Abstract
Comparative genomics, metagenomics, and single-cell technologies have shown that populations of microbial species encompass assemblages of closely related strains. This raises the question of whether individual bacterial lineages respond to the presence of their close relatives by modifying their gene expression or, instead, whether assemblages simply act as the arithmetic addition of their individual components. Here, we took advantage of transcriptome sequencing to address this question. For this, we analyzed the transcriptomes of two closely related strains of the extremely halophilic bacterium Salinibacter ruber grown axenically and in coculture. These organisms dominate bacterial assemblages in hypersaline environments worldwide. The strains used here cooccurred in the natural environment and are 100% identical in their 16S rRNA genes, and each strain harbors an accessory genome representing 10% of its complete genome. Overall, transcriptomic patterns from pure cultures were very similar for both strains. Expression was detected along practically the whole genome albeit with some genes at low levels. A subset of genes was very highly expressed in both strains, including genes coding for the light-driven proton pump xanthorhodopsin, genes involved in the stress response, and genes coding for transcriptional regulators. Expression differences between pure cultures affected mainly genes involved in environmental sensing. When the strains were grown in coculture, there was a modest but significant change in their individual transcription patterns compared to those in pure culture. Each strain sensed the presence of the other and responded in a specific manner, which points to fine intraspecific transcriptomic modulation.
INTRODUCTION
Bacterial species appear in nature as assemblages of closely related strains with extensive genetic heterogeneity, as revealed by comparative genomic analyses of isolates, metagenomics, and, more recently, single-cell genome analyses (1). However, data from genomic analyses of individual genomes from isolated representatives are inconclusive with respect to the ecological and functional significance of this variation, given that subtle genetic variations can lead to distinct ecological strategies (2, 3). In addition, genetic inventories per se are only lists of the capabilities of bacteria and do not provide information on their actual behavior in nature.
Salinibacter ruber is an extremely halophilic bacterium of the phylum Bacteroidetes that is present in hypersaline environments worldwide. Its discovery changed the paradigm that only extremely halophilic Archaea could thrive in close-to-saturation hypersaline environments and provided the first example of a member of the domain Bacteria for which ecological relevance in these systems could be proven. Studies of the large collection of strains of S. ruber isolated from around the world as well as metagenomic studies have shown that this species, while being highly homogeneous from a phylogenetic standpoint, harbors very wide genomic microdiversity, even for cooccurring strains (4, 5).
In a previous work (5), we addressed the level of coexisting intraspecific diversity among strains of this species by comparing the two most closely related strains (M8 and M31) at that time, which had been simultaneously isolated from the same crystallizer pond. The two genomes, with identical 16S rRNA genes, had an average nucleotide identity of 98.4% and a high degree of synteny but differed in their gene contents, with ∼10% of the genome of each strain harboring strain-specific accessory genes. Both genomes showed strain-specific areas of deviant GC content, which were highly enriched in transposases, cell wall genes, highly divergent orthologs, and strain-specific genes and were referred to as hypervariable regions (HVRs) and genomic islands (GIs), respectively, in M8 and M31 (HVR1 and HVR2 in M8 and GI1, GI2, and GI3 in M31 [we refer to them as islands 1, 2, and 3, respectively]). Both genomes shared a conserved region (CR) of 377 kb with a high degree of nucleotide sequence conservation (99.5%) in which there were no nonsynonymous changes in the predicted encoded proteins. Both strains had different metabolomic profiles, which could be correlated with the genes in hypervariable regions, as well as different phage susceptibilities, indicating that the genomic differences between both genomes could not be considered neutral from an ecological perspective (5).
Here, we took advantage of the power of transcriptome sequencing (RNA-seq) technology to explore the extent of differences at the transcriptomic level between strains M8 and M31 grown in pure and mixed cultures. Comparison of the transcriptomes in pure cultures can inform us on the direct impact of the differences found at the levels of genomic sequence and architecture on gene expression and help predict their ecological implications (6–8). Data from previous studies based on the observed patterns of strain-specific expression of orthologous genes suggest that regulatory mechanisms must play an essential role in microbial divergence and differentiation (3). Indeed, gene expression differences are likely the first manifestations of microbial divergence because regulatory elements probably evolve faster than the genes that they regulate (9). In this regard, RNA-seq allows for a deep and relatively fast way to characterize and compare transcription patterns of different strains under different conditions. Examples of the use of RNA-seq in bacterial intraspecific studies include, for instance, (i) characterization of the individual responses of different cooccurring strains of the marine bacterium Alteromonas macleodii under changing growth conditions (7), (ii) identification of quorum sensing-controlled genes in different strains of Pseudomonas aeruginosa (10), (iii) identification of differences in transcriptome architectures (transcription start sites and the nature of noncoding transcripts) between two strains of the marine cyanobacterium Prochlorococcus (8), and (iv) identification of differences in transcription among historic and recently emerged hypervirulent strains of the enteric pathogen Clostridium difficile (6).
However, given the high level of intraspecific diversity observed in simultaneously occurring strains for free-living microorganisms (1, 7, 11–13), it is very unlikely that a single lineage of S. ruber (or species of Prochlorococcus, Alteromonas, Pelagibacter, or Vibrio, etc.) will ever have the chance to be alone in its habitat, and thus, the results derived from analyses of pure cultures can hardly mirror the situation in nature. Thus, a key point to understanding the ecology of this species is to know if cooccurring S. ruber lineages in nature act as the “arithmetic” addition of the different strains or, instead, if they modify their activities due to the presence of close relatives (beyond well-known bacterial interaction systems such as quorum sensing). A way to address this issue is to investigate whether the presence of one strain influences the gene expression of the other and, if so, to which extent. Analyses of the (meta)transcriptome of a mixed culture of strains M8 and M31 served precisely this purpose; that is, by comparing the transcriptomes in pure and mixed cultures, we addressed the question of whether the combined transcriptome equals the addition of the individual transcriptomes or whether it rather reflects some direct or indirect regulatory cross talk between the two cocultured strains.
MATERIALS AND METHODS
S. ruber pure and mixed cultures.
Triplicates of pure and mixed cultures of S. ruber strains M8 and M31 were set by inoculating 1 × 109 cells, determined by 4′,6-diamidino-2-phenylindole (DAPI) staining (5), from an exponentially growing culture into 250 ml 25% salt medium supplemented with 0.2% yeast extract (5). Cultures were grown at 37°C in an orbital shaker (170 rpm) exposed to environmental sunlight until late stationary phase. Mixed cultures were inoculated with equal numbers of M8 and M31 cells (0.5 × 109 cells).
Growth was monitored by optical density at 600 nm (OD600) readings and DAPI staining. The relative amounts of M8 and M31 cells in mixed cultures were determined by quantitative (qPCR) with strain-specific primers, as described below. These primers were also used to check for cross-contamination in pure cultures.
PCR and qPCR with strain-specific primers.
M8 and M31 strain-specific PCR primers were designed by using Primer-BLAST (14) and Net Primer software (Premier Biosoft) for the strain-specific open reading frames (ORFs) SRM_00308 (M8 specific; coding for a hypothetical protein, putatively secreted) and SRU_1109 (M31 specific; coding for a predicted RNase H). The primer sequences were 308_Forward (5′-CGA GCC TGT GCC AAG AGA AT-3′), 308_Reverse (5′-CCG ACC GAC AAG CGT ACT G-3′), 1109_Forward (5′-GAG GGT CGC TAT CGC ATC TC-3′), and 1109_Reverse (5′-ACG GTT CTC ACT GGC ATT CC-3′).
These primers were used to quantify the relative abundance of cells of each strain in mixed cultures by qPCR, as follows. Template DNA was obtained by using the DNeasy blood and tissue kit (Qiagen, Netherlands), according to the manufacturer's recommendations. For the samples analyzed by RNA-seq, further quantification was carried out by using the DNA template obtained with the RNA extraction kit prior to DNA digestion. The amount and quality of the extracted DNA were checked by spectrophotometry (Nanodrop ND-1000 spectrophotometer) and in a 1% agarose gel (Seakem LE; FMC Bioproducts), respectively. qPCRs were performed in triplicate. Each reaction mixture contained 10 μl of FastStart Universal SYBR green Master Rox (Roche), 0.2 mM each primer, and 5 μl of the DNA template in a total volume of 20 μl. qPCRs were performed with 96-well plates by using an ABI Prism 7000 sequence detection system (Applied Biosystems, CA, USA) under the following conditions: 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 1 min, and extension at 72°C for 15 s, with a final dissociation step at 95°C for 15 s, 60°C for 15 s, and 90°C for 15 s. For quantification, cell-based calibration curves for M8 and M31 were included for each reaction. The DNA concentration for the calibration curve ranged from 0.003 to 3 ng/μl, which corresponded to 1 × 102 to 1 × 105 cells per reaction mixture. The averages of concentrations, amplification plots, and melting plots were visualized by using qPCR 700 system SDS software (Applied Biosystems). The specificity of the reaction was confirmed by obtaining a melting curve and visualizing the amplified products in a 2% agarose gel (Seakem LE; FMC Bioproducts). The efficiency of amplification for each pair of primers was determined from the standard curve, using serial dilutions of DNA.
The absence of cross-contamination in pure cultures was checked by conventional PCR using the strain-specific primers. Template DNA was obtained by boiling cell pellets from 2 ml of culture (at points in the growth curve showing OD600 values of <0.3) or 100 μl of culture (at points with OD600 values of >0.3). The amplification conditions were 94°C for 3 min followed by 35 cycles of denaturation (94°C for 30 s), annealing (60°C for 1 min), and extension (72°C for 2 min), with a final extension step at 72°C for 10 min. The PCR products were run in a 2% agarose gel (Seakem LE; FMC Bioproducts) and visualized by UV exposure after ethidium bromide staining.
Elementary composition of extracellular medium during exponential phase.
New batches of mixed M8 and M31 cultures were set as described above to analyze the ionic composition of the growth medium during mid-exponential phase. Ten milliliters of pure and mixed cultures was centrifuged (15 min at 3,900 × g at 4°C), and supernatants were filtered through 0.45-μm-pore-size filters (Millipore) and diluted 1:10 with Milli-Q water for the quantification of ionic compositions by employing inductively coupled plasma mass spectrometry (ICP-MS) (7700x; Agilent Technologies), which ranged between 0.5 and 10 ppb for Fe, Ni, Cu, and Mg and between 0.5 and 50 ppb for Zn.
RNA extraction and sequencing.
RNAs were obtained from two replicates of mixed cultures and one sample from each pure culture. Samples from the same time point, which corresponded to mid-exponential phase (82 h after inoculation), were used for RNA extraction. Cells were recovered by centrifugation (15 min at 3,900 × g at 4°C) and resuspended in TE buffer (100 mM Tris-HCl [pH 8], 100 mM EDTA [pH 8]) with 10 μl lysozyme (300 mg/ml). Total RNA was then extracted with the RNeasy minikit (Qiagen) according to the manufacturer's recommendations. The obtained nucleic acids were treated with Turbo DNase (Ambion) to remove residual DNA. rRNAs and tRNAs were partially eliminated with MICROBExpress and Mega Clear kits (Ambion), according to the manufacturer's protocol. The amount and quality of the resulting RNA extracts were determined with a BioAnalyzer instrument (Agilent) prior to sequencing.
cDNA synthesis and creation of libraries were performed with an Illumina mRNA sequencing Sample Prep kit according to the manufacturer's recommendations. RNA was fragmented (2 min at 94°C), and cDNA synthesis was done in two steps (first-strand synthesis and second-strand synthesis) with the SuperScript II enzyme. Sequencing was carried out by using Illumina Genome Analyzer II technology at the Center for Genomic Regulation (CRG) core facility, Barcelona Biomedical Research Park (PRBB). Obtained paired-end reads were 50 bp long and non-strand specific, with an insert size of 200 bp.
Validation of RNA-seq data.
RNA-seq reads obtained from the four samples analyzed presented similar high coverage and quality, enabling accurate comparisons between samples and the detection of very lowly expressed genes (see below). In fact, analyses with the two pure cultures indicated that >98% of the genes, in both cases, that showed significant coverage were expressed. Between 13.62% and 14.14% of the reads corresponded to mRNAs.
The relative amount of cells of each strain in mixed cultures was also estimated by means of qPCR, in order to further validate the RNA-seq data. In the case of mixed cultures, the relative abundance of each strain (M8/M31 ratio) was obtained by qPCR quantification (0.92 and 0.70 for two replicates of mixed cultures, MXA and MXB) (Table 1), validating the ratios of useful RNA-seq reads that mapped to the S. ruber M8 and M31 genomes (0.86 for MXA and 0.77 for MXB) (Table 1). In addition, the expression levels for the two biological replicates of mixed cultures showed a good correlation (R = 0.99 and R = 1 for M8 and M31, respectively) (see Fig. S1 in the supplemental material), supporting the reproducibility of the experimental data. These R values are higher than those for previously reported RNA-seq experiments: 0.95 to 0.98 (15), 0.93 to 0.95 (15), and 0.947 to 0.977 (6).
TABLE 1.
Summary of sequencing results for S. ruber samples sequenced by using the Illumina Genome Analyzer II technology platform
| Samplea | No. of processed paired reads | No. of filtered reads | % filtered reads | No. of rRNA reads | % rRNA reads | Final no. of useful reads | No. of kb sequencedb | Useful coverageb | % final useful readsb | No. of qPCR copiesb |
|---|---|---|---|---|---|---|---|---|---|---|
| M8-P | 43,071,950 | 7,088,806 | 16.45 | 25,838,764 | 65.45 | 10,144,380 | 507,219 | 132.33× | 14.09 | 3.63 × 108 |
| M31-P | 42,924,442 | 6,631,194 | 15.44 | 25,765,713 | 70.05 | 10,527,535 | 526,377 | 146.73× | 14.5 | 1.08 × 108 |
| MXA | 39,918,009 | 54,445,077 | 13.64 | 25,087,962 | 72.74 | M31, 5,044,060; M8, 4,340,910 | M31, 252,203; M8, 217,046 | M31, 70.30×; M8, 56.62× | M31, 7.32; M8, 6.30 | M31, 406.16; M8, 375.49 |
| MXB | 40,131,308 | 5,433,881 | 13.54 | 24,885,733 | 72.33 | M31, 5,549,600; M8, 4,262,094 | M31, 277,480; M8, 213,105 | M31, 77.35×; M8, 55.60× | M31, 8.00; M8, 6.14 | M31, 219.68; M8, 309.12 |
M8-P and M31-P, strains M8 and M31 in pure cultures, respectively. MXA and MXB are replicates of the mixed cultures.
The contribution of each strain is indicated for the mixed cultures.
Analysis of gene expression and detection of differentially expressed genes.
The four sequenced samples were first processed to remove poor-quality reads as follows. Reads with a PHRED quality score of <10 were trimmed at the first base. Subsequently, pairs with reads of <31 bases were discarded. The remaining reads were mapped against the genomes of M31 (chromosome [GenBank accession no. NC_007677.1] and pSR35 [accession no. NC_007678]) and M8 (chromosome [accession no. NC_014032.1], PSR_11 [accession no. NC_014026.1], PSR_11, PSR_35, PSR_56 [accession no. NC_014028.1], PSR_61 [accession no. NC_014030.1], and PSR_84 [accession no. NC_014157.1]) by using BWA v0.6.1-r104 (16). The expression profiles for pure cultures were normalized to the sample size and transcript length and are provided as RPKM (reads per kilobase per million mapped reads) values, as computed by Cufflinks v1.1.0 (17). We considered genes as being not expressed if their RPKM expression values estimated by Cufflinks were 0.
The complete sequences of M8 and M31 can be found under GenBank accession numbers FP565814.1 and CP000159.1, respectively. There is an update in the M8 nomenclature, but here, in order to make comparisons with previously reported data easier, we have kept the old locus tags. The equivalence between old and new tags can be found in the NCBI database (http://www.ncbi.nlm.nih.gov/genome/genomes/1400?) for the RefSeq sequences of each of the replicons.
For analyses of differential expression in mixed cultures of M8 and M31, the reads were aligned against a combined reference containing the chromosomes and plasmids of both strains. We ignored the paired reads that mapped in regions that were identical in both strains, as these reads cannot be assigned unambiguously to a specific strain. Orthologous genes between these strains were detected by using the best reciprocal Blast hit approach, applying an E value cutoff of 10−5 (18). Read counts were normalized to the sample size and orthologous gene length. Differentially expressed genes were detected by using Deseq (19) and Cufflinks, applying a P value cutoff of 0.05. A gene from a given strain was consider to be differentially expressed under both conditions (i.e., “from pure to mixed” cultures) when a P value of <0.05 was obtained with either software. The results obtained with Deseq and Cufflinks were pooled, yielding 354 and 446 genes that presented significant differences in comparisons of pure and mixed cultures of M8 and M31, respectively. Results from both programs were very similar, yielding overlaps of 77.79 and 98.28%, respectively.
Reannotation of genomes, mapping of metabolic pathways, and Fisher's enrichment test.
The functions of proteins encoded in the M8 and M31 genomes were reannotated with Blast2GO (20), obtaining annotations based on the Gene Ontology (GO) terms and Enzyme Classification (EC) numbers. All the genes selected as being differentially expressed in comparisons of (i) pure cultures of the strains and (ii) pure cultures versus mixed cultures were mapped against metabolic KEGG pathways (21). We tested whether differentially expressed genes were enriched functionally with respect to the corresponding S. ruber genomic background by using Fisher's test with a P value cutoff of 0.05.
Previously annotated extracytoplasmic sigma factors (ECFs) were confirmed with ECF Finder (http://ecf.g2l.bio.uni-goettingen.de:8080/ECFfinder/). The SignalP 4.1v server (22) and TAT Find 1.4v (23) were used to identify potential signal peptide cleavage sites and the presence of prokaryotic twin-arginine translocation (TAT) signal peptides, respectively.
Accession number.
The transcriptome sequencing data have been submitted to the European Nucleotide Archive (ENA) and are available under the project accession identifier PRJEB11409.
RESULTS AND DISCUSSION
Triplicates of pure cultures of M8 and M31 and mixed cultures (M8 plus M31) were inoculated with an equal amount of cells and grown until late stationary phase under standard conditions (see Materials and Methods). Growth was monitored by OD readings, DAPI counts, and, for the mixed cultures, qPCR. Growth curves for both pure and mixed cultures were highly reproducible (see Fig. S2 in the supplemental material). Growth rates were very similar for pure cultures of both strains (0.053 ± 0.002 and 0.055 ± 0.003 h−1 for M8 and M31, respectively) and, as expected, indicated that S. ruber has a relatively low growth rate (with doubling times of ∼13 h). Samples for transcriptomic analyses were taken at mid-exponential phase (82 h). At this point, the amounts of M8 and M31 cells in their respective pure cultures were almost identical, while the ratio of M8 to M31 cells in the mixed cultures, as determined by qPCR, was ∼8:10 (which was in good agreement with the amounts of mRNA recovered from M8 and M31 cells [see below]). Sequencing results (Table 1) showed that 3.9 × 107 to 4.3 × 107 raw read pairs were obtained for each of the four analyzed samples. After filtering of rRNA and tRNA reads, ∼107 useful reads were obtained for each culture, which resulted in a high level of coverage over the genome (>100 times). As described in Materials and Methods, RNA-seq data were highly reproducible for the biological replicates, with Pearson coefficient values above those reported previously for RNA-seq experiments (6, 14, 24).
Overall transcription patterns in M8/M31 pure cultures.
For both strains, gene transcription was detected throughout practically the entire genome (98% of annotated genes), including plasmids. Previous RNA-seq studies of bacterial transcriptomes reported a high proportion of expressed genes, such as 64% in Acidovorax (24), 83% in Listeria (25), and 89% in Pseudomonas (10). However, this level depends on sequencing depth and a threshold that is somehow arbitrary, given that transcript copy numbers for individual genes can differ by several orders of magnitude (26). Furthermore, we must keep in mind that individual cells within a population display different levels of gene expression, which can be detected only by single-cell approaches (27). In S. ruber pure cultures, only 55 and 30 genes (see Table S1 in the supplemental material), for M8 and M31, respectively, had expression levels below the detection threshold (see Materials and Methods). These genes are considered nontranscribed, although we cannot rule out the possibility that they do not correspond to real genes. A large proportion of these nonexpressed genes (87 and 67% for M8 and M31, respectively) were strain specific and coded for hypothetical proteins. Thus, the nontranscribed fraction was enriched in strain-specific ORFs with unassignable functions. This supports the idea that these genes have been acquired recently, although there are some remarkable deviations from this behavior, as discussed below.
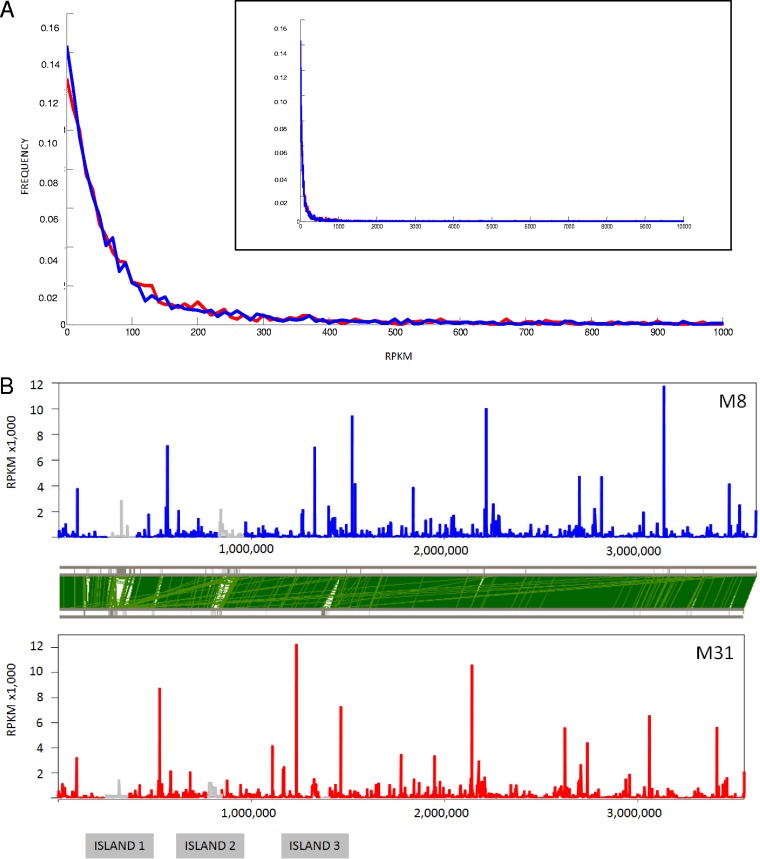
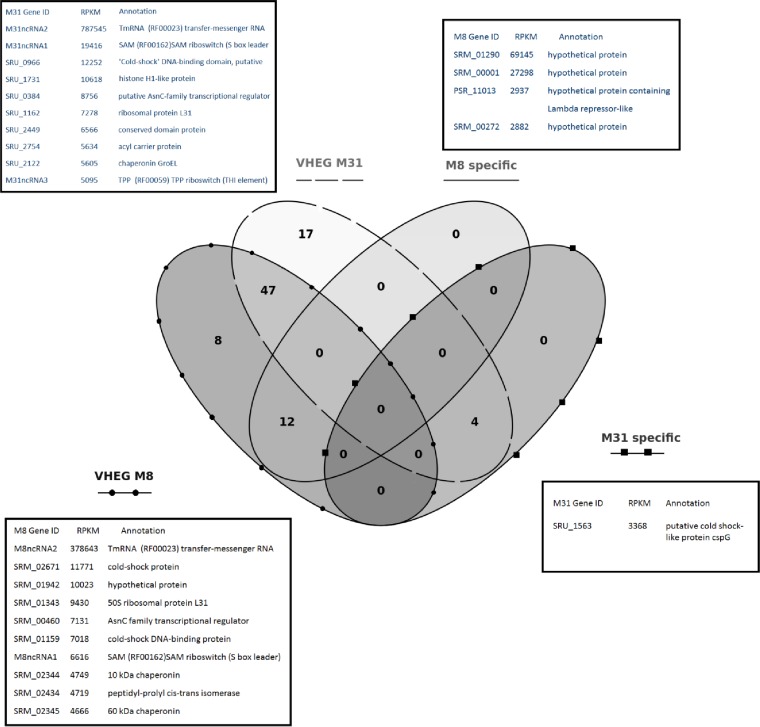
Both S. ruber strains showed similar overall chromosomal expression profiles (Fig. 1), with roughly 80% of the genes being expressed at levels below 200 RPKM and 97% of the genes being expressed at levels below 1,000 RPKM (see Fig. S3 in the supplemental material). The sets of very highly expressed genes (VHEGs) (with expression levels of >1,000 RPKM) (see Table S2 in the supplemental material) consisted of 64 and 65 ORFs (for M8 and M31, respectively) that seemed to be part of highly transcribed operons. Although there were also some strain-specific genes (see Table S2 in the supplemental material), ∼68% of the VHEGs were common to both strains (Fig. 2).
FIG 1.
Expression of S. ruber M8 and M31 chromosomes in pure culture. (A) Histogram showing normalized frequencies for expression levels of genes in the M8 (blue) and M31 (red) chromosomes. Bin size, 10 RPKM. Only genes with expression levels below 10,000 RPKM are shown. The inset shows a histogram up to 10,000 RPKM. (B) Aligned chromosomes with their gene expression levels. Expression of genes located in islands is represented in gray. The sequences have been aligned from the predicted replication origin. For clarity, ORFs SRM_0001 and SRM_1290 from M8, both coding for hypothetical proteins, with expression levels of 27,298 and 69,145 RPKM, respectively, are not included in the graph. The green bars linking both genomes represent ortholog matches identified by FASTA (fast nucleotide comparison) analysis with a 100-nucleotide window (5).
FIG 2.
Venn diagram showing the distribution of VHEGs in both genomes. The genes with expression levels of >2,800 RPKM for each of the four categories are shown in the corresponding tables.
Among the proteins encoded by VHEGs in both genomes, some were clearly related to exponential growth demands, like ribosomal proteins, TonB transporters, ATPases, cytochrome c oxidase, superoxide dismutase, and chaperones. However, there were intriguingly high expression levels of the genes coding for xanthorhodopsin (SRM_01698 and SRU_1500 in M8 and M31, respectively). This retinal binding proton pump transforms light into proton motive force and uses the carotenoid salinixanthin as a light-harvesting antenna (28). In addition, this VHEG set includes the three ORFs coding for the cold shock protein (CSP) analog CspC. CspC is a stress protein and a member of the CspA cold shock protein family (COG1278 [GenBank accession no. K03704] [transcription factors]). CSPs are thought to bind mRNA and regulate ribosomal translation, mRNA degradation, and the rate of transcription termination (29). For instance, several CSPs are upregulated in C. difficile following osmotic shock (6).
Among VHEGs, asnC, a transcriptional regulator related to the regulation of ectoine metabolism in Halomonas, is also worth mentioning (30). This is a specific gene regulator whose activity is triggered by asparagine binding, in a classic feedback mechanism (31). Other genes involved in transcriptional regulation are also highly expressed in both M8 and M31, such as noncoding RNA (ncRNA) riboswitches and the alternative sigma factor ECF (discussed below) as well as flagellar and ferric citrate sigma factors. Some of them displayed levels of expression higher than that of the standard sigma 70 factor (which is also highly expressed). Similarly, high expression levels of transcriptional regulators and ncRNAs were previously observed for Listeria monocytogenes by means of RNA-seq (32). Overall, there are 6 types of sigma factors annotated in S. ruber, most of which have similar expression patterns in both strains. The extracytoplasmic function alternative sigma factor E (also known as sigma 24) encoded by SRM_02973/SRU_2762 is the sigma factor displaying the highest expression level. According to ECF Finder and the MiST database, this factor belongs to ECF01 (envelope stress response, antimicrobial compound production, and detoxification), which is specialized in responses to the effects of heat shock and other stresses on membrane and periplasmic proteins. This finding, together with the high expression level of cspC mentioned above, raises the question of whether the conditions that we consider standard for S. ruber growth are in fact stressful.
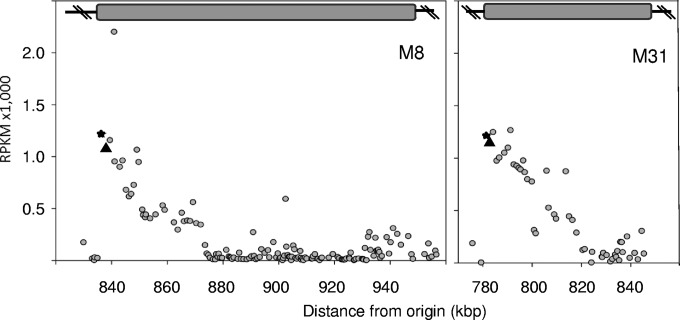
We noted a remarkably high expression level, in both genomes, of an ORF coding for a phage integrase-like protein. In both genomes, the integrase gene lies in the 5′ boundary of island 2 at the 3′ end of a VHEG involved in O-chain lipopolysaccharide (LPS) synthesis. Following this pair of genes, there was a cluster of ∼30 genes with expression levels remarkably higher than those of the rest of the genomic island (Fig. 3), with many of them being involved in cell wall synthesis. In other words, there was a clear polarity in the expression of the island. Remarkably, many of these highly expressed genes are frequently involved in virus recognition (5), although in this study, obviously, cells were grown in the absence of viral pressure.
FIG 3.
Gene expression polarity in the 5′ boundary of island 2 in S. ruber M8 (left) and M31 (right). ORFs coding for the LPS O-chain and phage integrase are labeled with stars and triangles, respectively. The gray bar represents the extension of the island.
Although it was not one of the main goals of this work, the transcriptomic data can also be used to shed light on the use of metabolic pathways by S. ruber. For instance, significant expression levels were detected for complete Embden-Meyerhof glycolysis, including fructose-1,6-bisphosphate aldolase, whose activity was not detected previously (33). Alternative glucose degradation by the Entner-Doudoroff pathway presented decreasing expression levels from the first reaction, supporting the lack of activity of 2-keto-3-deoxy-6-phosphogluconate aldolase reported previously (34).
Transcription and genome architecture.
For every individual genome, different expression profiles were observed in their corresponding replicons (see Fig. S4 in the supplemental material). Most genes carried by plasmids had very low expression levels, with the exception of pSR11 of M8, which included one of the genes with the highest transcription levels (pSR_11013, coding for a putative DNA-binding protein similar to transcriptional regulators) (see Table S2 in the supplemental material). The chromosomal gene transcription patterns were similar for both strains (Fig. 1), with no clear spatial clustering of genes according to their expression levels. In both cases, island 1 harbored the highest proportion of genes with very low expression levels. Island 2 also had a large proportion of genes with very low expression levels, although, as explained above, its 5′ region was highly expressed. Both islands have anomalously low GC contents (5), which is in good agreement with the general assumption that genes with deviant GC content are derived from recent lateral gene transfer (LGT) events and have expression levels below optimal levels (35, 36). Accordingly, for both genomes, genes with higher expression levels have GC contents close to that of the average for the genome (see Fig. S5 in the supplemental material).
In addition to the island boundary mentioned above, the regions containing most genes coding for ribosomal proteins (spanning ORFs SRM_01230 to SRM_01262 in M8 and ORFs SRU_1030 to SRU_1061 in M31) were also highly expressed in both genomes. The presence of a cluster of ∼60 genes (spanning ORFs SRM_2799 to SRM_2866 in M8 and ORFs SRU_2582 to SRU_2649 in M31) with low expression levels in both strains was also remarkable (Fig. 1A). All of these genes coded for proteins involved in cell motility functions (COG N [biosynthesis of flagella, flagellum structure, and chemotaxis factors]).
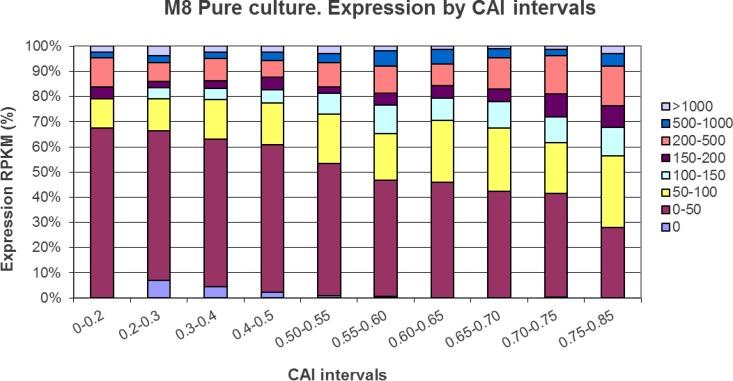
In our previous characterization of the S. ruber genome (5), codon adaptation indexes (CAIs) were used as a theoretical proxy for expression levels. Many genes involved in transport as well as in energy and nucleotide metabolism showed the highest CAIs, while genes with low CAIs encoded mostly hypothetical proteins and transposases. Most genes encoding ribosomal proteins had an average CAI, which is typical for many slow-growing microorganisms (37). Although we did not find a good correlation between CAIs and transcription levels for all the genes (r2 = 0.0012), it was patent (Fig. 4) that genes with lower CAIs had a higher proportion of poorly expressed genes, at least under the conditions analyzed.
FIG 4.
Gene distribution percentages based on CAIs and RPKM expression levels. The size of the interval increases with increasing RPKM levels since these categories harbor a smaller number of genes.
In the above-mentioned study (5), gene duplication events were identified and classified as occurring previous to and following M8 and M31 divergence, respectively. Here, we analyzed transcription differences between paralogs derived from these duplications (see Table S3 in the supplemental material). We found that, as expected, differences among the pairs of “ancestral” duplications were greater than those from more recent paralogs.
Differences in core genome expression between pure cultures.
The Pearson (r) correlation value for M8 and M31 transcriptomes was 0.975, below the values of 0.997 and 1 obtained for replicas of mixed cultures (see below). Around 165 genes shared by the M8 and M31 genomes presented significant (P < 0.05) differences in transcription between the two strains (see Data Set S1 in the supplemental material). The proportion of genes differentially transcribed between M8 and M31 was at least 5-fold higher in islands 1 and 2 than in the rest of the genome. Thus, the islands were enriched not only in strain-specific and divergent genes but also in genes displaying different levels of expression when M8 and M31 pure cultures are compared.
The set of 165 differentially expressed genes was enriched in three COG categories: N (cell motility), T (signal transduction mechanisms), and U (intracellular trafficking, secretion, and vesicular transport). A large proportion (88%) of the genes within these categories had higher expression levels in strain M8. In other words, a large part of the transcriptional differences between strains M8 and M31 seems to be related to responses to the environment, at the level of either chemotaxis transducers, two-component systems (which, in turn, can have an effect on gene expression), flagella, or secretion pathways. Conversely, the genes showing differential expression between the two pure cultures were depleted in genes of COG category J (translation, ribosomal structure, and biogenesis).
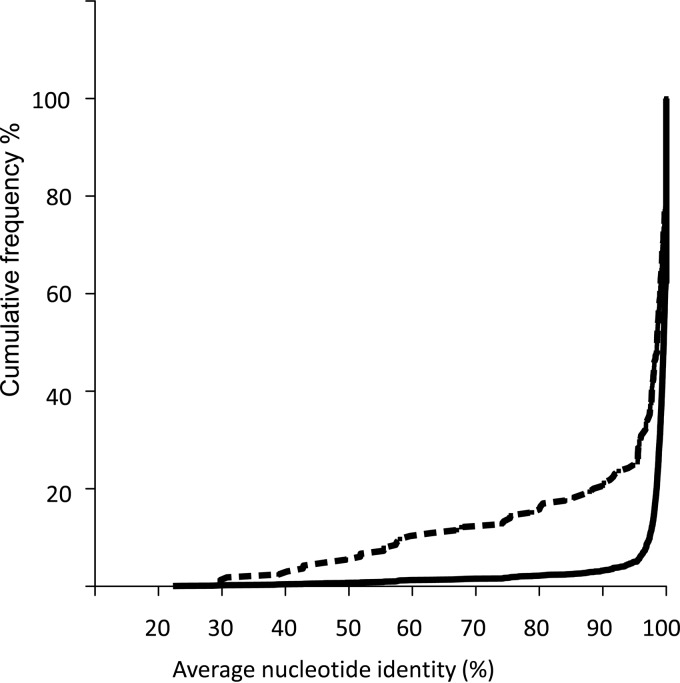
As expected, transcription differences were greater for more divergent orthologs (Fig. 5), and therefore, the fraction of genes differentially transcribed between the two strains was enriched in genes with lower average nucleotide identities (ANIs). However, there were also very similar genes that were being expressed at different levels in both strains, which pointed to differences in their regulatory networks. This was the case, for instance, for the 38 genes that were identical (i.e., 100% ANIs) (see Data Set S1 in the supplemental material). Among them are genes involved in transport across membranes, signal transduction, and gene regulation (i.e., responses to external signals). Thus, identity at the nucleotide level between two genes does not necessarily mean that they have similar levels of expression. As a result, estimates of intraspecific diversity based solely on gene sequences can underestimate the real levels of metabolic and physiological diversity between analyzed strains.
FIG 5.
Cumulative frequency distributions of ANI values between orthologous genes in the S. ruber M8 and M31 genomes. Solid line, all genes; dashed line, only genes with differential expression levels between both genomes.
Altogether, comparison of the transcriptomes of M8 and M31 in pure cultures reveals restricted but significant differences in how the shared set of genes is expressed under identical environmental conditions. Interestingly, a large part of the differences seem to be related to genes involved in environmental sensing, suggesting somewhat different strategies under identical conditions. Overall, however, the two strains showed similar transcription patterns in terms of both transcriptomic architecture and the nature of highly expressed genes.
Metatranscriptomes of M8/M31 mixed cultures.
Some studies indicate that bacteria alter their gene expression when confronted with another bacterial species (38). In this regard, there are a few examples of transcriptome analysis of cocultures of different bacterial species, such as studies of Acidovorax avenae subsp. avenae cultivated alone and in coculture with Burkholderia seminalis, which showed a slight response to the presence of other bacteria (24). However, the interaction between strains of the same species has not been previously explored through transcriptome analysis. This approach is particularly challenging for very closely related strains such as the ones studied here, given their high level of sequence similarity (see Materials and Methods).
According to the criteria explained in Materials and Methods, 354 and 446 genes of M8 and M31, respectively, showed differential expression in comparisons of pure and mixed cultures (Fig. 6). A total of 418 orthologous genes (i.e., present in both strains) were left out of this comparison, since, due to their high similarity, they could not be unambiguously assigned to either strain. The sets of genes that changed their expression levels when pure and mixed cultures were compared displayed very similar expression levels in the two strains when they were grown in pure cultures (Pearson correlation factor values of 0.989 and 0.984 for genes differentially expressed in M8 and M31, respectively).
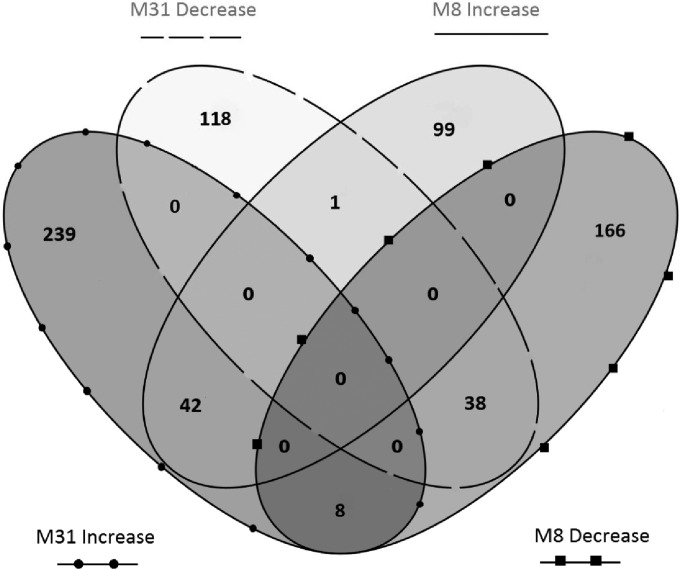
FIG 6.
Venn diagram showing the distribution of 446 and 364 genes with differential expression in M31 and M8, respectively. For each strain, the numbers of genes up- and downregulated are shown.
Changes in accessory versus core genomes.
According to the data shown in Table 2, the proportion of core genomes with differential expression ranged from 11.8% (if none of the “nonassignable” 418 genes changed) to 28.3% (if all of them changed) for M8 and from 16.6 to 33% for M31. Conversely, the fraction of accessory genome with differential expression was ∼7% for both strains. In other words, with the exception of genes in the CR (see below), the proportion of genes undergoing transcriptomic changes is higher in the core genome than in its accessory counterpart. This is expected, considering that accessory genes most likely have been acquired more recently than core genes and therefore have had less time to accommodate their regulatory mechanisms, at least under the conditions analyzed here. Previous studies on C. difficile strains grown under different conditions related to changes in virulence also showed a higher degree of change in the core than in the flexible genome (6). In any case, among the S. ruber genes with higher fold changes from pure to mixed cultures (fold change of >1) (see Data Set S1 in the supplemental material), there were also some accessory genes, most of them coding for hypothetical proteins, for which a putative function cannot be envisaged. As pointed out by Kimes et al. (7), these results highlight the need for further investigation of hypothetical proteins, which still constitute a large part of microbial genomes.
TABLE 2.
Comparison of transcription in M8 and M31 cells grown in pure and mixed culturesa
| Parameter | Value for indicated strain |
|
|---|---|---|
| M8 | M31 | |
| No. of ORFs in the genome | 3,303 | 2,898 |
| No. of ORFs in the accessory genome | 766 | 361 |
| No. of ORFs in islands | 280 | 128 |
| No. of ORFs analyzed in mixed cultures (% of ORFs in the genome) | 3,303 − 418 = 2,885 (87.34) | 2,898 − 418 = 2,480 (85.58) |
| No. of ORFs with differential expression from pure to mixed cultures | 354 | 446 |
| No. of ORFs with differential expression belonging to the core/accessory genome | 300/54 | 421/25 |
| No. (%) of ORFs upregulated from pure to mixed cultures | 142 (40) | 289 (65) |
| No. (%) of ORFs downregulated from pure to mixed cultures | 212 (60) | 157 (35) |
| No. of ORFs with differential expression located in islands | 18 | 17 |
| No. of ORFs with differential expression located in CR | 2 | 3 |
| No. of differentially expressed genes in category | ||
| Transport functions | 53 | 38 |
| Two-component systems | 7 | 7 |
| Kinases | 4 | 9 |
| Chemotaxis and flagella | 6 | 1 |
| Transcription regulation | 9 | 7 |
| Translation (translational factors/ribosome structural proteins/tRNA synthetases/tRNA rRNA methylases) | 0/0/3/0 | 6/20/6/3 |
| DNA replication and/or recombination | 10 | 9 |
| Secreted proteins (TAT signal peptide/P signal peptide) | 32/3 | 18/1 |
There were 89 ORFs with differential expression in both M8 and M31.
Architecture of changes.
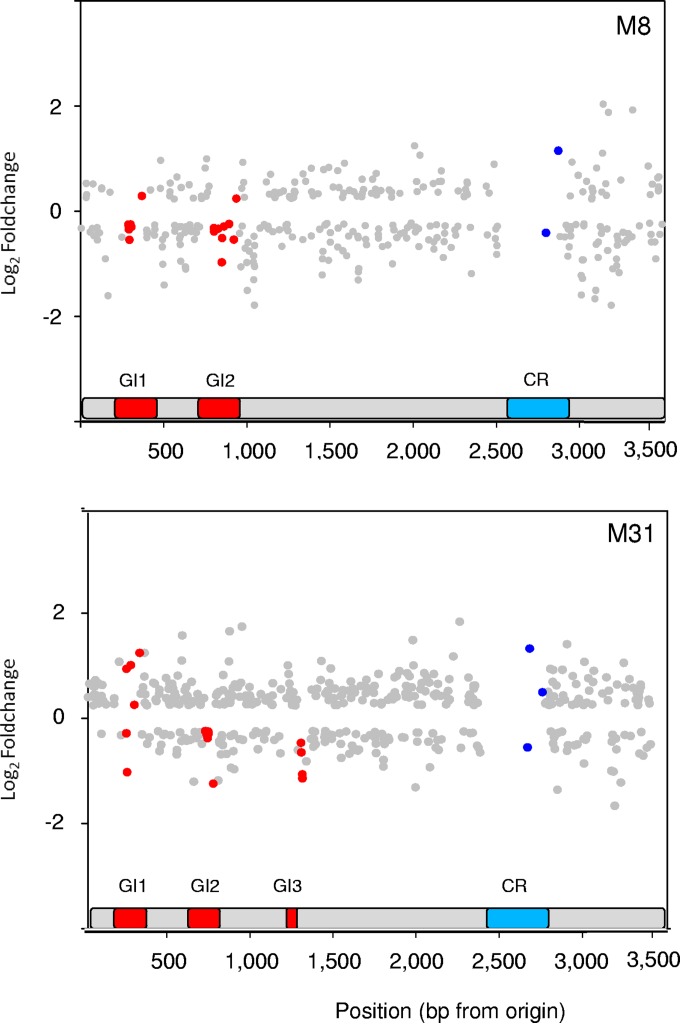
The so-called CR (5) showed a very small number of genes with differential expression in mixed cultures (Fig. 7). Although this area was depleted in genes that could be unambiguously assigned to either strain in the mixed culture, it still harbored 43 and 35 of such genes in M8 and M31, respectively. However, only 2 (in M8) and 3 (in M31) genes in these areas had differential expression when transferred from pure to mixed cultures, values that were low compared to the expected distribution in the whole genome (e.g., the probabilities of finding such a small number of differentially expressed genes in 43 and 35 contiguous genes in M8 and M31 are 0.077 and 0.055, respectively). This CR is enriched in genes involved in ionic transport (5) and contains the “hypersalinity island” first described in S. ruber M31 (39), which includes K+ uptake/efflux systems and cationic amino acid transporters of crucial importance to a hyperhalophilic life-style. These findings point to a strong control of CR expression that most likely depends on the salinity of the environment.
FIG 7.
Expression level changes in S. ruber M8 and M31 chromosomes in comparisons of pure and mixed cultures. Only genes that could be unambiguously assigned to either strain and that showed significant changes are displayed. Red, genes in islands; blue, genes in the conserved region.
Biological relevance of changes: the common and specific responses.
Overall, considering the proportion of genes with differential expression (Table 2 and Fig. 6) and the extent of changes (Fig. 4; see also Data Set S1 in the supplemental material), the changes in expression from pure to mixed cultures were rather modest but still significant. Each strain seemed to have a different response to the presence of the other, since only 89 (i.e., fewer than one-fourth) of the genes with differential expression were shared by M8 and M31. This proportion was even lower for the genes undergoing the largest changes. In other words, each strain reacted to the presence of the other one and did so in a specific way.
M31 displayed a higher ratio of upregulated genes (Table 2; see also Data Set S1 in the supplemental material), in good agreement with the higher proportion of M31 cells than of M8 cells at the analyzed time point. COGs J (translation, ribosomal structure, and biogenesis) and O (posttranslational modification, protein turnover, and chaperones) were the categories with higher proportions of upregulated genes in M31. Conversely, categories D (cell cycle control), N (cell motility and secretion), and V (defense mechanism) harbored many more downregulated genes in M8 than in M31.
There were 89 genes undergoing transcriptomic changes in both M8 and M31 when changed from pure to mixed cultures (see Data Set S1 in the supplemental material). The levels of most of them changed in both strains in the same direction (42 upregulated and 38 downregulated) (Table 2). Among these genes, it is worth mentioning the changes in the expression levels of 17 genes coding for transporters, including remarkable decreases in the expression levels of xanthorhodopsin- and bacteriorhodopsin-related coding genes and increases in the expression levels of the transporter of the compatible solute glycine betaine. The DprA-SMF gene, putatively involved in natural transformation, was also downregulated in the two strains. In addition, there were 9 genes with expression level changes in the opposite directions in both strains (8 of them upregulated in M31), 3 of which encoded secreted (hypothetical) proteins.
As mentioned above, the M31 cell density was higher than that of M8 in the mixed culture at the analyzed time point. In good agreement with this, there was an overexpression of M31 genes involved with several anaplerotic pathways, transcription factors, replisome machinery, lipid biosynthesis, and ribosomal proteins. In fact, the relative abundance of ribosomal protein transcripts for a given taxon has been proposed to be a metric for assessing in situ growth rates (40). In our case, this has proven true for pure cultures, in which the modest increase of the M31 growth rate with respect to that of M8 was mirrored by an increase in the number of normalized reads from ribosomal protein-encoding genes (53,777 and 50,247 RPKMs, respectively). Overall, there was an increase in the transcription of anabolic pathway genes in M31 in comparisons of pure and mixed cultures, which is also in agreement with its lower generation rate. There seemed to be a deviation of intermediates of the tricarboxylic acid cycle (see Fig. S6 in the supplemental material) due to growing demands, which was accompanied by increases of glycolysis and anaplerotic pathways.
For M8, the overexpression of genes coding for proteins related to stress responses, such as E and ECF sigma factors, and several subunits of NADH dehydrogenase is noteworthy. In addition, there were increased levels of transcription for multidrug resistance channels, beta-lactam resistance, penicillin and cephalosporin production, as well as RecR recombinase, involved in the repair of stress-induced DNA damage. It thus seems that, at least at the analyzed time point, the presence of M31 is somehow stressful for M8.
Antibiotics have been proposed (41) to be molecules mediating signaling between bacterial populations in nature. In this regard, β-lactam resistance and penicillin and cephalosporin biosynthesis are among the pathways with higher fold changes, when considered as a whole. In addition, genes related to efflux pups and drug resistance are among the ones affected by the change from a pure culture to a mixed culture. Indeed, all seven antibiotic-related pathways detected in S. ruber genomes include at least one gene whose expression changes in at least one of the strains when transferred from pure to mixed cultures. However, the small number of genes in these pathways hampers any statistical quantification of this phenomenon.
Does each strain sense the other directly or, rather, the ionic composition of the medium? As shown in Table 2 (see also Data Set S1 in the supplemental material), there were several transporters that changed their expression levels from pure to mixed cultures. This could have affected their activity and, in turn, the chemical composition of the growth medium. If this was the case, the effect of one strain on the other in the mixed culture would not be mediated by biochemical signals of specific activities but in an indirect way, mediated by the alteration of the medium composition. To explore this hypothesis, we analyzed the elementary composition of the medium in pure and mixed cultures. We chose elements (such as Fe, Co, Ni, Zn, Cu, and Co) corresponding to ions for which transporters displayed differential expression under both conditions. The concentrations of the analyzed elements in the mid-logarithmic phase were not significantly different (P < 0.05) between pure and mixed cultures, and therefore, it is unlikely that these components of the medium could trigger the (modest) differential expression levels observed for both strains.
In previous work (42), we showed that very closely related cooccurring S. ruber strains expressed different metabolomes when grown under the same laboratory conditions. Each strain seemed to have an exclusive extracellular chemistry (indeed, M8 and M31 expressed different extracellular metabolomes [5]). Thus, each strain has the capability to modify its environments in a specific way so that its presence can be sensed by the cooccurring microbes. Although we do not have cues on the nature of the compounds mediating M8 and M31 interactions, we observed different patterns for lipid metabolism and antibiotic-related compounds between S. ruber strains isolated from the same environmental sample (43). Considering that antibiotics at subinhibitory concentrations are known to play a regulatory function in nature (41, 43) and that we detected differential expression in some antibiotic-related genes, we speculate that the interactions that we have seen in this work are mediated by antibiotic-like molecules. Nevertheless, validation of this hypothesis would definitely need further experimental support, a goal that does not seem easy to achieve. This is especially true if we consider that, as pointed out by Traxler and Kolter, “the chemical landscape inhabited and manipulated by bacteria is vastly more complex and sophisticated than previously thought” (44).
Conclusions.
Here, we report the first genome-wide transcriptomic study of closely related bacterial strains grown in pure and mixed cultures. Altogether, our results present evidence that both strains react to the presence of the other at the transcriptional level. Notably, these transcriptomic alterations are specific for each strain, with only a small fraction of commonly induced/repressed genes. In other words, genetic differences result in variable transcriptomic profiles. The scope and direction of the transcriptional changes are consistent with the observed increase in the number of cells of M31 compared to the number of cells of M8 in the mixed culture at the time point analyzed.
The complex patterns of gene transcription found in nature respond to environmental cues and nutrient availability (as shown in reference 45). Here, we show that intraspecific interactions may also play a role in the regulation of such responses. Paraphrasing S. M. Friedman (46), we can assess that, from community-scale to intraspecific interactions, no microbe is an island.
Supplementary Material
ACKNOWLEDGMENTS
The group of J.A. is funded by grant CGL2012-39627-C03-01 from the Spanish Ministry of Economy and Competitiveness (MINECO), which is cofinanced with FEDER support from the European Union. P.G.-T. was an FPI-MINECO fellow. Research by the group of T.G. is funded in part by a grant from the Spanish Ministry of Economy and Competitiveness (BIO2012-37161), a grant from the Qatar National Research Fund (NPRP 5-298-3-086), and a grant from the European Research Council under the European Union's Seventh Framework Programme (FP/2007-2013)/ERC (grant agreement no. ERC-2012-StG-310325). L.P.P. was funded through the La Caixa-CRG international fellowship program.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02690-15.
REFERENCES
- 1.Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, Ding H, Marttinen P, Malmstrom RR, Stocker R, Follows MJ, Stepanauskas R, Chisholm SW. 2014. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 2.Denef VJ, VerBerkmoes NC, Shah MB, Abraham P, Lefsrud M, Hettich RL, Banfield JF. 2009. Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ Microbiol 11:313–325. doi: 10.1111/j.1462-2920.2008.01769.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilmes P. 2012. Genome-based and functional differentiation: hallmarks of microbial adaptation, divergence and speciation?, p 1–23. In Ogilvie LA, Hirsch PR (ed), Microbial ecological theory—current perspectives. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 4.Pašić L, Rodriguez-Mueller B, Martin-Cuadrado A-B, Mira A, Rohwer F, Rodriguez-Valera F. 2009. Metagenomic islands of hyperhalophiles: the case of Salinibacter ruber. BMC Genomics 10:570. doi: 10.1186/1471-2164-10-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peña A, Teeling H, Huerta-Cepas J, Santos F, Yarza P, Brito-Echeverría J, Lucio M, Schmitt-Kopplin P, Meseguer I, Schenowitz C, Dossat C, Barbe V, Dopazo J, Rosselló-Mora R, Schüler M, Glöckner FO, Amann R, Gabaldón T, Antón J. 2010. Fine-scale evolution: genomic, phenotypic and ecological differentiation in two coexisting Salinibacter ruber strains. ISME J 4:882–895. doi: 10.1038/ismej.2010.6. [DOI] [PubMed] [Google Scholar]
- 6.Scaria J, Mao C, Chen JW, McDonough SP, Sobral B, Chang YF. 2013. Differential stress transcriptome landscape of historic and recently emerged hypervirulent strains of Clostridium difficile strains determined using RNA-seq. PLoS One 8:e78489. doi: 10.1371/journal.pone.0078489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimes NE, López-Pérez M, Ausó E, Ghai R, Rodriguez-Valera F. 2014. RNA sequencing provides evidence for functional variability between naturally co-existing Alteromonas macleodii lineages. BMC Genomics 15:938. doi: 10.1186/1471-2164-15-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voigt K, Sharma CM, Mitschke J, Lambrecht J, Voß B, Hess WR, Steglich C. 2014. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J 8:2056–2068. doi: 10.1038/ismej.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vicente M, Mingorance J. 2008. Microbial evolution: the genome, the regulome and beyond. Environ Microbiol 10:1663–1667. doi: 10.1111/j.1462-2920.2008.01635.x. [DOI] [PubMed] [Google Scholar]
- 10.Chugani S, Kim BS, Phattarasukol S, Brittnacher MJ, Choi SH, Harwood CS, Greenberg EP. 2012. Strain-dependent diversity in the Pseudomonas aeruginosa quorum-sensing regulon. Proc Natl Acad Sci U S A 109:E2823–E2831. doi: 10.1073/pnas.1214128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peña A, Gomariz M, Lucio M, González-Torres P, Huertas-Cepa J, Martínez-García M, Santos F, Schmitt-Kopplin P, Gabaldón T, Rosselló-Móra R, Antón J. 2014. Salinibacter ruber: the never ending microdiversity?, p 37–53. In Papke T, Oren A, Ventosa A (ed), Genetics and genomics of halophiles. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 12.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salter I, Galand PE, Fagervold SK, Lebaron P, Obernosterer I, Oliver MJ, Suzuki MT, Tricoire C. 2015. Seasonal dynamics of active SAR11 ecotypes in the oligotrophic Northwest Mediterranean Sea. ISME J 9:347–360. doi: 10.1038/ismej.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabaldón T. 2008. Comparative genomics-based prediction of protein function. Methods Mol Biol 439:387–401. doi: 10.1007/978-1-59745-188-8_26. [DOI] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen T, Brunak S, Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 23.Rose RW, Brüser T, Kissinger JC, Pohlschröder M. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol 45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 24.Li B, Ibrahim M, Ge M, Cui Z, Sun G, Xu F, Kube M. 2014. Transcriptome analysis of Acidovorax avenae subsp. avenae cultivated in vivo and co-culture with Burkholderia seminalis. Sci Rep 4:5698. doi: 10.1038/srep05698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto AC, Melo-Barbosa HP, Miyoshi A, Silva A, Azevedo V. 2011. Application of RNA-seq to reveal the transcript profile in bacteria. Genet Mol Res 10:1707–1718. doi: 10.4238/vol10-3gmr1554. [DOI] [PubMed] [Google Scholar]
- 26.Filiatrault MJ. 2011. Progress in prokaryotic transcriptomics. Curr Opin Microbiol 14:579–586. doi: 10.1016/j.mib.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Nikel PI, Silva-Rocha R, Benedetti I, De Lorenzo V. 2014. The private life of environmental bacteria: pollutant biodegradation at the single cell level. Environ Microbiol 16:628–642. doi: 10.1111/1462-2920.12360. [DOI] [PubMed] [Google Scholar]
- 28.Balashov SP, Imasheva ES, Boichenko VA, Antón J, Wang JM, Lanyi JK. 2005. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science 309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dominy BN, Perl D, Schmid FX, Brooks CL. 2002. The effects of ionic strength on protein stability: the cold shock protein family. J Mol Biol 319:541–554. doi: 10.1016/S0022-2836(02)00259-0. [DOI] [PubMed] [Google Scholar]
- 30.Schwibbert K, Marin-Sanguino A, Bagyan I, Heidrich G, Lentzen G, Seitz H, Rampp M, Schuster SC, Klenk HP, Pfeiffer F, Oesterhelt D, Kunte HJ. 2011. A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581 T. Environ Microbiol 13:1973–1994. doi: 10.1111/j.1462-2920.2010.02336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, Rafferty JB. 2006. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res 34:1439–1449. doi: 10.1093/nar/gkl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour Filiatrault MJ, Wiedmann M, Boor KJ. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. doi: 10.1186/1471-2164-10-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oren A. 2013. Salinibacter: an extremely halophilic bacterium with archaeal properties. FEMS Microbiol Lett 342:1–9. doi: 10.1111/1574-6968.12094. [DOI] [PubMed] [Google Scholar]
- 34.Oren A, Mana L. 2003. Sugar metabolism in the extremely halophilic bacterium Salinibacter ruber. FEMS Microbiol Lett 223:83–87. doi: 10.1016/S0378-1097(03)00345-8. [DOI] [PubMed] [Google Scholar]
- 35.Hacker J, Carniel E. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep 2:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. 2014. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38:720–760. doi: 10.1111/1574-6976.12058. [DOI] [PubMed] [Google Scholar]
- 37.Carbone A, Zinovyev A, Képès F. 2003. Codon adaptation index as a measure of dominating codon bias. Bioinformatics 19:2005–2015. doi: 10.1093/bioinformatics/btg272. [DOI] [PubMed] [Google Scholar]
- 38.Garbeva P, Silby MW, Raaijmakers JM, Levy SB, de Boer W. 2011. Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors. ISME J 5:973–985. doi: 10.1038/ismej.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mongodin EF, Nelson KE, Daugherty S, Deboy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, Sharma A, Nesbø KCL, MacLeod D, Bapteste E, Doolittle WF, Charlebois RL, Legault B, Rodriguez-Valera F. 2005. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci U S A 102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gifford SM, Sharma S, Booth M, Moran MA. 2013. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J 7:281–298. doi: 10.1038/ismej.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies J. 2006. Are antibiotics naturally antibiotics? J Ind Microbiol Biotechnol 33:496–499. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 42.Antón J, Lucio M, Peña A, Cifuentes A, Brito-Echeverría J, Moritz F, Tziotis D, López C, Urdiain M, Schmitt-Kopplin P, Rosselló-Móra R. 2013. High metabolomic microdiversity within co-occurring isolates of the extremely halophilic bacterium Salinibacter ruber. PLoS One 8:e64701. doi: 10.1371/journal.pone.0064701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernier SP, Surette MG. 2013. Concentration-dependent activity of antibiotics in natural environments. Front Microbiol 4:20. doi: 10.3389/fmicb.2013.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traxler MF, Kolter R. 2012. A massively spectacular view of the chemical lives of microbes. Proc Natl Acad Sci U S A 109:10128–10129. doi: 10.1073/pnas.1207725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA, DeLong EF. 2013. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci U S A 110:E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman SM. 2013. No bacterium is an island. Microbe Mag 8:476. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.