Abstract
The LONI Image and Data Archive (IDA)1 is a repository for sharing and long-term preservation of neuroimaging and biomedical research data. Originally designed to archive strictly medical image files, the IDA has evolved over the last ten years and now encompasses the storage and dissemination of neuroimaging, clinical, biospecimen, and genetic data. In this article, we report upon the genesis of the IDA and how it currently securely manages data and protects data ownership.
Keywords: IDA, data repository, data sharing
Introduction
The IDA was initially created to de-identify and collect neuroimaging data for the International Consortium for Brain Mapping (ICBM) study in which Magnetic Resonance Imaging (MRI) and Positron-Emission Tomography (PET) scans from 850 normal adult subjects were collected at three North American sites (Mazziotta et al. 1995, 2009, Kochunov 2002). As interest in storing data in biomedical repositories grew, the number and size of studies utilizing the IDA expanded considerably. The IDA has become a global resource for storing and disseminating neuroimaging, clinical, biospecimen, and genetic data for a growing number of national and international consortia efforts and many smaller, single-center studies.
Background
Early IDA development proceeded in concert with new HIPAA (Health Insurance Portability and Accountability Act (US Department of Health and Human Services)) regulations that took effect in 2003, which put emphasis on maintaining patient confidentiality throughout data collection and data collaboration activities. Over time, attention on data sharing dynamics (Amari et al. 2002, Eckersley, et al. 2003, Gardner et al. 2003, Koslow 2000, Kulynych 2002, Toga and Dinov 2015) grew in parallel with the launch of increasingly complex multi-site consortia studies. The number, scale and scope of collaborations utilizing the IDA expanded and extended beyond data acquisition sites to involve organizations performing quality assessments and to external electronic data capture (EDC) systems. Consequently, IDA functionality and features were expanded to accommodate the specific needs of different types of studies and users.
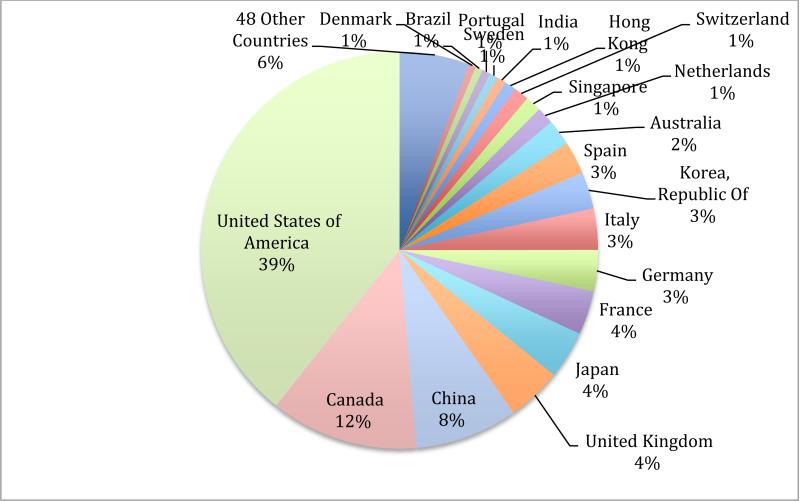
Today the IDA contains both raw (direct from scanner) and processed (output from processing programs) neuroimaging data, clinical data, and analysis results for dozens of studies on Alzheimer's disease (Toga and Crawford 2010), multiple sclerosis, Huntington's disease, Parkinson's disease (Marek et al. 2011), traumatic brain injury, normal development, HIV/AIDS, bipolar disorder, schizophrenia, and others. Sites across North America, Europe, Australia, and Asia have been actively uploading data since 2003 with the average number of newly added images reaching over 5,000 per month in 2014. For a growing number of studies, clinical data, image quality assessments, and analysis results are uploaded to the IDA on a daily basis. For many studies the IDA is the exclusive location for pooled data but for a small number of studies (Australian Imaging, Biomarkers and Lifestyle (Ellis et al. 2009), Autism Brain Imaging Data Exchange (Di Martino et al. 2014), Brain Genomics Superstruct Project (Buckner et al. 2012), and Human Connectome Project (Rosen et al. 2010)) the IDA mirrors data available in other repositories and this allows users to obtain data from all these studies within a single system. Worldwide, the number of image downloads from the IDA exceeds 7 million (Figure 1). The logo for each study, along with a link to the study's web site, appears at the top of each IDA web page in order to focus on the study rather than the IDA. There is no requirement to acknowledge the IDA in publications that use data obtained through the IDA.
Figure 1.
Users in 68 countries have downloaded over 7 million raw and processed images from the IDA since 2004.
Data collection
The IDA presently holds data from more than 70 studies and 125 different institutions, and is continually receiving new data. Table 1 lists a subset of research studies that are storing data in the IDA. On average, more than 120 raw scans are uploaded each weekday from sites located in the Americas, Europe, and Australia. There are currently over 350,000 neuroimaging scans (over 96 million files) archived in the IDA of which 64% are raw and 36% are processed scans. These scans consist of structural MRI, functional MRI (fMRI), diffusion MRI, magnetic resonance angiography (MRA), positron emission tomography (PET), computed tomography (CT), and single-photon emission computed tomography (SPECT) data from tens of thousands of human subjects (some followed longitudinally for over a decade) and hundreds of phantom scans used for image quality control.
Table 1.
Representative research studies utilizing the IDA.
| Study Name | Centers | Subjects | Deposit Activity | *Downloads |
|---|---|---|---|---|
| Aging & Dementia | ||||
| Alzheimer's Disease Neuroimaging Initiative (ADNI) | 58 | 2469 | 2005 - present | 7,400,000 |
| Australian Imaging, Biomarkers and Lifestyle (AIBL) | 1 | 810 | 2008 - present | 68,000 |
| Imaging & Genetic Biomarkers for AD | 1 | 177 | 2009 - present | Private |
| HIV | ||||
| Age Moderates HIV-Related CNS Dysfunction | 1 | 116 | 2006 - 2007 | Private |
| Cardiovascular & HIV/AIDS Effects on Brain & Cognition | 4 | 349 | 2009 - 2014 | Private |
| Huntington's Disease | ||||
| Huntington's Disease Neuro Imaging Initiative (HDNI) | 4 | 369 | 2007 - 2011 | Private |
| Track-On Huntington's Disease | 4 | 242 | 2012 - 2014 | Private |
| Brain Injury | ||||
| Effects of TBI & PTSD on Alzheimer's Disease in Vietnam Vets (DoD ADNI) | 17 | 115 | 2013 - present | 15,000 |
| Volumetrics in Brain Trauma | 1 | 393 | 2006 - present | Private |
| Transforming Research and Clinical Knowledge in TBI (Track-TBI) | 11 | 418 | 2014 - present | 7,000 |
| Normal & Development | ||||
| International Consortium for Brain Mapping (ICBM) | 3 | 852 | 2003 - 2009 | 123,000 |
| Genetic influences on the brain: A twin study | 1 | 1045 | 2007 - 2013 | Private |
| Multiple Sclerosis | ||||
| Hippocampal Volume Loss in Multiple Sclerosis | 1 | 58 | 2007 - 2010 | Private |
| Multi-center Estriol Study | 17 | 334 | 2007 - 2014 | Private |
| Parkinson's | ||||
| Parkinson's Progression Markers Initiative (PPMI) | 31 | 1230 | 2011 - present | 360,000 |
| Hippocampal atrophy in Parkinson's disease | 1 | 166 | 2008 - 2011 | Private |
| Schizophrenia | ||||
| North American Prodrome Longitudinal Study (NAPLS) | 8 | 845 | 2009 - present | Private |
| Studies Mirrored in the IDA | ||||
| Autism Brain Imaging Data Exchange (ABIDE) | 2012 | 48,000 | ||
| Brain Genomics Superstruct Project (GSP) | 1 | 1570 | 2014 | 50 |
Indicates the number of images, clinical and genetic datasets downloaded from the IDA for studies with open data sharing policies.
User interactions occur primarily through web-browsers that incorporate the Java2 plugin. Since minimal technical proficiency is required, new studies can quickly come online and begin to upload data without requiring lengthy training or devoting excessive time and resources at the participating sites.
Uploading and de-identifying data
Users can upload neuroimaging data files in many formats, including those listed in Table 2. During the data archiving process, the user opens a web-browser and logs into the web application. The web application deploys a Java applet that runs on the user's computer at the acquisition site. The applet automatically detects image file formats and invokes format-specific de-identification programs to remove patient-identifying information from the files before transferring the de-identified files to the IDA. Different de-identifications are used for different scanner types and different file formats in accordance with the individual needs of the studies that are involved. De-identification programs may be customized for the needs of the study, however the general approach involves the replacement of patient name and patient ID fields with the user-supplied research identifier, removal of all elements that are either not of a preserved type (e.g. numeric, code string) or in a set of preserved elements, and hashing of elements containing unique identifiers. Several scanner-specific private elements are also retained in order to preserve needed information, for example gradient information for diffusion scans. For fMRI scans, a paradigm file may be uploaded as an attachment. The fMRI attachment, which is linked to the fMRI image, will be provided whenever a user downloads the fMRI image files. Processed neuroimaging files are uploaded in conjunction with a structured processing provenance file. The provenance file contains information about the image processing workflow and identifies the image(s) from which the processed image was derived. A schema enforces the structure and content of the provenance files and the subject and image identifiers are validated at time of upload. Once the de-identified files are uploaded into the IDA, metadata from the image file headers (and provenance files) are extracted and used to detect duplicate data and classify images (Neu, Crawford et al. 2012) into subtypes (e.g., structural MRI, functional MRI, or diffusion MRI). This metadata is then stored in the database and combined with other information from the upload to support future queries on the data.
Table 2.
Summary of different types of data stored in the IDA and their formats.
| Category | Type | File Format |
|---|---|---|
| Neuroimaging | MRI, PET, CT, SPECT | ANALYZE 7.53, DICOM4, ECAT5, GE6, HRRT Interfile (Cradduck et al. 1989), FreeSurfer/MGH7, MINC8, NIfTI9, Varian FDF10 and NRRD11 |
| Subject characteristics | Demographics, health history, family history | Comma separated value (CSV) |
| Genetic | Genotype, SNPs, Indels, | Text, VCF, PLINK |
| Biospecimen | Lab procedures, analysis results | CSV |
| Study documents | CRFs, methods, reports | PDF, Text, Word document |
Uploaders may also send clinical data and analysis results to the IDA using a tool that transfers data in comma-separated value (CSV) files. Validity checks are performed on all CSV files before they are accepted to help ensure data quality. Many studies use the tool to copy the entire contents of clinical EDC systems to the IDA on a daily basis. The data transfer tool supports both incremental updates and full synchronization of entire data sets. Once this clinical data is made available, investigators may access all image and clinical data from a study through the IDA. This frees the EDC systems, which are focused on data collection, from having to manage access to the data.
Quality assessment
Beginning with the Alzheimer's Disease Neuroimaging Initiative (ADNI), the IDA offered an option for uploaded neuroimaging data to be quarantined (hidden from general users) until quality assessments by external reviewers are conducted. Newly uploaded image files can be automatically quarantined and assigned to modality-specific download queues where they are held until reviewers download them. Once the image files are downloaded and quality assessments are assigned, the IDA will update their quarantine status and make image files that have been rated as acceptable available to general users. Neuroimaging data can also be imported from the IDA directly into the LONI Quality Control system 12 for semi-automated QA processing. Results from the LONI Quality Control system can be returned to the IDA, triggering a status update. The LONI Quality Control system is built on the LONI Pipeline workflow environment (Rex, Ma et al. 2003, Dinov et al., 2009).
Data sharing and dissemination
The data ownership and access policies of the IDA have always stated that the data belongs solely to its owners and that all data access decisions remain under their direct control. Data access can be granted in three ways: 1) access to data from one or more sites in a study can be set in a user management web page, 2) a reviewer can grant guest-level (search and download) access to applicants through semi-automated data application web pages, and 3) a study can be made publicly accessible to everyone having an IDA user account. In addition, IDA role-based access controls provide different levels of access. For example, users may be granted access to upload and/or download data that is acquired only at their site or they can be given study-wide access for data from all sites in a study. This functionality is often needed by study managers to control access to the data that is being pooled from multiple sites. Permissions to edit and delete data may also be assigned as needed in order to support review, tracking, and other data management operations.
Widespread data sharing is supported by IDA web pages that allow study-designated reviewers to receive, evaluate, and approve/disapprove online data use applications. During the application process, applicants are presented with a data use agreement that is specific to the study for which the application is being submitted. After agreeing to the terms of the data use agreement, applicants must then provide their contact information and their intended use of the data. Once submitted, the application is sent to one or more reviewers who review and either approve or disapprove the applications. Applications for more than 11,000 investigators have been submitted and for most studies, applications are reviewed within 72 hours of submission.
There are several search interfaces in the IDA, including a visual web page for data exploration. Users may search across attributes drawn from subject characteristics, assessment scores, and imaging protocols. Search results are saved in user-specific collections that are used when downloading or passing the results to the LONI work flow environment for processing. Studies may also define preset collections that are shared among those with access to the study. This allows study leadership to group together data that meet specified criteria so that multiple users can access the same sets of data without first needing to conduct searches of the database. Since the IDA keeps extensive records of download activity, downloaders can avoid downloading the same data twice and can easily locate new data after it arrives. For some studies the IDA is used exclusively as a neuroimaging data repository with non-imaging data stored externally whereas for other studies the IDA stores both types of data. For studies that are using the IDA to store non-imaging data collected through an external EDC, a subset of data elements can be mapped into a common data model that enables searches across both imaging and clinical data as well as across multiple IDA studies. This includes, but is not limited to, searching demographic, genetic, cognitive assessments, and subject data. EDC data is matched against known study subject identifiers however additional data curation is not presently performed within the IDA.
The IDA incorporates integrated image file translators built on the LONI Debabeler engine (Neu, Valentino et al. 2005) so users can download image files in formats more suited to their viewing and processing environments. These formats can be different from the formats used when the image files were uploaded and this frees downloaders from having to perform common file format translations themselves. The IDA currently supports translating all the neuroimaging file formats in Table 2 to the ANALYZE 7.5, MINC, and NIfTI image file formats.
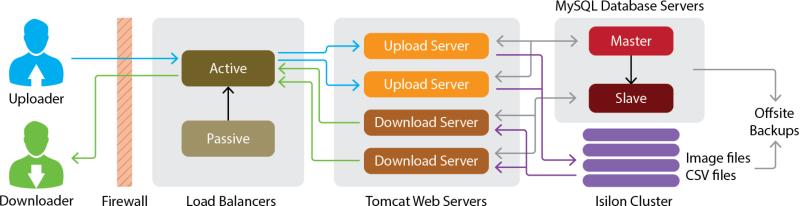
Infrastructure and architecture
High-availability network, server, storage, and electrical systems ensure that users have constant access to the IDA (Figure 2). In order to prevent download activity from impeding user uploads, load balancers split and send requests to separate sets of web servers. Multiple copies of archived data are backed up to different physical locations to secure against disaster and preserve data for future research. All user communications occur securely using the HTTPS protocol.
Figure 2.
System architecture of the IDA. Redundant load balancers distribute upload and download requests to different sets of web servers. Data files are stored in an Isilon13 clustered storage system and database servers manage system and neuroimaging metadata.
Multiple IDA machines manage download requests and start to throttle download speeds after a set number of unique users begin to download. Throttling involves tracking the number of simultaneous downloads per user and proportionally slowing downloads when throttling is active which facilitates equitable sharing of resources. Data are cyclic redundancy check (CRC) sum checked to ensure data are not corrupted during transfer. The Java download client run by the user will retry a download up to 10 times in order to complete data transfers over unreliable internet connections. Data is compressed on the server, sent to the client, and then decompressed by the client to reduce the time needed to send data. It can be difficult for users to manage multiple data downloads, especially for on-going longitudinal studies, so users are notified when they attempt to download data that they had previously downloaded.
Discussion
The sustainability of any informatics infrastructure rests not just on the financial support but also on the flexibility needed to preserve data in a manner that allows it to be useful in the future. The IDA has fully committed institutional, grant and foundation financial support and hence has broad, deep and continued stability and longevity. Further, neuroimaging files are stored in the original format with translations to other formats applied as needed. This retains nuances of the raw data thus preserving details that may become necessary in future as new investigative methods are devised.
Highlights.
A review of the genesis and development of the LONI IDA data repository
A description of the repository containing clinical, imaging and genetic data from more than 70 studies
For over 15 years the IDA has securely managed imaging, clinical, and genetic data.
10 neuroimaging file formats for MRI, fMRI, DTI, PET, CT, and SPECT data are handled.
Investigators in 68 countries have downloaded more that 7 million datasets from the IDA
Acknowledgments
This work was supported by National Institutes of Health grants 1U54EB020406-01, EB015922, W81XWH-12-2-0012, W81XWH-13-1-0259, U01 NS086090, the Parkinson's Progression Markers Initiative of the Michael J. Fox Foundation, and Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amari SI, Beltrame F, Bjaalie JG, Dalkara T, De Schutter E, Egan GF, Wrobel A. Neuroinformatics: the integration of shared databases and tools towards integrative neuroscience. Journal of integrative neuroscience. 2002;1(02):117–128. doi: 10.1142/s0219635202000128. [DOI] [PubMed] [Google Scholar]
- Borkin S. The HIPAA final security standards and ISO/IEC 17799. Collect. Information Security Reading Room. 2003 [Google Scholar]
- Buckner RL, Hollinshead M, Holmes AJ, Brohawn DG, Fagerness JA, O’Keefe T, Roffman JL. The Brain Genomics Superstruct Project. 2012 [Google Scholar]
- Cradduck TD, Bailey DL, Hutton BF, De Conninck F, Busemann-Sokole E, Bergmann H, Noelpp U. A standard protocol for the exchange of nuclear medicine image files. Nuclear medicine communications. 1989;10(10):703–714. doi: 10.1097/00006231-198910000-00002. [DOI] [PubMed] [Google Scholar]
- Dept of Health and Human Services [October 22, 2002];Administrative Simplification Standards. Available at: http://www.hhs.gov/ocr/hipaa.
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Milham MP. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular psychiatry. 2014;19(6):659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov ID, Petrosyan P, Liu Z, Eggert P, Hobel S, Vespa P, Toga AW. High-throughput neuroimaging-genetics computational infrastructure. Frontiers in neuroinformatics. 2014;8 doi: 10.3389/fninf.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov ID, Van Horn JD, Lozev KM, Magsipoc R, Petrosyan P, Liu Z, Toga AW. Efficient, distributed and interactive neuroimaging data analysis using the LONI pipeline. Frontiers in neuroinformatics. 2009;3 doi: 10.3389/neuro.11.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley P, Egan GF, De Schutter E, Yiyuan T, Novak M, Sebesta V, Toga AW. Neuroscience data and tool sharing. Neuroinformatics. 2003;1(2):149–165. doi: 10.1007/s12021-003-0002-1. [DOI] [PubMed] [Google Scholar]
- Ellis KA, Bush AI, Darby D, De Fazio D, Foster J, Hudson P, Ames D. The Australian Imaging, Biomarkers and Lifestyle (AIBL) study of aging: methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer's disease. International Psychogeriatrics. 2009;21(04):672–687. doi: 10.1017/S1041610209009405. [DOI] [PubMed] [Google Scholar]
- Gardner D, Toga AW, Ascoli GA, Beatty JT, Brinkley JF, Dale AM, Wong ST. Towards effective and rewarding data sharing. Neuroinformatics. 2003;1(3):289–295. doi: 10.1385/NI:1:3:289. [DOI] [PubMed] [Google Scholar]
- Java TM. Platform. (Standard Edition) 1(2):0. [Google Scholar]
- Kochunov P, Lancaster J, Thompson P, Toga AW, Brewer P, Hardies J, Fox P. An optimized individual target brain in the Talairach coordinate system. Neuroimage. 2002;17(2):922–927. [PubMed] [Google Scholar]
- Koslow SH. Should the neuroscience community make a paradigm shift to sharing primary data?. Nature neuroscience. 2000;3(9):863–865. doi: 10.1038/78760. [DOI] [PubMed] [Google Scholar]
- Kulynych J. Legal and ethical issues in neuroimaging research: human subjects protection, medical privacy, and the public communication of research results. Brain and cognition. 2002;50(3):345–357. doi: 10.1016/s0278-2626(02)00518-3. [DOI] [PubMed] [Google Scholar]
- Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Baca M. The parkinson progression marker initiative (PPMI). Progress in neurobiology. 2011;95(4):629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development the international consortium for brain mapping (ICBM). Neuroimage. 1995;2(2PA):89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Mazziotta JC, Woods R, Iacoboni M, Sicotte N, Yaden K, Tran M, Toga AW. The myth of the normal, average human brain—the ICBM experience:(1) subject screening and eligibility. NeuroImage. 2009;44(3):914–922. doi: 10.1016/j.neuroimage.2008.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu SC, Crawford KL, Toga AW. Practical management of heterogeneous neuroimaging metadata by global neuroimaging data repositories. Frontiers in neuroinformatics. 2012;6 doi: 10.3389/fninf.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu SC, Valentino DJ, Toga AW. The LONI Debabeler: a mediator for neuroimaging software. NeuroImage. 2005;24:1170–1179. doi: 10.1016/j.neuroimage.2004.10.035. [DOI] [PubMed] [Google Scholar]
- Rex DE, Ma JQ, Toga AW. The LONI pipeline processing environment. Neuroimage. 2003;19(3):1033–1048. doi: 10.1016/s1053-8119(03)00185-x. [DOI] [PubMed] [Google Scholar]
- Rosen B, Wedeen V, Van Horn JD, Fischl B, Buckner R, Wald L, Toga AW. The human connectome project.. Organization for Human Brain Mapping Annual Meeting; Barcelona, Spain. 2010. [Google Scholar]
- Toga AW, Crawford KL. The informatics core of the Alzheimer's Disease Neuroimaging Initiative. Alzheimer's & Dementia. 2010;6(3):247–256. doi: 10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Dinov ID. Sharing Big Biomedical Data. 2015 doi: 10.1186/s40537-015-0016-1. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Department of Health and Human Services HIPAA administrative simplification: Regulation text. 2006 [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Trojanowski JQ. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer's & Dementia. 2013;9(5):e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]