Abstract
Background:
Chinese tree shrew (Tupaia belangeri chinensis) is a small animal that possess many features, which are valuable in biomedical research, as experimental models. Currently, there are numerous attempts to utilize tree shrews as models for hepatitis C virus (HCV) infection.
Objectives:
This study aimed to construct a liver microRNA (miRNA) data of the tree shrew.
Materials and Methods:
Three second filial generation tree shrews were used in this study. Total RNA was extracted from each liver of the tree shrew and equal quality mixed, then reverse-transcribed to complementary DNA (cDNA). The cDNAs were amplified by polymerase chain reaction and subjected to high-throughput sequencing.
Results:
A total of 2060 conserved miRNAs were identified through alignment with the mature miRNAs in miRBase 20.0 database. The gene ontology and Kyoto encyclopedia of genes and genomes analyses of the target genes of the miRNAs revealed several candidate miRNAs, genes and pathways that may involve in the process of HCV infection. The abundance of miR-122 and Let-7 families and their other characteristics provided us more evidences for the utilization of this animal, as a potential model for HCV infection and other related biomedical research. Moreover, 80 novel microRNAs were predicted using the software Mireap. The top 3 abundant miRNAs were validated in other tree samples, based on stem-loop quantitative reverse transcription-polymerase chain reaction.
Conclusions:
According to the liver microRNA data of Chinese tree shrew, characteristics of the miR-122 and Let-7 families further highlight the suitability of tree shrew as the animal model in HCV research.
Keywords: MicroRNAs, Tree Shrew, Liver, High-Throughput Sequencing, Hepatitis C Virus
1. Background
The tree shrew (Tupaia belangeri chinensis) is a kind of bred small animal widely distributed in regions of Southeast Asia and Southwest China. Due to its high-affinity to primates, tree shrew has been proposed as animal model, alternative to primates, in human diseases and drug screening researches, for several decades (1). Currently, there are many attempts to employ tree shrew to create animal models for the research of viral disease. Previous studies have demonstrated that tree shrew could be infected by several hepatitis viruses, especially hepatitis C virus (HCV) (2). However, a great number of obstacles in the establishment of tree shrew hepatitis C animal model still remain, especially the low and unstable infection rates. Therefore, exploring the molecular mechanism on tree shrew’s susceptibility of HCV infection, in the genomic, transcriptional and post-transcriptional levels, is of great importance.
MicroRNAs (miRNAs) are endogenous non-coding RNA molecules found in plants and animals. They functions in transcriptional and post-transcriptional regulation levels, by targeting a diverse set of cellular genes, and regulate multiple biological processes, such as cell proliferation, cell cycle, apoptosis, immune response and cellular response to stress (3). There is increasing evidence that miRNAs play important roles in virus infection, through regulating gene expression in hosts. In vitro studies have found that miRNAs, including miR-122, miR-155, miR-27a, let-7b etc., were able to impact HCV replication, host’s post-infection response and related diseases progression (4). Although the genome and transcriptome data of tree shrew have been made available (5), the sRNA data for tree shrew has not been fully investigated. The data obtained from small RNA analysis may provide valuable information for further understanding of miRNAs in the regulation of HCV infection and will be a useful resource for researches taking tree shrew as hepatitis C animal model.
2. Objectives
To present the liver microRNA data of three second filial generation tree shrews.
3. Materials and Methods
3.1. Tissue Sample Collection and RNA Isolation
Three second filial generation tree shrews were used in this study, which were provided by the Institute of Medical Biology, in Yunnan, China. Those tree shrews selected were all six-month-old and in good physical health. The fresh liver tissues isolated from the three animals were placed immediately into liquid nitrogen and then stored at -80°C, until used. All procedures involving animals were approved by the Animal Care and Use Committee of Kunming University of Science and Technology, Kunming, China. Total RNA was extracted from the liver of each tree shrew using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The purity and concentration of isolated RNA were measured using a Thermo Scientific NanoDrop 2000c spectrophotometer (ThermoFisher Scientific, Waltham, MA, USA) and then the quality was confirmed by electrophoresis on a 1% agarose gel.
3.2. Small RNA Library Construction and Sequencing
All the high-quality RNA samples obtained were quantified and equalized, to guarantee equal amounts of RNA from each animal. The fragments of 18 - 30 nt in length were isolated and purified from the mixed RNA samples, using 15% denaturing polyacrylamide gel electrophoresis (PAGE). Subsequently, a 5’ RNA adaptor and 3’ RNA adaptor were ligated to the purified sRNA, using RNA ligase and reverse-transcribed to cDNA, using the adaptor primers. The cDNA was further amplified and fragments of around 75 bp (small RNA + adaptors) were collected from agarose gels and purified. The DNAs were confirmed by sequencing with Solexa sequencing technology (BGI, Shenzhen, China).
3.3. Data Analysis
All the raw datasets generated by the Solexa deep sequencing were analyzed, as follows. First, the low quality reads, contaminated reads involving 5’ primer contaminants, reads without 3’ primer, reads without the insert tag, reads with poly (A), and reads shorter than 18 nt were removed. The length distribution of the clean reads obtained was then summarized. Next, the clean reads were aligned against the tree shrew genome, using the SOAP (Short Oligonucleotide Alignment Program) (Beijing Genomics Institute, Beijing, China) to analyze their expression and distribution on the genome. Then, the clean reads were compared against the non-coding RNAs (ncRNAs), like ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA), deposited in the NCBI GenBank and the Rfam databases, using Basic Local Alignment Search Tool (BLAST) to annotate the small RNA sequences. Conserved miRNAs in the tree shrew were identified using BLAST at the miRBase (http://www.mirbase.org/). Finally, the unannotated sequences were used to predict potential novel miRNA candidates by Mireap (http://sourceforge.net/projects/mireap/). The potential novel miRNA candidates generated should meet the following criteria, as reported previously (6): minimal miRNA sequence length of 18 nt, maximal miRNA sequence length of 26 nt, minimal miRNA reference sequence length of 20 nt, maximal miRNA reference sequence length of 24 nt, minimal depth of Drosha/Dicer cutting site of three nt, maximal copy number of miRNAs on reference of 20 nt, maximal free energy allowed for a miRNA precursor of -18 kcal/mol, maximal space between miRNA and miRNA* of 35 nt, minimal base pairs of miRNA and miRNA* of 14 nt, maximal bulge of miRNA and miRNA* of four nt, maximal asymmetry of miRNA/miRNA* duplex of five nt and the flank sequence length of miRNA precursor of 10 nt. The secondary structure prediction of miRNA precursors was carried out using RNA Structure 4.6 software (Mathews Lab Software, University of Rochester Medical Center, Rochester, NY, USA).
3.4. Prediction and Functional Analysis of MicroRNA Targets
The targets of the differentially expressed miRNAs were predicted using the database miRecords (http://mirecords.umn.edu/miRecords). To infer the potential functions of the differentially expressed miRNAs, we performed the functional analysis of their target genes using the database for annotation, visualization, and integrated discovery (DAVID). Functional categories were clustered using the functional annotation clustering tool, and representative gene ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, from each clustered set, with a P < 0.05, were selected and taken into consideration for further analysis.
3.5. Novel MicroRNAs Validation
Stem-loop real time-polymerase chain reaction (RT-PCR) was used to validate novel miRNAs. Total RNA was extracted from 10 livers of tree shrews. The cDNA was synthesized from total RNA with specific stem-loop primers. All stem-loop reverse transcriptase-polymerase chain reaction (RT-PCR) primers and miRNA-specific PCR primers are listed in Table 1. Superscript reverse transcriptase was used to process reverse transcriptase reactions that contained 1.5 mg of total RNA and 10 mM stem-loop RT-primer. The 20 μL reactions were incubated in an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Waltham, MA, USA) at 25°C for 5 minutes, 42°C for 1 hour, and 70°C for 15 minutes. The cDNA were diluted 1:10 to perform PCR for validation. The PCR mixtures included 2 μL cDNA, 0.8 mM forward and reverse primers. The 20 μL reactions were incubated in an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Waltham, MA, USA) in a 96-well plate, at 95°C for 5 minutes, followed by 35 cycles of 95 for 15 seconds, 62°C for 30 swconds, and 30 seconds at 72°C. The PCR products were then gel-purified and T-A cloned, and at least three independent clones were sequenced to determine the potential novel miRNAs.
Table 1. The Primer Sequences of Stem-loop Real Time-Polymerase Chain Reactiona.
| miRNA ID and Primers | Sequences |
|---|---|
| tsh-miR-new1 | |
| RT | |
| ATCGGGAAAGTCATGACGCATT | |
| GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAATGCG | |
| Forward | CGCGCGATCGGGAAAGTC |
| Reverse | GTGCAGGGTCCGAGGT |
| tsh-miR-new2 | |
| RT | |
| AAATGCATCAGGACTTTTCA | |
| GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGAAA | |
| Forward | CGCGCGAAATGCATCA |
| Reverse | GTGCAGGGTCCGAGGT |
| tsh-miR-new3 | |
| RT | |
| CAGGTCCCTGTTCAGGCGCCA | |
| GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGGCGC | |
| Forward | CGCGCGCAGGTCCCTGT |
| Reverse | GTGCAGGGTCCGAGGT |
aAbbreviations: miRNA ID, MicroRNA Identification; RT, Reverse Transcriptase; Tsh, Tree Shrew.
4. Results
4.1. Expression Profile of MicroRNAs in Tree Shrew Liver
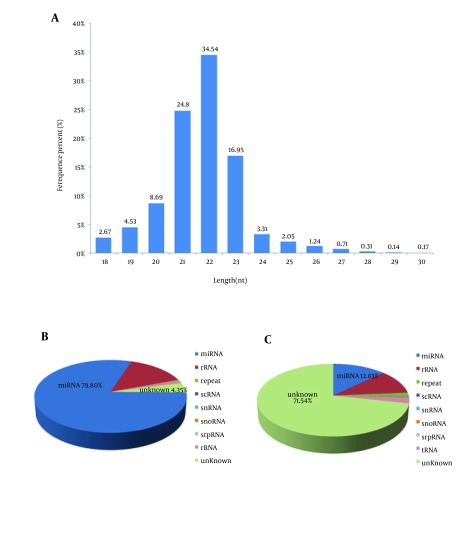
High-throughput sequencing was performed using small RNAs from the liver of the second filial generation tree shrew. After eliminating 3’ adapter null, 5’ adapter contaminants, insert null, smaller than 18 nucleotides (nts) and poly A sequences, we obtained 17503248 high-quality clean reads, accounting for 94.37% of the total raw reads, among which 14207005 reads were mapped to the Tupaia belangeri genome. Further analysis showed the majority of the small RNAs were 21 - 23 nt in size, accounting for 76.29% of the total clean reads (Figure 1A). MicroRNAs comprised 79.8% of the total small RNAs (Figure 1B), which accounted for only 12.8% of the total number of unique small RNAs (Figure 1C).
Figure 1. Profile of the Solexa Sequencing Data.
A, length distribution of sequenced small RNAs. Small RNA reads with a length of 22 nt were the most abundant, followed by 21 and 23 nt reads. B, total sRNA annotations. C, unique sRNA annotations.
4.2. The Similarity of HCV Related MicroRNAs From Tree Shrew and Human
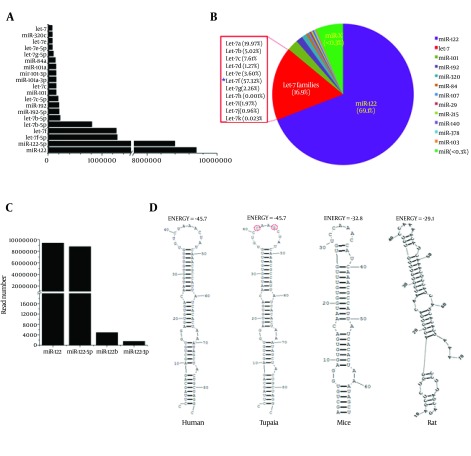
To identify conserved miRNAs in our data, the sorted clean sRNA reads were aligned with the mature miRNAs in miRBase 20.0 (http://www.mirbase.org/). As a result, 2060 unique small RNAs were identified to be matched with the known miRNAs. Expression levels of miRNAs that occupied the top 20 of all identified miRNAs are shown in Figure 2A. The most abundant conserved miRNA identified in the library was miR-122, with more than 9000000 reads, which accounted for 69.1% of the total miRNAs (Figure 2B). Four types of unique small RNA, belonging to the miR-122 family, were identified and nominated as tsh-miR-122, tsh-miR-122-5p, tsh-miR-122b and tsh-miR-122-3p, respectively. Expression levels of these four miRNAs were shown in Figure 2C. The abundance of tsh-miR-122 and tsh-miR-122-5p was much higher than that of tsh-miR-122b and tsh-miR-122-3p. The precursor of tsh-miR-122 was subjected to further analysis, by comparing with miR-122 precursors of 18 animal species, derived from the miRBase. It was shown that the sequence of the precursor of miR-122 of tree shrew was most identical with that of human (Figure 3). In addition, the secondary structure and the energy of primary miRNA of tree shrew for miR-122 were also similar with that of human (Figure 2D).
Figure 2. Characteristics of the Conserved MicroRNA and the MiR-122 Family in Tree Shrew.
A, expression of miRNAs of which the expression level occupied the top 20 of all conserved miRNAs. B, the pie chart of the percentage of miR-122 and Let-7 in the tree shrew liver miRNAs. C, expression of four members of miR-122 family in tree shrew. D, the secondary structures of the four precursors of miR-122 in human, tree shrew, mice and rat.
Figure 3. Sequences Alignment of the MiR-122 Precursors in 18 Tree Shrew Related Species.
The sequences variation compared with human miR-122 precursors was shown in capital letters.
The Let-7 family was ranked the second most abundant miRNA in tree shrew liver, with a presence of 16.9%. Among the Let-7 family, the Let-7f accounted for the largest proportion (57.32%), as shown in Figure 2B, while the Let-7a, Let-7c, Let-7b, Let-7e accounted for 19.91%, 7.61%, 5.02% and 3.60%, respectively. As reported in previous in vitro studies, Let-7b, miR-196, miR-199a and miR-130 were involved in inhibiting HCV infection, whereas miR-122, miR-141, miR-130a and miR-21were found to participate in promoting HCV infection (7). These HCV related miRNAs mentioned above were all found in miRNA data of the tree shrew liver, and the expression level is shown in Figure 4.
Figure 4. Tree Shrew MicroRNAs That Promote Hepacivirus Infection and Inhibit Hepacivirus Infection.
The detailed read number of miRNA sequences is shown in brackets.
4.3. Identification of Novel MicroRNAs in Tree Shrew Liver
Furthermore, the sRNA sequences were aligned to the genome of Tupaia belangeri to identify potential novel miRNAs. Using the miRNA prediction software Mireap (http://sourceforge.net/projects/mireap/), sequences that met the criteria for miRNA were considered to be miRNA precursors. Ultimately, 80 novel miRNAs were identified. However, the expression level of these predicted novel miRNAs was significantly lower than that of the conserved miRNAs. The novel miRNAs, whose read number was > 10, was listed in Table 2 The read number that occupied the top 3 of all the new miRNAs was 233,133,120, respectively (Table 2). The secondary structures of these three microRNAs were shown in Figure 5A. The stem-loop RT-PCR and sequencing methods were used to further validate the novel miRNAs, using liver samples of 10 adult tree shrews. The results showed that the new three miRNA could be amplified by PCR. Moreover, the sequences of the PCR products were consistent with the microRNA data (Figure 5B).
Table 2. Predicted Novel MicroRNAs Whose Read Number was Greater Than 10 in the Liver of Tree Shrewa.
| miRNA | Sequence | Read Number | Location in the Mapped reference Sequence |
|---|---|---|---|
| tsh-miR-new1 | AUCGGGAAAGUCAUGACGCAUU | 233 | GeneScaffold_1371:95107:95193:- |
| tsh-miR-new2 | AAAUGCAUCAGGACUUUUCA | 133 | GeneScaffold_1540:57607:57688:+ |
| tsh-miR-new3 | CAGGUCCCUGUUCAGGCGCCA | 120 | scaffold_111827:44617:44706:+ |
| tsh-miR-new4 | AAAUGCAUCAGGACUUUUCA | 95 | GeneScaffold_3954:101509:101576:+ |
| tsh-miR-new5 | AGAAAAGUCAGGACGUUUUU | 73 | GeneScaffold_1965:8472:8552:- |
| tsh-miR-new6 | UAGGAAAAGGCUUGAUGUAUU | 47 | GeneScaffold_3418:212285:212367:- |
| tsh-miR-new7 | AAAGACAUCAUGACCUUUCU | 44 | GeneScaffold_2862:20340:20417:- |
| tsh-miR-new8 | AAAUGCAUCAGGACUUUUC | 43 | scaffold_143150:13603:13679:+ |
| tsh-miR-new9 | AAAGACAUCAUGACCUUUCU | 42 | GeneScaffold_4000:66284:66361:- |
| tsh-miR-new10 | AUAUAUCAUGACCUUUCUGACA | 41 | GeneScaffold_3348:27549:27629:+ |
| tsh-miR-new11 | GGAAAAGUCAGGACAUAUUUUU | 32 | GeneScaffold_392:237217:237286:- |
| tsh-miR-new12 | CAAGUCCCUGUUCAGGCGCCA | 29 | scaffold_111037:39496:39573:+ |
| tsh-miR-new13 | UCGGAAAAGCCACGACACAUUU | 26 | GeneScaffold_2051:56933:57008:- |
| tsh-miR-new14 | GAAAAAGUCGUGACGCAUUUUU | 24 | scaffold_7441:31821:31888:+ |
| tsh-miR-new15 | UUCGGGAAUGUCGUGGCGUA | 23 | GeneScaffold_1984:111742:111825:+ |
| tsh-miR-new16 | UAGCACUGAGGUAAUGAAGCA | 22 | GeneScaffold_4320:407318:407398:- |
| tsh-miR-new17 | UGUAUCAUGGCUUUUCCGACA | 22 | scaffold_138507:126806:126884:+ |
| tsh-miR-new18 | UGGACAGUCAUGACACAUUUUU | 21 | scaffold_144204:767:841:- |
| tsh-miR-new19 | CAGGGUUUGGGGGUCGAGGGG | 20 | scaffold_49762:463:553:+ |
| tsh-miR-new20 | CAGGGUUUGGGGGUCGAGGGGG | 20 | scaffold_112869:124830:124919:- |
| tsh-miR-new21 | UGGACAGUCAUGACACAUUUUU | 20 | GeneScaffold_958:400490:400569:+ |
| tsh-miR-new22 | ACUGGAAACGUCGUGACAUUUU | 19 | GeneScaffold_39:143382:143461:- |
| tsh-miR-new23 | AAAGUCAUGACGUGUUUUUUC | 18 | GeneScaffold_2493:592697:592794:- |
| tsh-miR-new24 | UGGAAAGUCACGACACAUUUUU | 16 | scaffold_116786:19815:19891:+ |
| tsh-miR-new25 | AGAUGCAUCUCGACUUUCCU | 16 | scaffold_61753:113:191:+ |
| tsh-miR-new26 | AAAAAUACGUCAGGACUUUUC | 16 | GeneScaffold_951:129203:129286:+ |
| tsh-miR-new27 | AUUGGAAAAGUCGUGACAUAUU | 15 | GeneScaffold_1897:86780:86865:- |
| tsh-miR-new28 | AGGAAAGUCAUGCUGCAUUUUU | 14 | GeneScaffold_5176:213958:214037:- |
| tsh-miR-new29 | AAAGAUGCGUCAUGACUUUUC | 13 | GeneScaffold_3849:235632:235709:- |
| tsh-miR-new30 | CAGGUCCCUGUUCAGGCGCC | 13 | scaffold_71896:8426:8502:- |
| tsh-miR-new31 | AAAAAUACGUCAGGACUUUUC | 12 | scaffold_129971:5331:5411:+ |
| tsh-miR-new32 | AGAAAAGUCACGACAUGUUUUU | 12 | scaffold_128528:23932:24001:- |
| tsh-miR-new33 | UACGUCAUGUCUUUUCCGAUGA | 11 | GeneScaffold_2915:52587:52665:- |
| tsh-miR-new34 | AUUGGAAAAGUCGUGACAUAUU | 12 | GeneScaffold_4519:18599:18682:- |
| tsh-miR-new35 | AAAAUAUGUCUUGACUUUCCU | 11 | GeneScaffold_2467:83460:83537:+ |
| tsh-miR-new36 | AAAUACGUUAUGACUUUCCCAA | 11 | GeneScaffold_265:99552:99633:- |
| tsh-miR-new37 | GAGAAAGUCGUGAUGUAUUUUU | 11 | GeneScaffold_3303:423588:423666:- |
| tsh-miR-new38 | AAAGAUGCGUCAUGACUUUUC | 11 | GeneScaffold_383:83204:83279:- |
| tsh-miR-new39 | UUCUGUAGUCUUGGGGUCGCGCUG | 11 | GeneScaffold_4675:153676:153773:- |
| tsh-miR-new40 | AAGGAUGUGAGGCAGGCGAG | 11 | scaffold_11524:3240:3336:- |
| tsh-miR-new41 | AGAAGUCAUGACGUAUUUUUUU | 11 | scaffold_147786:4714:4783:- |
aAbbreviations: miR (NA), MicroRNA; Tsh, Tree Shrew.
Figure 5. The Secondary Structures of the Precursors of Three new MicroRNAs.
A, the secondary structure of the precursors of the new microRNAs was analyzed using the software RNA structure. B, the polymerase chain reaction and sequencing results of the top 3 abundant novel miRNAs in other tree shrew samples.
4.4. Functional Analysis of the MicroRNAs
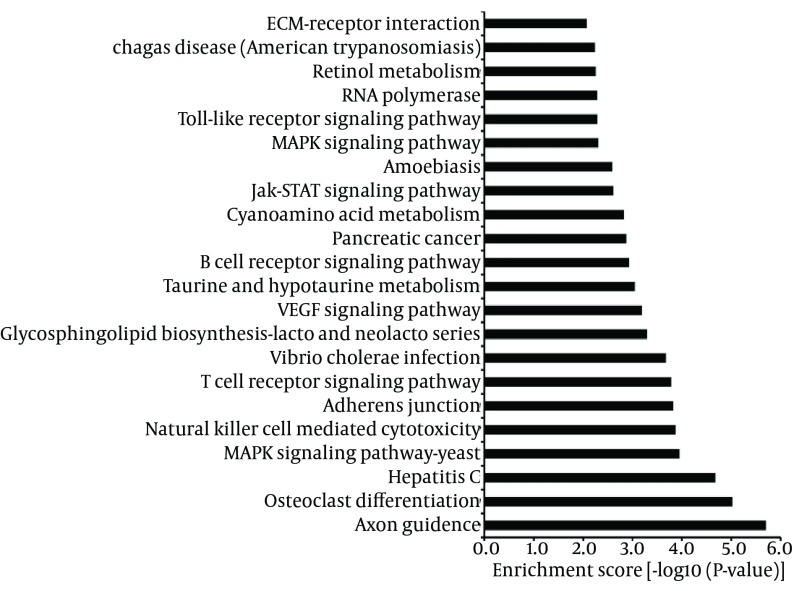
To identify the functions and pathways that the miRNAs and their targets may affect, GO and KEGG pathway analyses were performed. GO analysis showed that 32% of the genes were annotated to biological processes, 34.4% of the genes mapped to molecular function and 33.6% of the genes were composed of cellular component. These genes ranged from elevation of cytosolic calcium ion concentration to regulation of signaling pathway, genes regulating sodium channel activity to genes regulating phosphatase binding and genes regulating microvillus to genes regulating plasma membrane (Figure 6). The findings revealed that predicted targets of miRNAs were associated with a wide variety of biological processes. Furthermore, for KEGG pathway analysis, some classic pathways reported to involve in immunoregulation were found in this study, including Toll-like receptor signaling pathway, B cell receptor signaling pathway, T cell receptor signaling pathway, Natural killer cell mediated cytotoxicity and so on (Figure 7). Intriguingly, Hepatitis C was also enriched from the predicted targets, suggesting that these targets play important roles in the process of infection and inflammation. Taken together, GO and KEGG analyses supply a better understanding of the cellular components, molecular functions and biological processes of target genes, and provided a reference for future study.
Figure 6. The Gene Ontology Enrichment Analysis of Predicted Targets of Differentially Expressed MicroRNAs With P < 0.05.
A total of 36007 putative target genes were assigned to the GO annotations of Biological Process (11519), Cellular Component (12110), and Molecular Function (12378). The GO terms were sorted by the enrichment P value calculated by the calculating formula as previously described (8). A, biological process, B, molecular function, C, cellular component.
Figure 7. The Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of Predicted Targets Differentially Expressed MicroRNAs with P < 0.05.
The enrichment P value calculated by the calculating formula as previously described (8).
5. Discussion
It was estimated that HCV infection causes the liver-related death in more than 300000 people, annually (9). Once exposure to HCV occurs, only a minority of patients clear the acute infection by HCV-specific interferon-γ responses (10), whereas about 80% persist with life-long chronic viremia, if not successfully treated (11). Up to date, there is no vaccine for prevention of HCV infection. Interferon/ribavirin therapy is currently the mainstay in the management of HCV infection. However, the drug has been plagued with excessive toxicity, therapeutic limitations, and restricted patient availability (12). Recently, an oral antiviral agent sofosbuvir has been approved by the US food and drug administration (FDA) and European Union, for the treatment of chronic HCV infection. Although the new anti-HCV drug looks promising, the development of new therapy or preventative vaccines that definitely reduce the global HCV burden remains a formidable challenge.
Chimpanzee is the only animal model for studies of HCV infection and related innate and adaptive host immune responses (9). However, their large adult body size and long reproductive cycle create challenges for in-depth study on mechanism of HCV infection and pathogenicity. Most efforts have been directed towards the study of development of small animal models of HCV infection. Although T-cell and B-cell deficient mice grafted with human hepatocytes could support HCV infection robustly, it could not be used in studies of adaptive immunity (13). Recent studies in developing genetically humanized mice are in process, although these animal models only permit studies of specific steps in the HCV life cycle and have limited or no viral replication (14).
Genome sequencing of the Chinese tree shrew and comparison with 14 other species, including six primate species, showed that the tree shrew was clustered with primate species (5). Our previous studies had proved that CD81, scavenger receptor class B1, claudin-1 and occludin, the considered HCV receptors of tree shrew, have the mediating function to support HCV infection (15). Therefore, multiple attempts are made to employ tree shrew to create animal models for studying HCV infection (16, 17). However, although the tree shrew has the ability to be infected, the low infection rate and instable infection effect are still complicated issues, urgently needing to be addressed. Here, three second filial generation tree shrews were used to generate the liver miRNA data, which could help us in a future study to investigate the mechanisms of HCV infection in tree shrew.
MicroRNAs are important players in the establishment of HCV infection replication and its propagation in infected hepatocytes. Deregulated expression of miRNAs has been linked to the pathogenesis associated with HCV infection, by controlling the process of cell proliferation, apoptosis and migration (4). Therefore, the miRNA data will provide us more information for the utilization of tree shrew, as an animal for HCV research. The current study was designed to profile the general expression of miRNAs in the liver of tree shrew. A total of 2060 conserved miRNAs and 80 novel microRNAs were identified, and miR-122 was the most abundant miRNA in the liver of tree shrew. The miR-122 was also identified as a liver-specific, well-conserved and the most abundant microRNA in liver, accounting for about 70% in the adult liver. Many studies have previously demonstrated that miR-122 is bind to two different sites in the 5’ untranslated region (UTR) of the HCV genome and promotes viral RNA accumulation. The miR-122 plays an important positive role in the regulation of HCV replication and is a crucial target for anti-HCV therapy (18). Therefore, several miR-122 inhibitors become potential drug candidates for the treatment of HCV infection. However, miR-122 also has an essential role in maintaining liver homeostasis and differentiation, while a downregulated expression of miR-122 has been associated with liver disease. Recent studies demonstrated that decreased miR-122 have been associated with poor prognosis and metastasis of liver cancer and can even promote hepatocarcinogenesis (19). The findings mentioned above indicate that the role of miR-122 is controversial. To clarify the role of miR-122 in liver disease, it is imperative to find an applicable animal-model of HCV infection, to further research the molecular mechanisms of miR-122 in liver disease. In the current study, we demonstrated that the miRNA data generated in tree shrew was consistent with previous studies in humans. Further comparison of the secondary structures of miR-122 precursors, among tree shrew, human, mouse and rat, revealed the close relationship between tree shrew and human. Therefore, based on the evidences of the high expression level of miR-122 in the liver of tree shrew, the similarity of the secondary structure of miR-122 between tree shrew and human, and the link between miR-122 and HCV processing, it further supported tree shrew as an potential animal model for HCV infection. In addition, our findings also suggest that tree shrew may be a potential animal model to better understand the molecular mechanisms of miR-122 in liver disease caused by HCV infection in vivo.
Another interesting finding in our study was that half of the top 20 small RNAs, sorting by expression level, belonged to let-7 family. Previous studies found that the let-7 family was also strongly associated with HCV infection. Several members of let-7 family (has-let-7a, has-let-7b, has-let-7c, has-let-7d, has-let-7e, has-let-7f, has-let-7g, has-let-7i) had the potential to repress the expression of the HCV core protein in vitro (20). Another research found that Let-7b repressed HCV replication by interaction with the coding sequences of NS5B and 5’-UTR of HCV genome that were conserved among various HCV genotypes (21). Therefore, further studies on the roles of the let-7 family in HCV replication, in tree shrew, are helpful for the research on HCV infection.
Taken together, tree shrew has been proposed as a valid experimental animal to replace primates for studying HCV infection. However, there are limited usages of this animal in the field, because of the unclear mechanism on HCV infection. The miRNA data we generated and the recently annotated genome sequence of the Chinese tree shrew offer an opportunity to decipher the genetic basis of the tree shrews’ suitability, as an animal model in HCV research.
The second filial generation tree shrew, used in our study, underwent elaborate biological evolution. The indexes of microbiology, virology and parasitology can be precisely controlled. In addition, they have the advantages of relatively clear genetic background and strong disease resistance. However, there are several limitations in our current study. Due to the poor breeding potential, only three livers of tree shrews, which were 6-month-old, were used to generate the relatively limited miRNA data. To accurately characterize the expression profiles of miRNA in tree shrew, further studies, involving more liver tissue samples of different growth and development periods of tree shrew, would be necessary.
Acknowledgments
We thank Prof. Zhengming He from The National Institutes for Food and Drug Control, Beijing, China, for his kind suggestion.
Footnotes
Authors’ Contributions:All the authors gave final approval for this manuscript. Yue Feng, Yue-Mei Feng performed the experiments, analyzed sequences data and wrote the manuscript; Caixia Lu, Xiaomei Sun and Jiejie Dai breed animal and prepared the liver sample; Yang Feng, Lubiao Zhong and Li Liu attended the experiments or data analysis; Xueshan Xia and Jiejie Dai conceived the study and made the revision.
Funding/Support:This work was funded by research grants from The National Science and Key Technology Support Program (2014BAI01B01).
References
- 1.Cao J, Yang EB, Su JJ, Li Y, Chow P. The tree shrews: adjuncts and alternatives to primates as models for biomedical research. J Med Primatol. 2003;32(3):123–30. doi: 10.1034/j.1600-0684.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- 2.Xie ZC, Riezu-Boj JI, Lasarte JJ, Guillen J, Su JH, Civeira MP, et al. Transmission of hepatitis C virus infection to tree shrews. Virology. 1998;244(2):513–20. doi: 10.1006/viro.1998.9127. [DOI] [PubMed] [Google Scholar]
- 3.Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184(5):2369–76. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrivastava S, Mukherjee A, Ray RB. Hepatitis C virus infection, microRNA and liver disease progression. World J Hepatol. 2013;5(9):479–86. doi: 10.4254/wjh.v5.i9.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan Y, Huang ZY, Cao CC, Chen CS, Chen YX, Fan DD, et al. Genome of the Chinese tree shrew. Nat Commun. 2013;4:1426. doi: 10.1038/ncomms2416. [DOI] [PubMed] [Google Scholar]
- 6.Yuan C, Wang X, Geng R, He X, Qu L, Chen Y. Discovery of cashmere goat (Capra hircus) microRNAs in skin and hair follicles by Solexa sequencing. BMC Genomics. 2013;14:511. doi: 10.1186/1471-2164-14-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta P, Cairns MJ, Saksena NK. Regulation of gene expression by microRNA in HCV infection and HCV-mediated hepatocellular carcinoma. Virol J. 2014;11:64. doi: 10.1186/1743-422X-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song Y, An X, Zhang L, Fu M, Peng J, Han P, et al. Identification and profiling of microRNAs in goat endometrium during embryo implantation. PLoS One. 2015;10(4):e0122202. doi: 10.1371/journal.pone.0122202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houghton M. The long and winding road leading to the identification of the hepatitis C virus. J Hepatol. 2009;51(5):939–48. doi: 10.1016/j.jhep.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Flynn JK, Sacks-Davis R, Higgs P, Aitken C, Moneer S, Suppiah V, et al. Detection of HCV-Specific IFN-gamma Responses in HCV Antibody and HCV RNA Negative Injecting Drug Users. Hepat Mon. 2014;14(1):e29053. doi: 10.5812/hepatmon.14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoofnagle JH, di Bisceglie AM. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336(5):347–56. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 12.Kayali Z, Schmidt WN. Finally sofosbuvir: an oral anti-HCV drug with wide performance capability. Pharmgenomics Pers Med. 2014;7:387–98. doi: 10.2147/PGPM.S52629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kneteman NM, Toso C. In vivo study of HCV in mice with chimeric human livers. Methods Mol Biol. 2009;510:383–99. doi: 10.1007/978-1-59745-394-3_29. [DOI] [PubMed] [Google Scholar]
- 14.Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142(6):1279–1287 e3. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Tong Y, Zhu Y, Xia X, Liu Y, Feng Y, Hua X, et al. Tupaia CD81, SR-BI, claudin-1, and occludin support hepatitis C virus infection. J Virol. 2011;85(6):2793–802. doi: 10.1128/JVI.01818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amako Y, Tsukiyama-Kohara K, Katsume A, Hirata Y, Sekiguchi S, Tobita Y, et al. Pathogenesis of hepatitis C virus infection in Tupaia belangeri. J Virol. 2010;84(1):303–11. doi: 10.1128/JVI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Chen H, Cao X, Ben K. Efficient infection of tree shrew (Tupaia belangeri) with hepatitis C virus grown in cell culture or from patient plasma. J Gen Virol. 2007;88(Pt 9):2504–12. doi: 10.1099/vir.0.82878-0. [DOI] [PubMed] [Google Scholar]
- 18.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84(13):6615–25. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122--a key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–57. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J, et al. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013;87(17):9707–18. doi: 10.1128/JVI.00802-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng JC, Yeh YJ, Tseng CP, Hsu SD, Chang YL, Sakamoto N, et al. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci. 2012;69(15):2621–33. doi: 10.1007/s00018-012-0940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]