Abstract
Background:
Germ-line mutations of BRCA1 and BRCA2 genes are responsible for approximately 25–30% of dominantly inherited familial breast cancers; still a big part of genetic component is unknown. The aim of this study was to investigate genetic causes of familial breast cancer in a pedigree with recessive pattern of inheritance.
Methods:
We applied exome sequencing as a useful approach in heterogeneous diseases gene identification in present study for familial breast cancer. Sanger sequencing was applied for validation and segregation analysis of mutations.
Results:
Here, we describe a family with three affected sisters of early-onset invasive ductal carcinoma due to heterozygous frame shift mutation rs80359352 in BRCA2 gene as the first report in Iranian patients in association with a novel missense SNP of STK11 (p.S422G). These mutations are inherited from their normal father.
Conclusion:
Despite apparent recessive pattern of inheritance a dominant gene (here BRCA2) can be involved in pathogenesis of hereditary breast cancer which can be explained by incomplete penetrance of BRCA2 mutations.
Keywords: BRCA2, Familial breast cancer, rs80359352, STK11, Iran
Introduction
Breast cancer is the most common malignancy affecting women worldwide and its incidence rate is increasing in both developed and developing countries in recent years (1). Hereditary breast cancer is defined by significant familial aggregation of breast cancer, which includes approximately 5% to 10% of cases diagnosed with breast cancer (2). In hereditary breast cancer, BRCA1 and BRCA2 germline mutations explain ∼25 % of the cases (3) and thus over 70 % of the cases are not associated with such mutations (4).
Both BRCA1 and BRCA2 genes are involved in DNA double strand breaks (DSBs) repair. In eukaryotes, two major pathways exist to repair DSB: non-homologous end joining (NHEJ) and homology-directed recombination (HR). BRCA2 is involved mainly, if not solely, in HR by “gene conversion” (GC), through its interaction with the essential DSB repair protein RAD51 (5).
“In BRCA2 gene, most of mutations occur in exons 10 and 11 and usually include insertions or deletions, which raise the missense alterations and premature stop codon ending in truncated and nonfunctional protein” (6). Three main functional segments of BRCA2 protein including double strand break domain (DBD), nuclear localization signal (NLS), and Rad-51 binding motif are located in C-terminal of the protein (7).
Massively parallel sequencing is a good strategy for the identification of causative genes of monogenic diseases (8–10) or diseases with a high degree of genetic heterogeneity (11). This technology has provided a new revolution for genetic analysis and has begun to allow resequencing at single-nucleotide resolution of entire normal human and cancer genomes at a relatively low cost (12).
We aimed to investigate genetic causes of familial breast cancer in a pedigree with recessive pattern of inheritance.
Materials and Methods
Family description
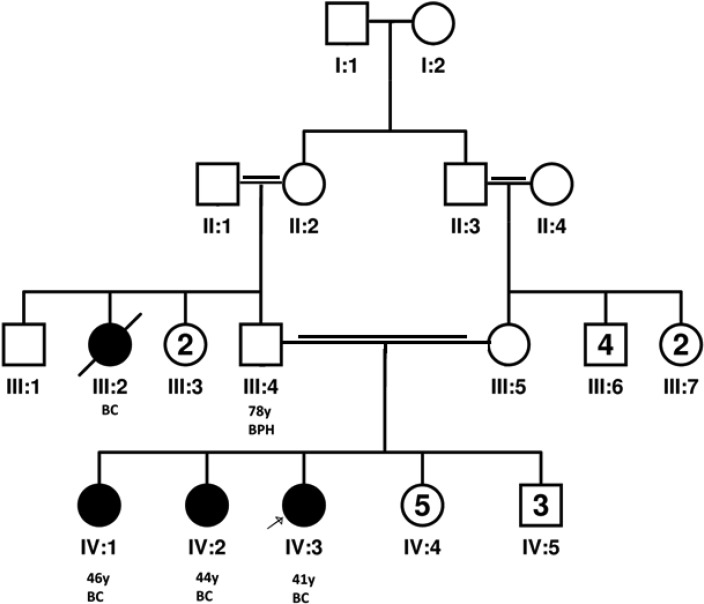
This study was conducted in Tarbiat Modares University Tehran, Iran in 2013. The proband of the family was a 41 year-old Arab female, the sixth daughter of a consanguineous marriage. The clinical signs in the proband began at age 38, and clinical investigations led to the diagnosis of Invasive medullary and ductal carcinoma, and the patient underwent left breast mastectomy. The tumor was a grade II lesion, with positive receptors for estrogen and progesterone, stage T3N1M1, and with negative P53 and Ki67 positivity of about 20% in both tumor types.
Positive family history of cancer in proband consists of her aunt who was deceased and two elder sisters, 46 and 44 year-old women respectively, both had left invasive ductal carcinoma combined with medullary features in elder sister. Tumor tissues were surgically removed at age 38 and 36 respectively.
The family pedigree is illustrated in Fig.1. In this family, who live in South-west Iran, there is no family history of cancer in the maternal branch. Our research has been approved by the Ethical Committee of Tarbiat Modares University, Tehran, Iran. Written informed consent was obtained from all participants involved in this study.
Fig.1:
Family pedigree
Sample preparation
Peripheral blood sampling was performed from three affected and two healthy siblings and their parents. DNA was extracted using the conventional salting-out method. TheOD260/OD280 values of the extracted samples were measured using a UV spectrophotometer. A value of approximately 1.8 indicates DNA of high purity; a value greater than 1.9 suggests contaminating RNA; and a value less than 1.6 indicates the presence of proteins and other contaminants.
Whole exome sequencing
Whole exome sequencing of the subjects was performed at McGill University and Genome Quebec Innovation Centre. Briefly, 3 μg of genomic DNA was used to perform exome capture with Agilent SureSelect kit following the manufacturer’s instructions. 100bp paired-end sequencing was performed on Illumina HiSeq2000 platform.
Bioinformatic analysis
Sequencing reads were aligned to hg19 reference genome using the BWA software (13). Variant discovery and genotype calling of multi-allelic substitutions, insertions and deletions was performed on all individuals globally using the Unified Genotyper module from Genome Analysis Toolkit (GATK) (14) with the minimum call quality parameter set to 30. SAMtools and Annovar were used to call and annotate variants (15–16). Variants with an allele frequency higher than 5% in the 1000 genomes database (http://www.1000genomes.org), higher than 5% in the NHLBI exomes (Exome Variant Server, NHLBI GO Exome Sequencing Project, Seattle, WA; URL: http://evs.gs.washington.edu/EVS/, v.0.0.14, June 20, 2012) or higher than 1% in in-house control database (which consists of about 1000 non-cancer exomes) were filtered out. We applied a recessive genetic model which required the variant to be present in all affected and healthy people in homozygous and heterozygous state respectively, within the family. We used computational tools including PolyPhen-2, SIFT and Mutation Taster to predict the potential impact of sequence variants on protein function. We also obtained conservation scores using GERP and Phast Cons to predict mutation impact based on evolutionary constraint analyses.
Validation of mutations and Segregation analysis
To validate the causal mutations and segregation of the mutations with the disease in the family, direct Sanger sequencing was performed in family members (affected and unaffected) from whom DNA was available. Primers were designed using GENERUNR v3.4.0.0 to surround the candidate mutation and the amplified targets (forward and reverse) were sequenced by standard Sanger sequencing technique using BigDye® Terminator (Invitrogen, ABI, Foster City, CA). Primer sequences are available upon request.
Results
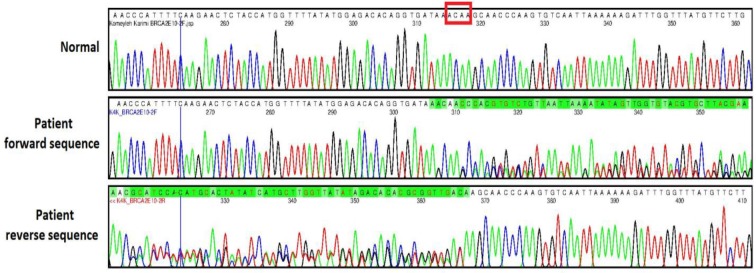
Whole Exome Sequencing revealed, a 4bp frameshift deletion (BRCA2:NM_000059:exon11-:c.2808_2811del:p.936_937del) at position chr13:32-911297 that results in the insertion of 20 novel amino acids before a premature stop codon. Sanger sequencing confirmed that the three affected sisters and their father shared the same mutation which has been shown in Fig.2 . In addition we identified a novel missense heterozygous missense variant p.S422G in STK11 gene. This latter variant is predicted to be benign by Polyphen-2 but it segregates along with BRCA2 mutation in the family.
Fig.2:
Sanger sequencing of normal homozygous sibling and heterozygous patient for BRCA2 mutation with forward and reverse primers
Discussion
In this study, we identified rs80359352 in BRCA2 gene in heterozygous form and report it for the first time in Iranian patients. Both BRCA1 and BRCA2 are implicated in the repair of DNA by homologous recombination. BRCA2 can interact directly with RAD51, both through its BRC repeats and through a domain in its carboxyl terminus (17). The detected mutation in this family result in premature stop codon, which follows by non-sense, mediated mRNA decay. Since BRCA2 is a tumor suppressor gene the second hit must have occurred in tumor tissue.
rs80359352 is a frequent mutation of BRCA2 gene which has been previously reported in several populations but we report it for the first time in Iranian patients. Also we showed the segregation of this mutation in a family with hereditary breast cancer through paternal line.
Studies of families with BRCA2 mutations suggest that the spectrum of cancer manifestation is wide for male carriers. In particular, the total cancer risk to male carriers of BRCA2 mutations is high before age 65 years, largely attributable to excesses of breast, prostate, and pancreatic cancers (18). However, the father of this family is a healthy 78 years old man just affected with Benign Prostatic Hyperplasia (BPH) probably due to his age. In this regard it is suggested that modifying genetic and environmental factors may significantly influence the penetrance of male breast cancer and female breast cancer in individuals carrying germline BRCA2 mutations in some populations (19). Besides, paternal origin of mutation affects breast cancer penetrance and confers an earlier age at breast cancer diagnosis compared with maternal origin (20). This can be seen in our family with three affected patients under the age of 40 years.
Interestingly, despite we have detected BRCA2 mutation as the cause of hereditary breast cancer in this family, the pedigree has apparent recessive pattern where susceptibility to familial breast cancer is thought not to be conferred by a highly penetrant dominant gene (like BRCA2). As a result, one cannot have appropriate assessment of transmitting genetic factors just based on the family pedigree.
In addition we have identified a missense variant p.S422G in STK11 gene. Peutz-Jeghers syndrome (PJS) is a rare autosomal dominant disease due to mutations in the tumor suppressor gene STK11. PJS is associated with a significant increase in cancer risk (relative risk of 89% over the life according to the most recent series). Digestive cancers are the most frequent with cumulative incidences of 55% for gastro-intestinal cancer. There is also an increased risk of non-digestive cancers. In particular, the risk of breast cancer is similar to that of patients carrying deleterious BRCA1 or BRCA2 mutations (cumulative incidence of 45%) (21).
Although, the identified novel variant of STK11 gene is predicted to be benign by Polyphen-2 but it segregates along with BRCA2 mutation in the family. Thus, investigation of allele frequency of this novel variant in control group can be imperative if this gene is involved in disease process.
Conclusion
The present results confirm incomplete penetrance of BRCA2 mutations in male carriers. In addition the co-segregation of STK11 novel variant alongside BRCA2 mutation in the family suggests a possible cooperation of these two genes in disease pathogenesis.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
The authors thank the patients for their assistance. The authors declare that there is no conflict of interests.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. (2011). Global cancer statistics. CA Cancer J Clin, 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Arason A, Gunnarsson H, Johannesdottir G, Jonasson K, Bendahl PO, Gillanders EM, Agnarsson BA, Jonsson G, Pylkas K, Mustonen A, Heikkinen T, Aittomaki K, Blomqvist C, Melin B, Johannsson OT, Moller P, Winqvist R, Nevanlinna H, Borg A, Barkardottir RB. (2010). Genome-wide search for breast cancer linkage in large Icelandic non-BRCA1/2 families. Breast Cancer Res, 12: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Didraga MA, van Beers EH, Joosse SA, Brandwijk KI, Oldenburg RA, Wessels LF, Hogervorst FB, Ligtenberg MJ, Hoogerbrugge N, Verhoef S, Devilee P, Nederlof PM. (2011). A non-BRCA1/2 hereditary breast cancer sub-group defined by aCGH profiling of genetically related patients. Breast Cancer Res Treat, 130: 425–36. [DOI] [PubMed] [Google Scholar]
- 4. Sabatier R, Adelaide J, Finetti P, Ferrari A, Huiart L, Sobol H, Chaffanet M, Birnbaum D, Bertucci F. (2010). BARD1 homozygous deletion, a possible alternative to BRCA1 mutation in basal breast cancer. Genes Chromosomes Cancer, 49: 1143–51. [DOI] [PubMed] [Google Scholar]
- 5. Lacroix M, Leclercq G. (2005). The “portrait” of hereditary breast cancer. Breast Cancer Res Treat, 89( 3): 297–304. [DOI] [PubMed] [Google Scholar]
- 6. Karami F, Mehdipour P. (2013). A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int, 2013; 2013: 928562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pisano M, Mezzolla V, Galante MM, Alemanno G, Manca C, Lorusso V, Malvasi A, Tinelli A. (2011). A new mutation of BRCA2 gene in an Italian healthy woman with familial breast cancer history. Fam Cancer, 10: 65–71. [DOI] [PubMed] [Google Scholar]
- 8. Heron SE, Smith KR, Bahlo M, Nobili L, Kahana E, Licchetta L, Oliver KL, Mazarib A, Afawi Z, Korczyn A, Plazzi G, Petrou S, Berkovic SF, Scheffer IE, Dibbens LM. (2012). Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet, 44( 11): 1188–90. [DOI] [PubMed] [Google Scholar]
- 9. Horani A, Druley TE, Zariwala MA, Patel AC, Levinson BT, Van Arendonk LG, Thornton KC, Giacalone JC, Albee AJ, Wilson KS, Turner EH, Nickerson DA, Shendure J, Bayly PV, Leigh MW, Knowles MR, Brody SL, Dutcher SK, Ferkol TW. (2012). Whole-exome capture and sequencing identifies HEATR2 mutation as a cause of primary ciliary dyskinesia. Am J Hum Genet, 91: 685– 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campeau PM, Lu JT, Sule G, Jiang MM, Bae Y, Madan S, Hogler W, Shaw NJ, Mumm S, Gibbs RA, Whyte MP, Lee BH. (2012). Whole-exome sequencing identifies mutations in the nucleoside transporter gene SLC29A3 in dysosteosclerosis, a form of osteopetrosis. Hum Mol Genet, 21: 4904– 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Comino-Méndez I, Gracia-Aznárez FJ, Schiavi F, et al. (2011). Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet, 43( 7): 663–7. [DOI] [PubMed] [Google Scholar]
- 12. Natrajan R, Reis-Filho JS. (2011). Next-generation sequencing applied to molecular diagnostics. Expert Rev Mol Diagn, 11( 4): 425–44. [DOI] [PubMed] [Google Scholar]
- 13. Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res, 20: 1297– 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. (2009). Genome Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang K, Li M, Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res, 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Narod SA, Foulkes WD. (2004). BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer, 4: 665–76. [DOI] [PubMed] [Google Scholar]
- 18. Liede A, Karlan BY, Narod SA. (2004). Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. J Clin Oncol, 22: 735– 42. [DOI] [PubMed] [Google Scholar]
- 19. Syrjakoski K, Kuukasjarvi T, Waltering K, Haraldsson K, Auvinen A, Borg A, Kainu T, Kallioniemi OP, Koivisto PA. (2004). BRCA2 mutations in 154 finnish male breast cancer patients. Neoplasia, 6: 541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bernholtz S, Laitman B, Fau-Kaufman Y, Kaufman S, Fau-Paluch Shimon B, Paluch Shimon E, Fau – Friedman S, Friedman E. (2011). Cancer risk in Jewish BRCA1 and BRCA2 mutation carriers: effects of oral contraceptive use and parental origin of mutation. Breast Cancer Res Treat, 129( 2): 557–63 [DOI] [PubMed] [Google Scholar]
- 21. Turpin A, Cattan S, Leclerc J, Wacrenier A, Manouvrier-Hanu S, Buisine MP, Lejeune-Dumoulin S. (2014). Hereditary predisposition to cancers of the digestive tract, breast, gynecological and gonadal: Focus on the Peutz-Jeghers. Bull Cancer, 101 ( 9): 813–822. [DOI] [PubMed] [Google Scholar]