Abstract
Insulin resistance is associated with obesity but mechanisms controlling this relationship in humans are not fully understood. Studies in animal models suggest a linkage between adipose reactive oxygen species (ROS) and insulin resistance. ROS oxidize cellular lipids to produce a variety of lipid hydroperoxides that in turn generate reactive lipid aldehydes that covalently modify cellular proteins in a process termed carbonylation. Mammalian cells defend against reactive lipid aldehydes and protein carbonylation by glutathionylation using glutathione-S-transferase A4 (GSTA4) or carbonyl reduction/oxidation via reductases and/or dehydrogenases. Insulin resistance in mice is linked to ROS production and increased level of protein carbonylation, mitochondrial dysfunction, decreased insulin-stimulated glucose transport, and altered adipokine secretion. To assess protein carbonylation and insulin resistance in humans, eight healthy participants underwent subcutaneous fat biopsy from the periumbilical region for protein analysis and frequently sampled intravenous glucose tolerance testing to measure insulin sensitivity. Soluble proteins from adipose tissue were analyzed using two-dimensional gel electrophoresis and the major carbonylated proteins identified as the adipocyte and epithelial fatty acid–binding proteins. The level of protein carbonylation was directly correlated with adiposity and serum free fatty acids (FFAs). These results suggest that in human obesity oxidative stress is linked to protein carbonylation and such events may contribute to the development of insulin resistance.
INTRODUCTION
The inverse correlation between insulin sensitivity and adiposity is well known (1) and overweight is the single most important predictor of type 2 diabetes mellitus (2). Although the mechanisms controlling this relationship is not fully understood, substantial evidence exists from animal model systems (3) and human studies to suggest that obesity-related oxidative stress and adipose tissue inflammation may be contributing factors (4,5). Adipose tissue plays an important role in the insulin responsiveness of the whole organism directly through lipolysis and the production of free fatty acid (FFA), increased levels of which are known to potentiate hepatic glucose output, inhibit muscle glucose uptake (6) and indirectly through secretion of adipokines influence insulin sensitivity in liver and muscle (7).
During the development of obesity, expansion of adipose mass has been related to a variety of factors including localized tissue hypoxia, infiltration of inflammatory cells, and alteration in cytokine profile (8–10). Consistent with this, hypoxia in 3T3-L1 adipocytes is associated with inhibition of insulin receptor signaling, increased lipolysis and the induction of reactive oxygen species (ROS) (11). In other experiments, increased levels of ROS in adipocytes have been causally linked to development of insulin resistance in model systems (3).
ROS such as superoxide anion, hydrogen peroxide, and hydroxyl radicals react with a variety of cellular biomolecules leading to modification of intracellular proteins, DNA, RNA, carbohydrates, and lipids. Of the various ROS, hydroxyl radicals are perhaps the most deleterious and lead to the peroxidation of polyunsaturated fatty acyl chains of membrane phospholipids and triglycerides. These peroxidated acyl chains undergo non-enzymatic Hock cleavage forming a family of short and medium chain reactive aldehydes, the most abundant of which are trans-4-hydroxy-2-nonenal and trans-4-oxo-2-nonenal (12). These reactive aldehydes covalently modify the side chains of histidine, cysteine and lysine residues in a process generically referred to as protein carbonylation. Since the side chains of histidine, lysine and cysteine are frequently components of enzyme active sites or participate in protein–protein interactions, carbonylation commonly leads to loss of function. Moreover, modification frequently targets the proteins for selective degradation by the 26S proteasome (13) leading not only to loss of activity, but also to selective loss of protein. Recent studies in adipose tissue from obese insulin-resistant C57Bl/6J mice have revealed two- to threefold increased levels of total protein carbonylation when compared to lean controls. Among the panel of modified proteins are key factors involved in carbohydrate and lipid metabolism, insulin signaling and a variety of enzymes involved in the antioxidant response (14).
Of the mechanisms used to inactivate reactive lipid species, glutathionylation via glutathione-S-transferase A4 is well studied (15). The expression of glutathione-S-transferase A4 (GSTA4) in murine systems is downregulated in part by tumor necrosis factor-α leading to decreased expression in the adipose tissue of high fat fed or genetically obese mice (14,16) while expression levels of other inactivating enzymes such as acetaldehyde dehydrogenase 2 (ALDH2) and fatty aldehyde dehydrogenase (FALDH) are unchanged (17). Microarray analysis of human subcutaneous adipose tissue (SAT) indicates that GSTA4 expression was reduced in obese insulin resistant, but not obese insulin sensitive individuals (17) relative to lean controls. Silencing of GSTA4 in 3T3-L1 adipocytes results in increased total protein carbonylation and multiple metabolic changes including increased basal lipolysis, altered glucose transport, and decreased β-oxidation of fatty acids (13). Both GSTA4-silenced 3T3-L1 adipocytes and adipose tissue from GSTA4 knockout mice showed significant impairment in mitochondrial oxidative respiration and increased mitochondrial ROS (17). Other studies in 3T3-L1 adipocytes have shown that insulin receptor substrate-1 and 2 are carbonylated by reactive lipid species leading to reduced insulin-stimulated tyrosine phosphorylation and also targeted degradation (18).
The present study was designed to assess human adipose tissue measures of oxidative stress including protein carbonylation and expression of antioxidant genes and their relationship to body size, body composition and insulin sensitivity. We report herein for the first time protein carbonylation in human adipose tissue is positively correlated with adiposity and serum FFAs.
METHODS AND PROCEDURES
Subjects
This study was approved by the institutional review board of University of Minnesota (Minneapolis, MN) and informed consent was obtained from participants. A total of eight subjects (six men, two women; seven white, one African American) were recruited by letter and telephone call from among a cohort of 357 individuals who had previously participated in a longitudinal study of insulin resistance and cardiovascular risk factors from childhood (age 11–14) into young adulthood, last studied at average age 22 (19,20). From the subjects who responded positively (13 males and 10 females), participants were selected according to availability during the study period. Exclusion criteria included pregnancy and chronic disease. A bariatric surgery patient, not part of the longitudinal study, was recruited only for assessment of visceral adipose tissue (VAT) obtained at the time of bariatric surgery from the omental fat. No subjects were on medications affecting insulin sensitivity.
Clinical measures
Anthropometric measures were obtained by trained research personnel. Body composition was determined by dual energy X-ray absorptiometry with a GE Lunar Prodigy scanner (GE Healthcare Biosciences, Pittsburgh, PA). Abdominal computed tomography at the L4/L5 level was used to measure visceral fat distribution as previously described (21). Venous blood samples were taken after overnight fast for analyses of glucose, insulin and lipids.
Frequently sampled intravenous glucose tolerance test
Following a 10–12 h overnight fast, participants were admitted to the Clinical Research Center. An intravenous catheter was placed in each arm for (i) infusion of a glucose bolus (0.3 g/kg body weight) at time 0 followed by an infusion of insulin (4 mU/kg/min) over 5 min starting at 20 min and (ii) blood samples for plasma glucose and insulin measurements at −10, −1, 2, 4, 8, 19, 22, 30, 40, 50, 70, 90, and 180 min as previously described (22). The data were analyzed according to the minimal model, using the computer program MINMOD to generate insulin sensitivity index (SI), glucose disposition index (DI) and the acute insulin response to glucose (23,24).
Adipose tissue sampling
Under standard sterile technique and local anesthesia (1% lidocaine with epinephrine), a curvilinear ellipse of skin along with the underlying adipose tissue was excised from the inferior periumbilical abdominal area and a total of 2–5 g of fat was obtained from each subject. The specimens were dissected to remove stromal tissue, immediately frozen with liquid nitrogen in aliquots of 0.1–0.2 g, and stored at −70 °C until further study.
Analysis of protein carbonylation
To evaluate protein carbonylation, adipose tissue was minced and homogenized on ice in coupling buffer (100 mmol/l sodium acetate at pH 5.5, 20 mmol/l NaCl and 0.1 mmol/l EDTA supplemented with 0.1 mmol/l phenylmethanesulfonyl fluoride, 2 mg/ml pepstatin, 2 mg/ml aprotinin, 2 mg/ml leupeptin). Homogenates were centrifuged at 600g at 4 °C for 5 min to separate the lipid cake, the infranatant was removed and sodium dodecyl sulfate (SDS; Sigma-Aldrich, St Louis, MO) was added to a final concentration of 1%. The lysate was centrifuged at 100,000g for 1 h at 4 °C to remove insoluble residue and the supernatant recovered and assayed for protein content (BCA assay; Sigma-Aldrich). A total of 25 µg of soluble protein was coupled with EZ-link biotin hydrazide (Pierce Biotechnology, Rockford, IL) to covalently label free aldehydes as previously (14) described. Proteins were separated by SDS-polyacrylamide gel electrophoresis (10–20% acrylamide gradient gel), transferred to polyvinylidene fluoride (Millipore, Billerica, MA) membranes and blocked in Odyssey Blocking Buffer (Li-Cor, Lincoln, NE). Biotinylated proteins were detected with DyLight 800 Conjugated Streptavidin (Pierce). As a loading control, the biotin hydrazide blot was probed with mouse anti β-actin antibody and IRDye 680 conjugated goat (polyclonal) anti-mouse secondary antibody. Biotinylated proteins and β-actin loading control bands were visualized using a LiCor Odyssey Infrared Imager. Both total protein carbonylation as well as intensity of the β-actin band were determined by quantification of LiCor signal intensity. Protein carbonylation is reported normalized to β-actin band.
Two-dimensional gel electrophoresis
Total soluble protein was isolated from the visceral fat sample and coupled with biotin hydrazide. One hundred micrograms of sample was allowed to rehydrate into a 13 cm Immobline DryStrip, pH 3–10 (GE Healthcare) overnight. After overnight rehydration, the Immobline DryStrip sample was resolved with an Ettan IPGphor IEF apparatus (Amersham (GE Healthcare), Waukesha, WI) until a total 19.255 kVh was reached. The IEF strip was equilibrated for 0.5 h in SDS equilibration buffer (50 mmol/l Tris, pH 8.8, 8 mol/l urea, 30% glycerol, 4% SDS, 1% dithiothreitol (Sigma-Aldrich) followed by separation on 10–20% Tris-HCl SDS-PAGE Criterion gel (BioRad). After SDS-polyacrylamide gel electrophoresis, the gel was stained with Deep Purple total protein stain (GE Healthcare) according to the manufacturer’s instructions. The gel image was visualized with the Typhoon 8610 scanner (GE Healthcare). Separated protein from parallel gels were transferred to polyvinylidene fluoride membranes and probed for carbonylated proteins with IR800-streptavidin as described. Additionally, membranes were incubated with either rabbit anti-mouse A-FABP polyclonal antibody or rabbit anti-mouse E-FABP polyclonal antibody at 1:10,000 dilution at 4 °C overnight. Membranes were washed with phosphate-buffered saline containing 0.05% Tween (phosphate-buffered saline tween) and incubated for 1 h with IR-680 goat anti-rabbit secondary antibody at 1:10,000 and scanned using an Odyssey Infrared Imager in both the 700 nm and 800 nm channels.
RNA extraction, reverse transcription and real-time PCR
Total RNA was extracted from about 0.2 g of adipose tissue using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. After DNase treatment, cDNA was synthesized using iScript cDNA synthesis kit (BioRad, Hercules, CA). Relative quantification of mRNAs was performed by real-time PCR using iQ SYBR green Supermix and the MyiQ detection system (BioRad). Primers for the target genes were hGSTA4: forward 5′-GGA GTC CGT GAG ATG GGT TTT AG-3′ and reverse 5′-TGC TAT GTA GTG GAG AAT GCT TCG-3′; hALDH2: forward 5′-CACTTCGCCCTGTTCTTCAACC-3′ and reverse 5′-CC TGCTCGGTCTTGCTATCAAAG-3′; hFALDH: forward 5′-TACGG GAAACACTGCGGTTG-3′ and reverse 5′-GCAAACAATGTCCA GGTCACAATC-3′; and the reference gene TATA binding protein: forward 5′-AGC GGT TTG CTG CGG TAA TC-3′ and reverse 5′-ACT GTT CTT CAC TCT TGG CTC CTG-3′. All primers were validated in human subcutaneous and VAT, using TATA binding protein as the reference gene.
Data analysis
All comparisons were done using Pearson’s correlations. Statistical analyses were conducted using the statistical package SAS, version 9.2 for Windows (SAS Institute, Cary, NC).
RESULTS
Subject characteristics
There was a narrow age range (25–27 years), but a broad range in BMI, with two subjects of normal weight (BMI <25), three were overweight (BMI 25–30), and three with BMI measurements greater than 40 kg/m2 (Table 1). None had diabetes and none had impaired fasting glucose (≥100 mg/dl) as defined by the American Diabetes Association (25). The subjects exhibited a wide range of fasting insulin, acute insulin response to glucose, Sg (glucose mediated glucose uptake), SI and DI. Characteristics for men and women in the study were similar, with the exception of the ratio of VAT to SAT, which was significantly higher in males relative to females (0.39 ± 0.07 vs. 0.24 ± 0.03, P = 0.031), and serum high-density lipoprotein which was significantly higher in females relative to males (58 ± 3 vs. 37 ± 8, P = 0.012).
Table 1.
Subject characteristics, mean ± s.d.
| Clinical characteristics | All (n = 8) | Females (n = 2) | Males (n = 6) | Range |
|---|---|---|---|---|
| Age (years) | 26 ± 1 | 26 ± 1 | 26 ± 1 | 25–27 |
| BMI (kg/m2) | 32 ± 11 | 31 ± 14 | 33 ± 11 | 22–51 |
| Body fat (%) | 35 ± 14 | 41 ± 25 | 33 ± 11 | 20–59 |
| VAT volume (cm3) | 258 ± 197 | 142 ± 124 | 296 ± 210 | 55–645 |
| SAT volume (cm3) | 724 ± 510 | 647 ± 616 | 750 ± 534 | 211–1,708 |
| VAT/SAT ratio | 0.34 ± 0.10 | 0.24 ± 0.03a | 0.39 ± 0.07a | 0.21–0.52 |
| Cholesterol (mg/dl) | 168 ± 43 | 178 ± 47 | 165 ± 46 | 110–232 |
| LDL (mg/dl) | 105 ± 39 | 100 ± 36 | 107 ± 43 | 51–168 |
| HDL (mg/dl) | 42 ± 12 | 58 ± 3a | 37 ± 8a | 27–60 |
| Triglycerides (mg/dl) | 103 ± 37 | 98 ± 45 | 104 ± 39 | 62–163 |
| FFA (mmol/l) | 0.57 ± 0.22 | 0.65 ± 0.20 | 0.54 ± 0.24 | 0.10–0.79 |
| SBP (mm Hg) | 118 ± 16 | 107 ± 9 | 121 ± 16 | 101–152 |
| DBP (mm Hg) | 68 ± 13 | 63 ± 12 | 69 ± 14 | 54–93 |
| Fasting glucose (mg/dl) | 88 ± 3 | 86 ± 3 | 89 ± 3 | 84–93 |
| Fasting insulin (mU/l) | 7.0 ± 5.0 | 4.5 ± 1.4 | 7.8 ± 5.6 | 3–18 |
| AIRg (mU/l/min) | 490 ± 340 | 370 ± 310 | 520 ± 360 | 140–1,130 |
| SI (min·mU/l)−1 | 4.0 ± 2.5 | 4.7 ± 0.7 | 3.8 ± 2.9 | 0.8–7.3 |
| Sg (min−1) | 0.020 ± 0.007 | 0.024 ± 0.001 | 0.018 ± 0.008 | 0.004–0.028 |
| DI | 1,450 ± 1,050 | 1650 ± 1210 | 1380 ± 1200 | 605–3,560 |
AIRg, acute insulin response to glucose; DBP, diastolic blood pressure; DI, disposition index; FFA, free fatty acid; HDL, high-density lipoprotein; LDL, low-density lipoprotein; SAT, subcutaneous adipose tissue; SBP, systolic blood pressure; Sg, glucose effectiveness; SI, sensitivity index; VAT, visceral adipose tissue.
Denotes significant difference between female and male subjects (P < 0.05).
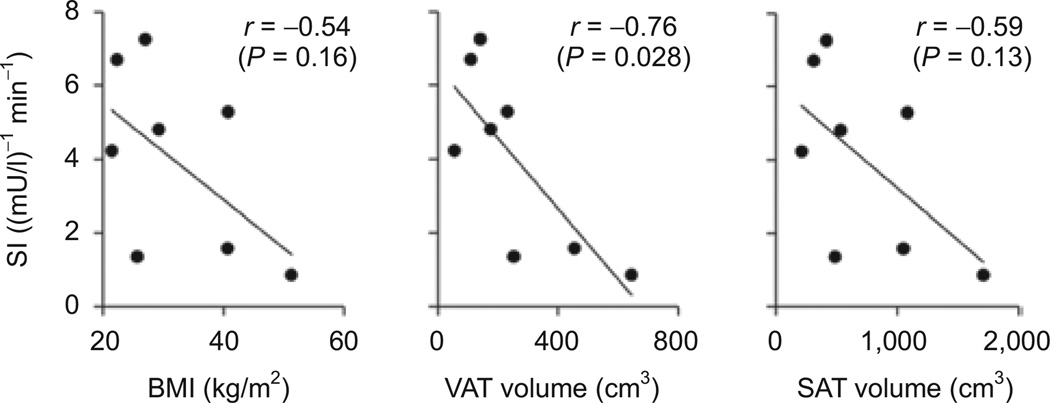
There were strong trends for an inverse correlation between SI and BMI (r = −0.54), VAT (r = −0.71) and SAT (r = −0.59) (Figure 1), but only the correlation with visceral adiposity was significant (P = 0.028) most likely due to the small sample size. DI was inversely related to BMI (r = −0.53, P = 0.17) and FFA levels (r = −0.83, P = 0.011).
Figure 1.
Sensitivity index correlates negatively with BMI, visceral, and subcutaneous fat volume. Visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT) volumes determined by single-slice CT at midabdomen (level of L2–L4). Pearson’s correlation (r) and P value are indicated for each graph.
Carbonylation profile of adipose tissue protein
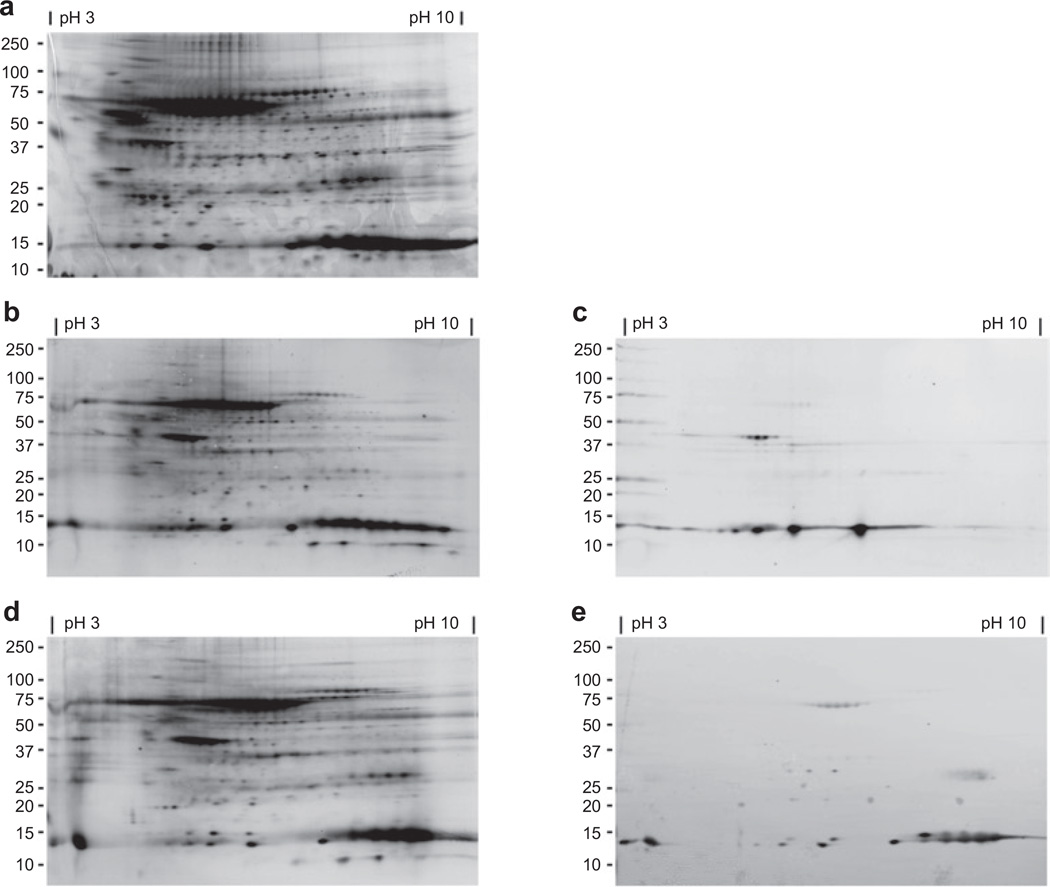
Figure 2a shows the profile of total soluble proteins in adipose tissue separated by both mass (vertically) and isoelectric point (horizontally). Figure 2b,d shows the subset of carbonylated adipose proteins with a prominent species noted at about 67 kDa, likely representing albumin, as well as multiple species at about 15 kDa. Two members of the fatty acid–binding protein multigene family are expressed in adipose tissue: adipocyte fatty acid–binding protein (A-FABP) found primarily in adipocytes, and epidermal fatty acid–binding protein (E-FABP) expressed abundantly in macrophages. Both A-FABP and E-FABP are carbonylated in murine adipose tissue suggesting that the 15 kDa human carbonylated proteins may be FABPs. In order to verify the identity of the carbonylated 15 kDa proteins, the membranes were immunoblotted with anti–A-FABP or anti–E-FABP antibodies (Figure 2c,e). The FABP immunoreactive protein exactly matched the 15 kDa carbonylated protein indicating that both A-FABP and E-FABP are carbonylated in human adipose tissue and that multiple charged species for each protein are modified.
Figure 2.
Two-dimension gel electrophoresis of total soluble protein from human visceral fat. All blots and gel have molecular weight in kilodaltons (kDa) noted along left hand side and pH of range of isoelectric focusing is noted along the top. (a) Total soluble protein stained with Deep Purple. (b) Total carbonylated protein and corresponding immunoblot (c) using anti–A-FABP antibody. (d) Total carbonylated protein and corresponding immunoblot (e) using anti–E-FABP antibody. A-FABP, adipocyte fatty acid–binding protein; E-FABP, epidermal fatty acid–binding protein.
Carbonylation of adipose proteins from study subjects
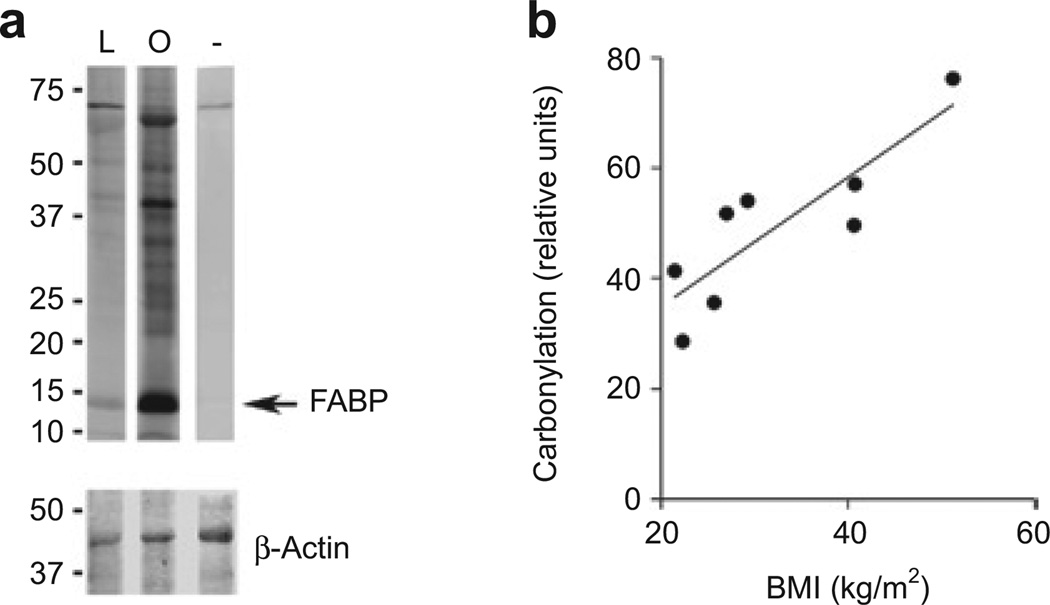
Figure 3a shows a representative blot from one lean and one obese individual where the fatty acid–binding protein band is denoted by an arrow. For each individual, total carbonylation was quantified by analysis of the signal intensity for the entire lane. There was a highly significant positive correlation between total protein carbonylation and BMI (r = 0.86, P = 0.0067) (Figure 3b), VAT (r = 0.71, P = 0.048) and SAT (r = 0.84, P = 0.0098). When carbonylation was measured excluding the 67 kDa albumin band, these relationships were essentially unchanged, with significant positive correlations between carbonylation and BMI (r = 0.85, P = 0.0080), VAT (r = 0.74, P = 0.036) and SAT (r = 0.83, P = 0.010).
Figure 3.
Carbonylation correlates positively with adiposity. (a) Carbonylation of total soluble proteins (upper panel) from a representative lean individual (L) and obese individual (O). Negative control is shown on the right (−). Molecular masses (kDa) of proteins are indicated on the left. The same membrane was blotted for β-actin (lower panel) as a loading control. Image shown is from the same gel taken at the same exposure but composed from lanes not adjacent to each other. (b) Total carbonylated protein was quantified for each individual, normalized to β-actin and plotted relative to BMI (kg/m2). FABP, fatty acid–binding protein.
Relationship of carbonylation to metabolic parameters
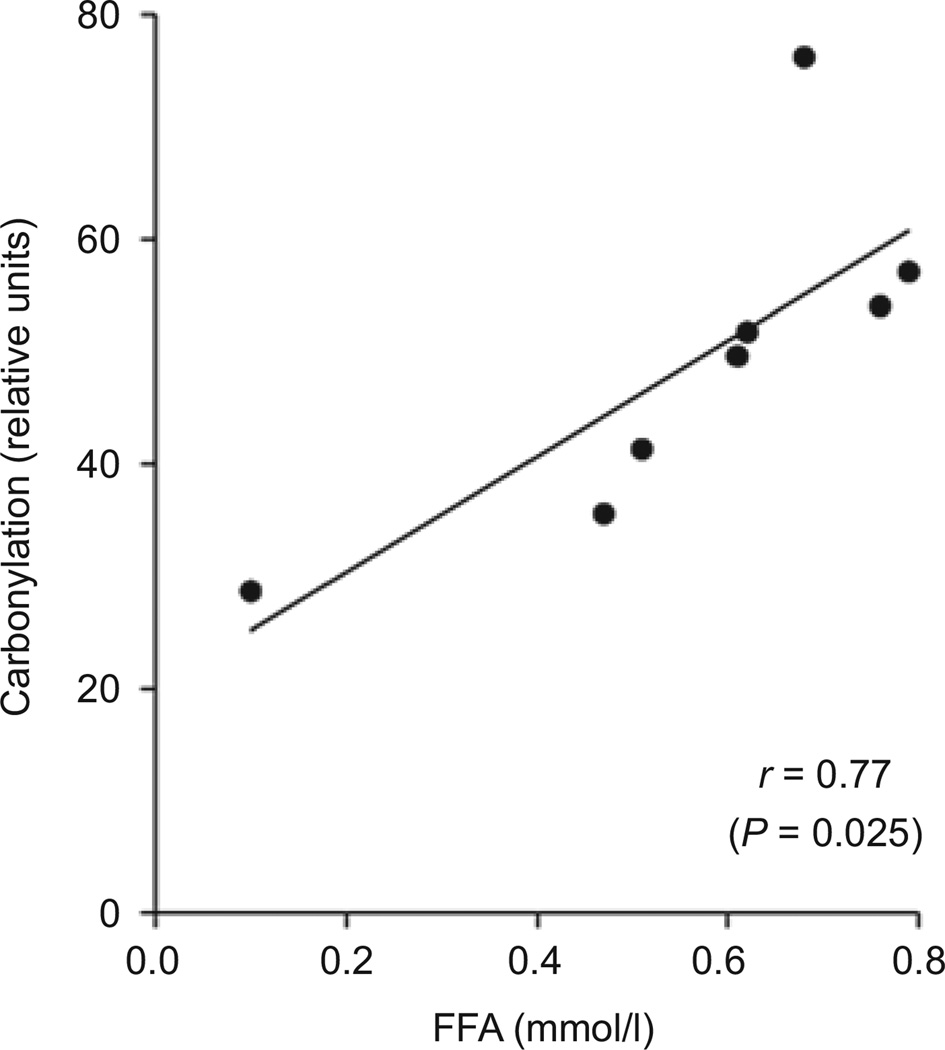
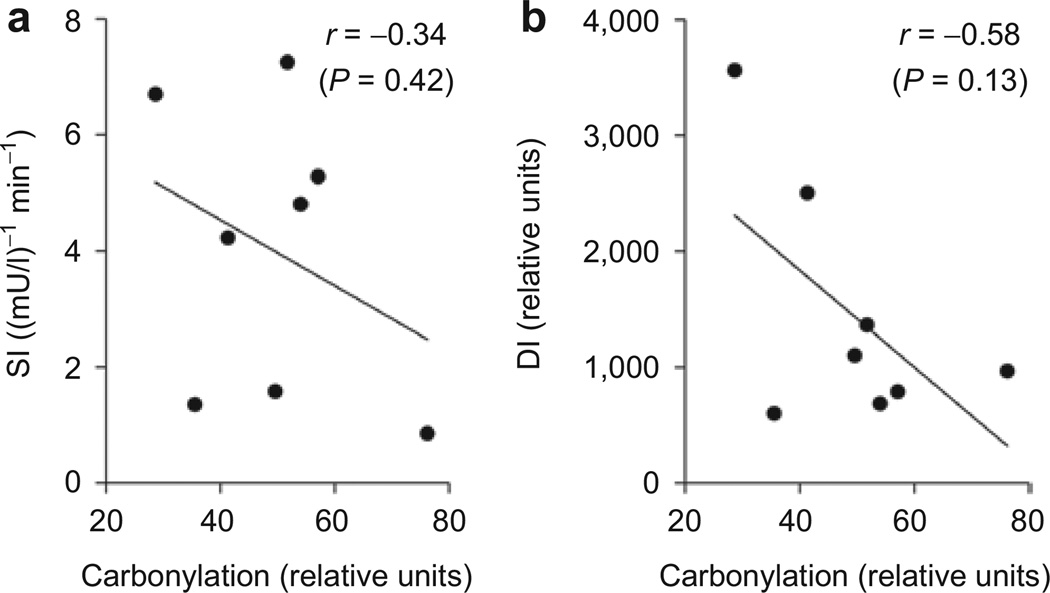
Total carbonylation was significantly related to FFA (r = 0.77, P = 0.025) (Figure 4) and remained significant after adjusting for visceral adiposity (r = 0.80, P = 0.033) and VAT/SAT ratio (r = 0.76, P = 0.048); however the correlation was slightly attenuated and non-significant after adjusting for BMI (r = 0.66, P = 0.11). Fasting insulin also was significantly related to carbonylation (r = 0.74, P = 0.034). It was significant after adjusting for VAT/SAT ratio (r = 0.85, P = 0.014) but was no longer significant after adjusting for visceral adiposity (r = 0.32, P = 0.49) or BMI (r = −0.01, P = 0.98). As seen in Figure 5, the relationship of total carbonylation to DI and SI showed a negative trend but was not statistically significant (r = −0.58, P = 0.13 and. r = −0.34, P = 0.42, respectively). The relationship of DI to total carbonylation became more significant when adjusted for the ratio of visceral to SAT volume (r = −0.67, P = 0.099).
Figure 4.
Relationship of protein carbonylation to serum free fatty acid (FFA) levels. Total protein carbonylation was quantified for each individual, normalized to β-actin, and plotted relative to serum FFA (mmol/l).
Figure 5.
Relationship of protein carbonylation to insulin sensitivity. Total protein carbonylation was quantified for each individual, normalized to β-actin, and plotted relative to (a) insulin sensitivity index (SI) and (b) disposition index (DI). Pearson’s correlation r and P value are indicated for each graph.
Expression of antioxidant genes involved in reactive aldehyde metabolism
The expression of GSTA4, ALDH2 and FALDH, three enzymes linked to oxidative stress and reactive aldehyde metabolism were strongly related to each other and were related to measures of adiposity (Table 2). FALDH significantly correlated with BMI (r = 0.67) and % body fat (r = 0.59). ALDH2 was significantly correlated with % body fat (r = 0.73). GSTA4 was not significantly correlated with any of the three measures of body fatness. The expression of GSTA4, ALDH2 or FALDH was not significantly related to carbonylation (Table 3).
Table 2.
Pearson’s coefficients of correlation between gene expression and BMI, visceral adipose tissue (VAT) volume and total body fat
| Gene | BMI | VAT volume (cm3) | % Body fat |
|---|---|---|---|
| GSTA4 | 0.25 | −0.07 | 0.59 |
| ALDH2 | 0.57 | 0.36 | 0.73* |
| FALDH | 0.67* | 0.45 | 0.83** |
ALDH2, acetaldehyde dehydrogenase 2; FALDH, fatty aldehyde dehydrogenase; GSTA4, glutathione S-transferase A4.
P < 0.10,
P < 0.05.
Table 3.
Pearson’s coefficients of correlation between carbonylation and expression of antioxidant genes
ALDH2, acetaldehyde dehydrogenase 2; FALDH, fatty aldehyde dehydrogenase; GSTA4, glutathione S-transferase A4.
P < 0.05,
P < 0.01,
P < 0.001.
DISCUSSION
Obesity is associated with an increase in inflammation and oxidative stress in adipose tissue (26). Oxidative stress-induced lipid peroxidation results in production of reactive aldehydes that diffuse within cells and across membranes to covalently modify the side chains of histidine, lysine and cysteine residues, a process known as protein carbonylation. Studies in mice have shown that the level of carbonylation is significantly associated with obesity and insulin resistance. The present study shows for the first time the relationship of carbonylation to obesity in humans and demonstrates that multiple protein targets are modified by reactive aldehydes in human adipose tissue and carbonylation occurs to a greater degree in the adipose tissue of obese relative to lean individuals. Moreover, protein carbonylation is strongly related to serum FFA levels, even after adjusting for adiposity. Although not statistically significant, the study also showed r values of −0.34 and −0.58 between carbonylation and DI and SI, respectively, suggesting the need for studies conducted in a larger number of subjects. It should be noted that carbonylation does not necessarily imply a pathogenic process. Increased carbonylation in adipocytes might represent a homeostatic response to increased adipocyte fat and oxidative stress limiting nutrient influx to large cells and diverting carbohydrates and lipids to smaller adipocytes.
As shown in Figure 2, a wide variety of adipose proteins are carbonylated suggesting that modification may result in alteration of multiple metabolic processes. Moreover, since the extent of carbonylation on any one specific target protein is not likely to be stoichiometric, changes in metabolism or signaling are likely to be the result of multiple small changes common to a pathway or process. For example, in aged rat muscle mitochondria (27) carbonylation was detected in multiple enzymes of the electron transport system and even within a particular complex. Multiple carbonylation events on targets within a single pathway, even if each is sub-stoichiometric, may be sufficient to alter overall metabolic flux and/or function. This may be particularly true of targets that are rate-limiting enzymes or regulatory (e.g., protein kinases or phosphatases) in nature.
In humans, defects of mitochondrial metabolism in subcutaneous and VAT have been associated with insulin resistance (28,29). It is known that production of ROS by mitochondria inhibits the proliferation of preadipocytes without inducing apoptosis (reviewed in ref. 30). This could lead to adipocyte hypertrophy resulting in larger, dysfunctional fat cells. Carbonylation may also lead to decreased mitochondrial biogenesis as evidenced by changes in key factors in adipocytes at both the mRNA and protein levels (17) or there may be direct carbonylation of mitochondrial proteins, causing dysfunction or targeting for degradation.
Increased adipose protein carbonylation has been associated with impaired insulin signaling (18) and increased lipolysis in cell culture models (17). Consistent with this, in this study there was a positive association of carbonylation with serum FFA levels that persisted after adjustment for adiposity. Elevated serum FFAs are associated with both peripheral insulin resistance as well as impaired β-cell function in humans (26,31). A-FABP and E-FABP, proteins that mediate lipolysis in adipocytes (14,32), were identified by this study as carbonylation targets in human adipose tissue. Modification of both fatty acid–binding proteins has been shown in mice to occur at a conserved cysteine residue that lies within the lipidbinding cavity (32,33). This cysteine residue is also conserved in human A-FABP (34) and occupation of the lipid-binding site by trans-4-hydroxy-2-nonenal may affect lipolytic rate (14). Beyond lipolysis, there are other potential reasons why carbonylation of FABPs may affect cellular function. As some of the most abundant proteins in adipose tissue, FABPs may play an important role as a scavenger for the reactive aldehyde products of oxidative stress, sequestering them and removing the reactive lipids from the cell. In addition, A-FABP is known to interact with other metabolically important adipocyte proteins, such as hormone sensitive lipase and peroxisome proliferator-activated receptor-γ (31,32). It should be noted that these studies are of whole adipose tissue that consists of inflammatory macrophages, T-cells, stromal-vascular cells and preadipocytes in addition to mature adipocytes. A-FABP is primarily adipocyte in origin while E-FABP is expressed more highly in macrophages. Further study will be needed to determine whether carbonylation is playing an important role in the various cell types found in adipose tissue.
Increased intracellular oxidative stress, possibly related to localized tissue hypoxia, occurs in the adipose tissue of obese individuals (26). The increased levels of ROS likely reflect production of ROS by NADPH oxidase and/or the mitochondria as well as decreased detoxification of ROS by the antioxidant enzyme system. In the present study, the expression of three antioxidant genes, GSTA4, ALDH2 and FALDH, was investigated with the expectation that decreased expression of some or all of these genes would be associated with increased carbonylation as was previously reported in mouse and cell culture studies (13,14). Although there was no significant change in GSTA4 expression with increasing adiposity, the expression of both ALDH2 and FALDH increased with fat mass, possibly representing a counter-regulatory response to increased lipid aldehyde production. However, none of the three was strongly associated with total carbonylation level despite being previously shown to be inversely related to insulin resistance and carbonylation in animal model systems (18,35,36). It should be noted however that none of the patients in the study had diabetes or impaired fasting glucose. While all three enzymes have antioxidant functions, their regulation may be quite different, making simple associations in humans difficult. For example, FALDH, but not GSTA4 or ALDH2, is upregulated by insulin (36).
In summary, the results show for the first time in humans that protein carbonylation in adipose tissue is directly related to BMI and is directly related to serum free fatty levels. These findings are in concert with prior studies in animal models that suggest protein carbonylation is associated with adipose mitochondrial dysfunction and insulin resistance. Further studies will be needed to determine whether there is a significant relationship between carbonylation and disposition index and insulin sensitivity. Additionally, the identification of carbonylation targets and their regulation by oxidative stress may further elucidate the role of this process in obesity-related adipose dysfunction.
ACKNOWLEDGMENTS
This work was supported by National Institute of Health Grants DK084669 (to D.A.B.) and HL52851 (to A.R.S.), the Minnesota Obesity Center (NIH DK050456) and Vikings Pediatric Fellow’s Grant (to B.I.F.). We thank Lori Schafer for her invaluable work in study coordination. We thank the members of the Bernlohr laboratory for helpful comments and suggestions during the preparation of this manuscript. We also thank Todd Markowski at the Center for Mass Spectrometry and Proteomics and the University of Minnesota Supercomputing Institute.
Footnotes
Disclosure
The authors declared no conflict of interest.
REFERENCES
- 1.DeFronzo RA, Bonadonna RC, Ferrannini E. Pathogenesis of NIDDM. A balanced overview. Diabetes Care. 1992;15:318–368. doi: 10.2337/diacare.15.3.318. [DOI] [PubMed] [Google Scholar]
- 2.Hu FB, Manson JE, Stampfer MJ, et al. Diet, lifestyle, and the risk of type 2 diabetes mellitus in women. N Engl J Med. 2001;345:790–797. doi: 10.1056/NEJMoa010492. [DOI] [PubMed] [Google Scholar]
- 3.Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- 4.Sinaiko AR, Steinberger J, Moran A, et al. Relation of body mass index and insulin resistance to cardiovascular risk factors, inflammatory factors, and oxidative stress during adolescence. Circulation. 2005;111:1985–1991. doi: 10.1161/01.CIR.0000161837.23846.57. [DOI] [PubMed] [Google Scholar]
- 5.Steffes MW, Gross MD, Lee DH, Schreiner PJ, Jacobs DR., Jr Adiponectin, visceral fat, oxidative stress, and early macrovascular disease: the Coronary Artery Risk Development in Young Adults Study. Obesity (Silver Spring) 2006;14:319–326. doi: 10.1038/oby.2006.41. [DOI] [PubMed] [Google Scholar]
- 6.Abdul-Ghani MA, DeFronzo RA. Pathogenesis of insulin resistance in skeletal muscle. J Biomed Biotechnol. 2010;2010:476279. doi: 10.1155/2010/476279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bays H, Mandarino L, DeFronzo RA. Role of the adipocyte, free fatty acids, and ectopic fat in pathogenesis of type 2 diabetes mellitus: peroxisomal proliferator-activated receptor agonists provide a rational therapeutic approach. J Clin Endocrinol Metab. 2004;89:463–478. doi: 10.1210/jc.2003-030723. [DOI] [PubMed] [Google Scholar]
- 8.Ye J, Gao Z, Yin J, He Q. Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. Am J Physiol Endocrinol Metab. 2007;293:E1118–E1128. doi: 10.1152/ajpendo.00435.2007. [DOI] [PubMed] [Google Scholar]
- 9.Rausch ME, Weisberg S, Vardhana P, Tortoriello DV. Obesity in C57BL/6J mice is characterized by adipose tissue hypoxia and cytotoxic T-cell infiltration. Int J Obes (Lond) 2008;32:451–463. doi: 10.1038/sj.ijo.0803744. [DOI] [PubMed] [Google Scholar]
- 10.Hosogai N, Fukuhara A, Oshima K, et al. Adipose tissue hypoxia in obesity and its impact on adipocytokine dysregulation. Diabetes. 2007;56:901–911. doi: 10.2337/db06-0911. [DOI] [PubMed] [Google Scholar]
- 11.Trayhurn P, Wood IS. Adipokines: inflammation and the pleiotropic role of white adipose tissue. Br J Nutr. 2004;92:347–355. doi: 10.1079/bjn20041213. [DOI] [PubMed] [Google Scholar]
- 12.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 13.Carini M, Aldini G, Facino RM. Mass spectrometry for detection of 4-hydroxy-trans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spectrom Rev. 2004;23:281–305. doi: 10.1002/mas.10076. [DOI] [PubMed] [Google Scholar]
- 14.Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Hubatsch I, Ridderström M, Mannervik B. Human glutathione transferase A4-4: an alpha class enzyme with high catalytic efficiency in the conjugation of 4-hydroxynonenal and other genotoxic products of lipid peroxidation. Biochem J. 1998;330(Pt 1):175–179. doi: 10.1042/bj3300175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moraes RC, Blondet A, Birkenkamp-Demtroeder K, et al. Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by microarray and reverse transcription-polymerase chain reaction analyses. Endocrinology. 2003;144:4773–4782. doi: 10.1210/en.2003-0456. [DOI] [PubMed] [Google Scholar]
- 17.Curtis JM, Grimsrud PA, Wright WS, et al. Downregulation of adipose glutathione S-transferase A4 leads to increased protein carbonylation, oxidative stress, and mitochondrial dysfunction. Diabetes. 2010;59:1132–1142. doi: 10.2337/db09-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demozay D, Mas JC, Rocchi S, Van Obberghen E. FALDH reverses the deleterious action of oxidative stress induced by lipid peroxidation product 4-hydroxynonenal on insulin signaling in 3T3-L1 adipocytes. Diabetes. 2008;57:1216–1226. doi: 10.2337/db07-0389. [DOI] [PubMed] [Google Scholar]
- 19.Moran A, Jacobs DR, Jr, Steinberger J, et al. Insulin resistance during puberty: results from clamp studies in 357 children. Diabetes. 1999;48:2039–2044. doi: 10.2337/diabetes.48.10.2039. [DOI] [PubMed] [Google Scholar]
- 20.Sinaiko AR, Jacobs DR, Jr, Steinberger J, et al. Insulin resistance syndrome in childhood: associations of the euglycemic insulin clamp and fasting insulin with fatness and other risk factors. J Pediatr. 2001;139:700–707. doi: 10.1067/mpd.2001.118535. [DOI] [PubMed] [Google Scholar]
- 21.van der Kooy K, Seidell JC. Techniques for the measurement of visceral fat: a practical guide. Int J Obes Relat Metab Disord. 1993;17:187–196. [PubMed] [Google Scholar]
- 22.Steil GM, Volund A, Kahn SE, Bergman RN. Reduced sample number for calculation of insulin sensitivity and glucose effectiveness from the minimal model. Suitability for use in population studies. Diabetes. 1993;42:250–256. doi: 10.2337/diab.42.2.250. [DOI] [PubMed] [Google Scholar]
- 23.Muniyappa R, Lee S, Chen H, Quon MJ. Current approaches for assessing insulin sensitivity and resistance in vivo: advantages, limitations, and appropriate usage. Am J Physiol Endocrinol Metab. 2008;294:E15–E26. doi: 10.1152/ajpendo.00645.2007. [DOI] [PubMed] [Google Scholar]
- 24.Pacini G, Bergman RN. MINMOD: a computer program to calculate insulin sensitivity and pancreatic responsivity from the frequently sampled intravenous glucose tolerance test. Comput Methods Programs Biomed. 1986;23:113–122. doi: 10.1016/0169-2607(86)90106-9. [DOI] [PubMed] [Google Scholar]
- 25.Genuth S, Alberti KG, Bennett P, et al. Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care. 2003;26:3160–3167. doi: 10.2337/diacare.26.11.3160. [DOI] [PubMed] [Google Scholar]
- 26.de Ferranti S, Mozaffarian D. The perfect storm: obesity, adipocyte dysfunction, and metabolic consequences. Clin Chem. 2008;54:945–955. doi: 10.1373/clinchem.2007.100156. [DOI] [PubMed] [Google Scholar]
- 27.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 28.Sears DD, Hsiao G, Hsiao A, et al. Mechanisms of human insulin resistance and thiazolidinedione-mediated insulin sensitization. Proc Natl Acad Sci USA. 2009;106:18745–18750. doi: 10.1073/pnas.0903032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlman I, Forsgren M, Sjögren A, et al. Downregulation of electron transport chain genes in visceral adipose tissue in type 2 diabetes independent of obesity and possibly involving tumor necrosis factor-alpha. Diabetes. 2006;55:1792–1799. doi: 10.2337/db05-1421. [DOI] [PubMed] [Google Scholar]
- 30.De Pauw A, Tejerina S, Raes M, Keijer J, Arnould T. Mitochondrial (dys) function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol. 2009;175:927–939. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffer JE. Lipotoxicity: when tissues overeat. Curr Opin Lipidol. 2003;14:281–287. doi: 10.1097/00041433-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 32.Bennaars-Eiden A, Higgins L, Hertzel AV, et al. Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J Biol Chem. 2002;277:50693–50702. doi: 10.1074/jbc.M209493200. [DOI] [PubMed] [Google Scholar]
- 33.Hellberg K, Grimsrud PA, Kruse AC, et al. X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2- nonenal. Protein Sci. 2010;19:1480–1489. doi: 10.1002/pro.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baxa CA, Sha RS, Buelt MK, et al. Human adipocyte lipid-binding protein: purification of the protein and cloning of its complementary DNA. Biochemistry. 1989;28:8683–8690. doi: 10.1021/bi00448a003. [DOI] [PubMed] [Google Scholar]
- 35.Xu F, Chen Y, Lv R, et al. ALDH2 genetic polymorphism and the risk of type II diabetes mellitus in CAD patients. Hypertens Res. 2010;33:49–55. doi: 10.1038/hr.2009.178. [DOI] [PubMed] [Google Scholar]
- 36.Demozay D, Rocchi S, Mas JC, et al. Fatty aldehyde dehydrogenase: potential role in oxidative stress protection and regulation of its gene expression by insulin. J Biol Chem. 2004;279:6261–6270. doi: 10.1074/jbc.M312062200. [DOI] [PubMed] [Google Scholar]