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wilson R, Norris EL, Brachvogel B, Angelucci C, Zivkovic S, Gordon L, et al. Changes in the Chondrocyte and Extracellular Matrix Proteome during Post-natal Mouse Cartilage Development. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wilson R. The extracellular matrix: an underexplored but important proteome. Expert Rev Proteomics. 2010;7:803–6. doi: 10.1586/epr.10.93. [DOI] [PubMed] [Google Scholar]
- [29].Wilson R, Diseberg AF, Gordon L, Zivkovic S, Tatarczuch L, Mackie EJ, et al. Comprehensive Profiling of Cartilage Extracellular Matrix Formation and Maturation Using Sequential Extraction and Label-free Quantitative Proteomics. Mol Cell Proteomics. 2010;9:1296–313. doi: 10.1074/mcp.M000014-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].de Castro Bras LE, Ramirez TA, DeLeon-Pennell KY, Chiao YA, Ma Y, Dai Q, et al. Texas 3-step decellularization protocol: looking at the cardiac extracellular matrix. J Proteomics. 2013;86:43–52. doi: 10.1016/j.jprot.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–90. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- [32].Welham NV, Chang Z, Smith LM, Frey BL. Proteomic analysis of a decellularized human vocal fold mucosa scaffold using 2D electrophoresis and high-resolution mass spectrometry. Biomaterials. 2013;34:669–76. doi: 10.1016/j.biomaterials.2012.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Naba A, Clauser KR, Whittaker CA, Carr SA, Tanabe KK, Hynes RO. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer. 2014;14:518. doi: 10.1186/1471-2407-14-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Naba A, Clauser KR, Lamar JM, Carr SA, Hynes RO. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. eLife. 2014;3:e01308. doi: 10.7554/eLife.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med. 2012;9:866–76. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- [37].Uygun BE, Price G, Saeidi N, Izamis M-L, Berendsen T, Yarmush M, et al. Decellularization and Recellularization of Whole Livers. J Vis Exp. 2011;48:2394. doi: 10.3791/2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gilpin SE, Guyette JP, Gonzalez G, Ren X, Asara JM, Mathisen DJ, et al. Perfusion decellularization of human and porcine lungs: Bringing the matrix to clinical scale. J Heart Lung Transplant. 2014;33:298–308. doi: 10.1016/j.healun.2013.10.030. [DOI] [PubMed] [Google Scholar]
- [39].Guyette JP, Gilpin SE, Charest JM, Tapias LF, Ren X, Ott HC. Perfusion decellularization of whole organs. Nat Protoc. 2014;9:1451–68. doi: 10.1038/nprot.2014.097. [DOI] [PubMed] [Google Scholar]
- [40].Sheynkman GM, Shortreed MR, Frey BL, Smith LM. Discovery and Mass Spectrometric Analysis of Novel Splice-junction Peptides Using RNA-Seq. Mol Cell Proteomics. 2013;12:2341–53. doi: 10.1074/mcp.O113.028142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wiśniewski JR, Zielinska DF, Mann M. Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal Biochem. 2011;410:307–9. doi: 10.1016/j.ab.2010.12.004. [DOI] [PubMed] [Google Scholar]
- [42].Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, et al. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D8. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- [44].Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- [45].Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res. 2011;10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- [46].Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics. 2005;21:3448–9. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- [47].Kersteen EA, Higgin JJ, Raines RT. Production of human prolyl 4-hydroxylase in Escherichia coli. Protein Expr Purif. 2004;38:279–91. doi: 10.1016/j.pep.2004.09.008. [DOI] [PubMed] [Google Scholar]
- [48].Pflieger D, Chabane S, Gaillard O, Bernard BA, Ducoroy P, Rossier J, et al. Comparative proteomic analysis of extracellular matrix proteins secreted by two types of skin fibroblasts. Proteomics. 2006;6:5868–79. doi: 10.1002/pmic.200402040. [DOI] [PubMed] [Google Scholar]
- [49].Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- [50].Shin JB, Krey JF, Hassan A, Metlagel Z, Tauscher AN, Pagana JM, et al. Molecular architecture of the chick vestibular hair bundle. Nat Neurosci. 2013;16:365–74. doi: 10.1038/nn.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ekblom P, Thesleff I, Saxen L, Miettinen A, Timpl R. Transferrin as a Fetal Growth Factor: Acquisition of Responsiveness Related to Embryonic Induction. Proc Natl Acad Sci U S A. 1983;80:2651–5. doi: 10.1073/pnas.80.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Matheson S, Larjava H, Hakkinen L. Distinctive localization and function for lumican, fibromodulin and decorin to regulate collagen fibril organization in periodontal tissues. J Periodontal Res. 2005;40:312–24. doi: 10.1111/j.1600-0765.2005.00800.x. [DOI] [PubMed] [Google Scholar]
- [53].Kessler E, Mould AP, Hulmes DJS. Procollagen Type I C-Proteinase Enhancer Is a Naturally Occurring Connective Tissue Glycoprotein. Biochem Biophys Res Commun. 1990;173:81–6. doi: 10.1016/s0006-291x(05)81024-1. [DOI] [PubMed] [Google Scholar]
- [54].Challa AA, Stefanovic B. A Novel Role of Vimentin Filaments: Binding and Stabilization of Collagen mRNAs. Mol Cell Biol. 2011;31:3773–89. doi: 10.1128/MCB.05263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- [56].Arber S, Caroni P. Thrombospondin-4, an Extracellular Matrix Protein Expressed in the Developing and Adult Nervous-System Promotes Neurite Outgrowth. J Cell Biol. 1995;131:1083–94. doi: 10.1083/jcb.131.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Andrews NW, Abrams CK, Slatin SL, Griffiths G, A T. cruzi-secreted protein immunologically related to the complement component C9: evidence for membrane pore-forming activity at low pH. Cell. 1990;61:1277–87. doi: 10.1016/0092-8674(90)90692-8. [DOI] [PubMed] [Google Scholar]
- [58].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The Matrisome: In Silico Definition and In Vivo Characterization by Proteomics of Normal and Tumor Extracellular Matrices. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014647. M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hynes RO, Naba A. Overview of the Matrisome-An Inventory of Extracellular Matrix Constituents and Functions. Cold Spring Harbor Perspectives in Biology. 2012;4:a004903. doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Keene DR, Engvall E, Glanville RW. Ultrastructure of Type-Vi Collagen in Human-Skin and Cartilage Suggests an Anchoring Function for This Filamentous Network. J Cell Biol. 1988;107:1995–2006. doi: 10.1083/jcb.107.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Atkinson JC, Ruehl M, Becker J, Ackermann R, Schuppan D. Collagen VI regulates normal and transformed mesenchymal cell proliferation in vitro. Mol Biol Cell. 1996;7:283–91. doi: 10.1006/excr.1996.0328. [DOI] [PubMed] [Google Scholar]
- [62].Ricard-Blum S. The Collagen Family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chang JW, Lee SH, Jeong JY, Chae HZ, Kim YC, Park ZY, et al. Peroxiredoxin-I is an autoimmunogenic tumor antigen in non-small cell lung cancer. FEBS Lett. 2005;579:2873–7. doi: 10.1016/j.febslet.2005.04.028. [DOI] [PubMed] [Google Scholar]
- [64].Karasawa R, Ozaki S, Nishioka K, Kato T. Autoantibodies to peroxiredoxin I and IV in patients with systemic autoimmune diseases. Microbiol Immun. 2005;49:57–65. doi: 10.1111/j.1348-0421.2005.tb03640.x. [DOI] [PubMed] [Google Scholar]
- [65].Iwata Y, Ogawa F, Komura K, Muroi E, Hara T, Shimizu K, et al. Autoantibody against peroxiredoxin I, an antioxidant enzyme, in patients with systemic sclerosis: possible association with oxidative stress. Rheumatology. 2007;46:790–5. doi: 10.1093/rheumatology/kem010. [DOI] [PubMed] [Google Scholar]
- [66].Xu YK, Harder KW, Huntington ND, Hibbs ML, Tarlinton DM. Lyn tyrosine kinase: Accentuating the positive and the negative. Immunity. 2005;22:9–18. doi: 10.1016/j.immuni.2004.12.004. [DOI] [PubMed] [Google Scholar]
- [67].Kajita M, Sugimura K, Ohoka A, Burden J, Suganuma H, Ikegawa M, et al. Filamin acts as a key regulator in epithelial defence against transformed cells. Nat Commun. 2014;5:4428. doi: 10.1038/ncomms5428. [DOI] [PubMed] [Google Scholar]
- [68].Schreiber VR, Moog-Lutz C, Regnier CH, Chenard MP, Boeuf H, Vonesch JL, et al. Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol Med. 1998;4:675–87. [PMC free article] [PubMed] [Google Scholar]
- [69].Tsuruta D, Jones JCR. The vimentin cytoskeleton regulates focal contact size and adhesion of endothelial cells subjected to shear stress. J Cell Sci. 2003;116:4977–84. doi: 10.1242/jcs.00823. [DOI] [PubMed] [Google Scholar]
- [70].Marcal H, Ahmed T, Badylak SF, Tottey S, Foster LJR. A comprehensive protein expression profile of extracellular matrix biomaterial derived from porcine urinary bladder. Regen Med. 2012;7:159–66. doi: 10.2217/rme.12.6. [DOI] [PubMed] [Google Scholar]
- [71].Hill RC, Calle EA, Dzieciatkowska M, Niklason LE, Hansen KC. Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol Cell Proteomics. 2015;14:961–73. doi: 10.1074/mcp.M114.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Moghe PV, Berthiaume F, Ezzell RM, Toner M, Tompkins RG, Yarmush ML. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials. 1996;17:373–85. doi: 10.1016/0142-9612(96)85576-1. [DOI] [PubMed] [Google Scholar]
- [73].Sellaro TL, Ranade A, Faulk DM, McCabe GP, Dorko K, Badylak SF, et al. Maintenance of Human Hepatocyte Function In Vitro by Liver-Derived Extracellular Matrix Gels. Tissue Eng Part A. 2010;16:1075–82. doi: 10.1089/ten.tea.2008.0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zeisberg M, Kramer K, Sindhi N, Sarkar P, Upton M, Kalluri R. De-differentiation of primary human hepatocytes depends on the composition of specialized liver basement membrane. Mol Cell Biochem. 2006;283:181–9. doi: 10.1007/s11010-006-2677-8. [DOI] [PubMed] [Google Scholar]
- [75].Lecluyse EL, Audus KL, Hochman JH. Formation of Extensive Canalicular Networks by Rat Hepatocytes Cultured in Collagen-Sandwich Configuration. Am J Physiol. 1994;266:C1764–C74. doi: 10.1152/ajpcell.1994.266.6.C1764. [DOI] [PubMed] [Google Scholar]
- [76].Lin YM, Zhang A, Rippon HJ, Bismarck A, Bishop AE. Tissue Engineering of Lung: The Effect of Extracellular Matrix on the Differentiation of Embryonic Stem Cells to Pneumocytes. Tissue Eng Part A. 2010;16:1515–26. doi: 10.1089/ten.TEA.2009.0232. [DOI] [PubMed] [Google Scholar]
- [77].Schuppan D, Ruehl M, Somasundaram R, Hahn EG. Matrix as a modulator of hepatic fibrogenesis. Semin Liver Dis. 2001;21:351–72. doi: 10.1055/s-2001-17556. [DOI] [PubMed] [Google Scholar]
- [78].Zhu CX, Coombe DR, Zheng MH, Yeoh GCT, Li LJ. Liver progenitor cell interactions with the extracellular matrix. J Tissue Eng Regen Med. 2013;7:757–66. doi: 10.1002/term.1470. [DOI] [PubMed] [Google Scholar]
- [79].Badylak SF, Weiss DJ, Caplan A, Macchiarini P. Engineered whole organs and complex tissues. Lancet. 2012;379:943–52. doi: 10.1016/S0140-6736(12)60073-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.