Abstract
The ability of bacteria to monitor their metabolism and adjust their behavior accordingly is critical to maintain competitiveness in the environment. The motile microaerophilic bacterium Azospirillum brasilense navigates oxygen gradients by aerotaxis in order to locate low oxygen concentrations that can support metabolism. When cells are exposed to elevated levels of oxygen in their surroundings, motile A. brasilense cells implement an alternative response to aerotaxis and form transient clumps by cell-to-cell interactions. Clumping was suggested to represent a behavior protecting motile cells from transiently elevated levels of aeration. Using the proteomics of wild-type and mutant strains affected in the extent of their clumping abilities, we show that cell-to-cell clumping represents a metabolic scavenging strategy that likely prepares the cells for further metabolic stresses. Analysis of mutants affected in carbon or nitrogen metabolism confirmed this assumption. The metabolic changes experienced as clumping progresses prime cells for flocculation, a morphological and metabolic shift of cells triggered under elevated-aeration conditions and nitrogen limitation. The analysis of various mutants during clumping and flocculation characterized an ordered set of changes in cell envelope properties accompanying the metabolic changes. These data also identify clumping and early flocculation to be behaviors compatible with the expression of nitrogen fixation genes, despite the elevated-aeration conditions. Cell-to-cell clumping may thus license diazotrophy to microaerophilic A. brasilense cells under elevated oxygen conditions and prime them for long-term survival via flocculation if metabolic stress persists.
INTRODUCTION
The ability of bacteria to monitor their metabolism and adjust their behavior is critical to maintain competitiveness in the environment. A common metabolic stress for bacteria is starvation. Chemotaxis toward sources of limiting nutrients represents an adaptive behavior of motile cells to these conditions. If conditions limiting metabolism persist, most bacteria implement a variety of additional strategies, such as biofilm formation, sporulation, and cyst formation (1–3), which involve significant changes in cell physiology and morphology.
Azospirillum brasilense is an alphaproteobacterium isolated from various soils and the rhizosphere of plants worldwide. These bacteria have a versatile oxidative metabolism, with maximum intracellular energy being produced when cells are grown under conditions of very low oxygen concentrations corresponding to about 0.4% dissolved oxygen. The microaerophilic conditions that promote the optimum growth of A. brasilense are also the best suited for free-living nitrogen fixation (4, 5). Motile A. brasilense cells actively seek microaerophilic conditions by aerotaxis (or taxis in a gradient of oxygen) to accumulate in metabolically favorable regions (5, 6). When high oxygen concentrations persist, motile cells form clumps by cell-to-cell interactions (7, 8). Clumps correspond to motile bacteria that transiently adhere at their nonflagellated poles. Clumps are not observed unless the cells are grown with vigorous shaking (7). Persisting deleterious conditions cause the clumps to become more stable, with cells remaining adherent for longer times (8). Mathematical modeling predicts that cell-to-cell clumping limits the surface area-to-volume ratio, thereby reducing oxygen diffusion through the cells (7). Clumping is also a prerequisite to flocculation, a process during which motile vibroid A. brasilense cells transition to nonmotile round cells encased in a dense matrix of exopolysaccharides (EPS) (7, 9). Consistent with this notion, mutants unable to clump do not flocculate and precociously clumping mutants flocculate more than the wild type (7, 8, 10, 11). Flocculation is induced under conditions of high aeration, similar to the conditions that induce clumping, but it also requires a limitation in the availability of a combined nitrogen source (9). Flocculated cells are better able to endure various environmental stresses (9, 12), and some cells eventually become cysts (9). Clumping is thus an uncommitted and reversible response of motile cells to elevated oxygen concentrations, with cells being able to return to free swimming or to flocculate, depending on the evolution of the surrounding oxygen concentrations. Clumping and flocculation are likely triggered by some metabolic stress, the nature of which has not been described in much detail. Clumping and flocculation are also directly relevant to the ability of cells to colonize and establish within the rhizospheres of host plants. Microcolonies and microaggregates of A. brasilense cells formed during colonization of plant root surfaces, and strains unable to flocculate are impaired in plant root colonization (12, 13).
Initial cell-to-cell clumping in A. brasilense is caused by the impaired function of the Che1 chemotaxis signaling pathway, which controls the increase in swimming speed that occurs during chemotaxis in this species (8, 11, 14, 15). A. brasilense cells moving along an oxygen gradient not only suppress changes in the swimming direction to run for longer times and move up the gradient, but they also swim at a significantly but transiently increased speed. Both this increase in speed and smooth swimming combined are essential for aerotaxis, and mutants unable to increase their swimming speed are impaired in aerotaxis (8). Che1 controls this transient increase in swimming velocity in oxygen gradients (8). Transient changes in the swimming speed in response to changes in oxygen levels during aerotaxis also correlate with another transient behavior, which we have named clumping (8, 16). Mutants unable to increase their swimming speed in an oxygen gradient, such as strains lacking the chemotaxis response regulator CheY1 (strain AB104), clump quantitatively more than the wild type under high aeration conditions. In contrast, a mutant lacking both adaptation proteins CheB1 and CheR1 (strain BS106) swims at a constant higher velocity and is significantly impaired in its inability to clump compared to the wild-type strain, which clumps at about 9 to 10 h postinoculation under clumping conditions. The BS106 mutant may start clumping weakly, if at all, after at least 48 h of incubation (7, 8). Mathematical modeling supports the hypothesis that defects in swimming speed alone can account for the difference in the propensity for clumping (16). Consistent with the functional link between clumping and flocculation, strain AB104 flocculates more than the wild-type strain, while strain BS106 does not flocculate at all, unless it is incubated for extended periods over 48 h (7). Similarly, mutations in genes that code for receptors regulating aerotaxis (AerC and Tlp1) also result in impaired clumping and flocculation phenotypes (8, 11, 14). The Che1 signal transduction pathway also regulates cell size at division; although the underlying mechanism has not yet been elucidated, it does not seem to be functionally related to clumping (7).
The transition from free swimming to clumping and then flocculation represents an ordered set of events in response to aeration stress and nitrogen limitation. Metabolic stress is thus a determinant of clumping and flocculation. However, the physiology of clumping cells that makes them relate to this stress and how this response ensures the transition to flocculation have not been characterized. Here, we show that specific metabolic changes prime clumping cells for long-term survival, with some of the metabolic adaptations being compatible with nitrogen fixation under otherwise prohibitive oxygen conditions. These findings, in turn, provide a rationale for how some nonflocculating A. brasilense mutants have enhanced nitrogen fixation (17–19). Further, we use mutagenesis and behavioral assays to identify and implicate additional functions in the modulation of clumping and the transition between clumping and flocculation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. A. brasilense strains were grown at 28°C with shaking in tryptone-yeast extract (TY) or MMAB medium (6). We used a previously described procedure (7) in order to induce clumping behavior and flocculation. Briefly, 100 μl of an overnight culture of cells grown in TY (optical density at 600 nm [OD600], 1) was used to inoculate 5 ml of flocculation medium (MMAB medium containing 0.5 mM sodium nitrate as the sole nitrogen source and 8 mM fructose as the sole carbon source) and then grown with shaking at 28°C (7). For growth under nitrogen fixation conditions, cells were grown in MMAB medium supplemented with malate (10 mM) but lacking a nitrogen source and incubated at 28°C without any shaking to ensure microaerophilic conditions. To select for transposon mutants impaired in flocculation, we took advantage of the previous observation that changes in the properties of the cell surfaces characterize clumping and flocculation (7, 13, 17, 20, 21). We thus screened colonies for changes in the ability to bind two different dyes, Congo red and trypan blue, that target flocculation-specific surface sugars. After transposon mutagenesis, colonies were plated on MMAB medium flocculation plates supplemented with either Congo red (final concentration, 40 μg/ml) or trypan blue (final concentration, 50 μg/ml). The antibiotic used was kanamycin (30 μg/ml for A. brasilense, 50 μg/ml for Escherichia coli). The motility of the isolated mutants compared to that of the wild type was evaluated in semisoft TY (0.3% agar) after 24 h of incubation at 28°C.
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Description | Reference or source |
|---|---|---|
| Plasmids | ||
| pRL27 | Tn5-RL27 (Kmr-oriR6K); Kmr | 22 |
| pCR2.1 | TOPO cloning vector | Invitrogen |
| Strains | ||
| Escherichia coli | ||
| TOP10 | General cloning strain | Invitrogen |
| One Shot PIR1 | General cloning strain, pir-116 (used for rescue cloning) | Invitrogen |
| EA145 | 2,4-Diaminopimelic acid auxotroph, RP4 (used for mutagenesis using pRL27) | Gift from A. Buchan, The University of Tennessee |
| Azospirillum brasilense | ||
| Sp7 | Wild-type strain (ATCC 29145) | Laboratory collection |
| AB7001 | Sp7 exoC::Tn5; Kmr | 51 |
| AB104 | Sp7 ΔcheY1::Km; Kmr | 7 |
| BS106 | Sp7 Δ(cheB1-cheR1)::Km; Kmr | 52 |
| phbC | Sp7 Δ(phbC)::Km; Kmr | 43 |
| Sp72002 (flcA) | Sp7 flcA::Tn5; Kmr | 13 |
| EJ1 (rmlD) | Sp7 rmlD::Tn5; Kmr | 53 |
| FAJ310 | Sp7 amtB::Km; Kmr | 54 |
| 7628 (glnB) | Sp7 glnB::Km; Kmr | 42 |
| 7611 (glnZ) | Sp7 glnZ::Ω; Spr Smr | 42 |
| 7148 (ntrC) | Sp7 ntrC::Tn5-148; Kmr | 55 |
Transposon mutagenesis and transposon mutant complementation.
Transposons were inserted into A. brasilense strain Sp7 via biparental conjugation with an E. coli strain carrying the suicide vector pRL27 essentially as described previously (22). After incubation, the cells were resuspended in MMAB liquid medium with kanamycin (30 μg/ml) but lacking nitrogen to select against the E. coli donor strain. This transposon library was then used to isolate mutants impaired in flocculation after enrichment on the basis of flocculation, using an approach modified from that of Katupitiya et al. (17). The transposon library was first grown in TY supplemented with kanamycin to a high density (OD600, at least 1) and then transferred to a flocculation liquid medium supplemented with kanamycin. After 18 h, the cells that flocculated early were collected, and the culture was then grown for another 24 h, at which time the cells remaining in the supernatant (the nonflocculated fraction) were collected. The early-flocculated fraction of the library and the nonflocculated fraction were then separately reinoculated into TY liquid medium with kanamycin to initiate another round of flocculation and isolation of the early and nonflocculated fractions of the library. Five rounds of enrichment were performed before each fraction was serially diluted and plated on MMAB medium plates lacking nitrogen supplemented with kanamycin and Congo red or trypan blue. The colonies selected (54 total) exhibited altered staining with Congo red or trypan blue (pattern, intensity) after 72 h of growth compared to the staining of the wild type on a reference plate inoculated under similar conditions. Selected colonies were individually retested for flocculation to confirm their mutant phenotype and checked for growth impairment under conditions conducive to flocculation. We used a method described by Burdman et al. (23) to quantitatively determine the extent of flocculation. This selection strategy resulted in the identification of 32 transposon mutants. The presence of the transposon in a mutant genome was confirmed via PCR using the inwardly directed and transposon-specific primers tpnRL-13-1 (5′-AACAAGCCAGGGATGTAACG) and tpnRL-17-2 (5′-CAGCAACACCTTCTTCACGA) (22). Rescue cloning was used to isolate the DNA regions flanking the insertion of the transposon essentially as previously described (22). The transposon insertions were identified by detecting the transposon sequence from the DNA sequences obtained from sequencing of the transposon flanking regions from the rescue clones, as described previously (22).
For functional complementation of transposon mutants, parental genes were cloned into the pBBR1MCS5 broad-host-range vector and downstream of the nptII promoter available on this vector (24). Each gene locus was amplified from the genomic DNA of the wild-type strain by PCR, using a set of forward and reverse primers (Table 2) that included engineered restriction sites and optimized ribosome binding sites, as we have previously described (7), to ensure expression. PCR products were first cloned into pCR2.1 by TOPO cloning according to the manufacturer's instructions (Invitrogen, CA), verified by sequencing, and digested with KpnI and XhoI to isolate the genes whose sequences had been verified. Next, the isolated DNA fragments were cloned into the pBBR1MCS5 vector that had been linearized by KpnI and XhoI digestion. Recombinant plasmids were isolated from transformed E. coli TOP10 cells, prior to being transferred into E. coli S17-1 for biparental mating with A. brasilense transposon mutant recipient strains, as described previously (7). An empty pBBR1MCS5 vector was transferred into A. brasilense Sp7 and its transposon mutant derivatives and used as a control for functional complementation. Restoration of clumping and flocculation was tested by growing the cells carrying empty control vectors or expressing the parental genes to be rescued under clumping and flocculation conditions, as described above.
TABLE 2.
Primers used for functional complementation of transposon mutant strains
| Primer name | Restriction site | Sequencea |
|---|---|---|
| mannosyl_For | KpnI | GGTACCAGGAGGGGCCCGTATGACCGTTGAAGCCCTTGAG |
| mannosyl_Rev | XhoI | CTCGAGTCAGAACGCCTTCAGCTTCTTCCA |
| cheA3_For | KpnI | GGTACCAGGAGGGGCCCGTATGGCTGTCGCTGGTCGCCCGCCTG |
| cheA3_Rev | XhoI | CTCGAGTCATGCGGCACCTTTCTGCTCGGA |
| transp_For | KpnI | GGTACCAGGAGGGGCCCGTATGCTCCACGTCTTCGCCTGC |
| transp_Rev | XhoI | CTCGAGTTACAGCCAGCGCCGCCGCTTGAAGTA |
| noeL_For | KpnI | GGTACCAGGAGGGGCCCGTATGACCGCGCTCCTCTTACGC |
| noeL_Rev | XhoI | CTCGAGTTATGGCTCGCGCATGCCTTCCGCACG |
| phosp_For | KpnI | GGTACCAGGAGGGGCCCGTATGACCGACATCGTCCAGCTT |
| phosp_Rev | XhoI | CTCGAGTTACCAGCGCAGGCTGGGGAG |
| glycgp1_For | KpnI | GGTACCAGGAGGGGCCCGTAATGGACGTCGAATCCCAGCT |
| glycgp1_Rev | XhoI | CTCGAGTCAGCGGCCTTGCGCGGCCCG |
| tonB_For | KpnI | GGTACCAGGAGGGGCCCGTATGCTGATCGTGCCGAGCCTTC |
| tonB_Rev | XhoI | CTCGAGTCAGAAATCGACGGACACCGACAG |
| amyl_For | KpnI | GGTACCAGGAGGGGCCCGTATGAGGCGCGGCGGCACGGCATCG |
| amyl_Rev | XhoI | CTCGAGTTAAACTTCAGGTTCGTGAACTTATGA |
| ybgC_For | KpnI | GGTACCAGGAGGGGCCCGTATGGCGAATCTCTCGGGCTGGT |
| ybgC_Rev | XhoI | CTCGAGTCAGTCTCGCTTCTGCCTTTTGTACAA |
Engineered restriction sites are underlined. Engineered ribosome binding sites in forward primers are indicated in italics.
Proteome preparation and liquid chromatography/liquid chromatography-tandem mass spectrometry.
Cell pellets were collected from cultures grown under clumping conditions (∼10 h postinoculation, high aeration, MMAB medium), weighed, and frozen at −80°C for later proteome preparation. Under these conditions, the cells divide at similar rates and the number of cells is similar between the wild type and the mutants (7). Cells were harvested and lysed by sonication. Cleared lysate was centrifuged at 10,000 rpm in a Sorvall ultracentrifuge, and the pellet was resolubilized in 50 mM Tris-HCl, pH 7.5, to a protein concentration of 1 mg/ml. Cell pellets were resuspended at a rate of 0.1 g (wet weight) cell pellet per 500 μl buffer in 6 M guanidine hydrochloride and lysed by sonication. The lysate was then cleared of debris by centrifugation, and the cleared lysate was diluted for subsequent digestion. After addition of 10 mM dithiothreitol and 1 h of incubation at 60°C, 20 μg sequencing-grade trypsin (Promega) was added, and the solution was incubated overnight at 37°C. An additional 20 μg trypsin was added to each sample on the following morning, and the samples were incubated for an additional 6 h. Digestion was halted by addition of 0.1% formic acid. The digested samples were then desalted using Sep-Pak Plus C18 solid-phase extraction (Waters, Milford, MA) following the manufacturer's recommendations. Eluates were then solvent exchanged via vacuum centrifugation into buffer A (high-pressure liquid chromatography-grade water [Burdick-Jackson], 0.1% formic acid). Samples were aliquoted into 40-μl volumes and frozen at −80°C for later analysis. Analysis of cleaned, digested samples was accomplished using the multidimensional protein identification technology (25, 26), with each sample being analyzed in duplicate. Data were acquired using an LTQ mass spectrometer (Thermo Scientific). Data were collected under the control of Xcalibur software in a data-dependent mode, with one parent ion scan being followed by six tandem mass spectrometry scans. Dynamic exclusion was enabled, with a repeat count of 1, a repeat duration of 60 s, an exclusion list size of 300, and an exclusion duration of 180 s being used.
Proteomics data analysis.
Raw sequence files were searched, using the SEQUEST (version 27) program (27), against the A. brasilense Sp245 genome sequence (28) (version of 19 September 2008, http://genome.ornl.gov/microbial/abra/19sep08/) since an A. brasilense Sp7 species-specific database is not yet available. Sequences of common contaminants were appended to the database, as were reversed sequences of each protein in the database, to allow estimation of false discovery rates. The database contained data for 6,927 A. brasilense proteins, 52 contaminants, and 6,979 reversed sequences, for a total of 13,958 entries. Search parameters were as follows: tryptic digestion, a peptide mass tolerance of 3 m/z, and a fragment ion tolerance of 0.5 m/z. Additionally, search parameters included two dynamic modifications: (i) methylation was represented by a mass shift of +14 m/z on glutamate residues, and (ii) deamidation followed by methylation was represented by a mass shift of +15 m/z on glutamine residues. The results were then filtered and collated using the DTASelect tool (29) with the following parameters: XCorr (cross-correlation) values of ≥1.8 for peptides with a charge state of +1, ≥2.5 for those with a charge state of +2, and ≥3.5 for those with a charge state of +3, a delta CN (specificity of fit) value of ≥0.08, and at least 2 peptides (or 1 peptide in two different charge states) per protein identification. False discovery rates were 1% at the peptide level and 4% at the protein level for the Sp7 and BS106 strains and 4% and 12%, respectively, for the AB104 strain. Raw mass spectrometry data files are available at chorusproject.org under the project name “Azospirillum brasilense.” Complete lists of the identified proteins are provided in Table S1 in the supplemental material.
Search results were imported into Microsoft Access software for analysis. Two technical replicates of each sample were pooled for all data analyses. Raw values of the spectrum count for each protein were adjusted to account for shared tryptic peptides (30). Missing spectrum count values were replaced with a value of 0.3 to prevent divide-by-zero errors when ratios were computed. The relative abundance of each protein detected within a sample was determined through calculation of a normalized spectral abundance factor (NSAF) for each identified protein (30). The levels of upregulation and downregulation were determined by calculating the NSAF value of each protein in the mutant cultures divided by the NSAF value of the same protein in Sp7 control cultures. Comparisons of protein abundance across samples were performed only for proteins with an average spectrum count of ≥4 across all measurements, after adjustment and normalization as described above. The G test (31) was applied to pairwise comparisons of mutant and control strains, and the Benjamini-Hochberg correction (32) was further applied with a cutoff value of 0.05 to limit the false discovery of protein abundance changes. Further, filter levels of a 2.0-fold change in expression were applied to these data sets.
RT-PCR analysis.
RNA was extracted from cultures grown in TY liquid medium at 28°C with shaking to log phase (OD600 of <0.5) or stationary phase (OD600 of >0.8) using a Qiagen RNeasy kit (Qiagen, Inc.), according to the manufacturer's instructions. cDNA was produced using random hexamers and a ThermoScript reverse transcription-PCR (RT-PCR) system (Invitrogen) following the manufacturer's recommendations. PCRs were performed using the cDNA produced as the templates, primers specific for the transcripts to be detected, and a Failsafe PCR system (Epicentre Biotechnologies). The primers used were specific for internal regions of the moaA transcripts (primers moaA-for [5′-TGAACGACGACGAGTTCC] and moaA-rev [5′-TCATCCGCCGGTCACGCT]), of the Fe-S ferredoxin transcripts (primers Fe-S-for [5′ CGGAGGGCTGCACGCTCT] and Fe-S-rev [5′-TCAGTGGACGCTTCCGCC]), and of the 16S rRNA gene, used as a control (primers 16S-for [5′ GGTCTGAGAGGATGATCA] and 16S-rev [5′-TGCACCCCAGCGTCTAGC]). PCR conditions consisted of an initial denaturation at 95°C for 3 min, followed by 40 cycles consisting of subsequent steps of denaturation at 95°C for 45 s, annealing at 55.5°C for 30 s, and elongation at 72°C for 1 min, and these steps were followed by a final elongation at 72°C for 10 min. The PCR products were analyzed on a 1% agarose gel. The experiment was repeated at least twice on independently prepared cDNA samples.
Western blotting.
Cell pellets were obtained from cultures of the wild-type strain grown in MMAB medium supplemented with ammonium chloride (concentration, 10 mM) under flocculation conditions in flocculation medium or under nitrogen fixation conditions. Whole-cell protein extracts were prepared by sonication of cell pellets in phosphate-buffered saline (PBS) buffer, adjusted to equivalent cell densities, and resuspended and washed by centrifugation in PBS buffer. Protein concentrations were determined using the bicinchoninic acid method (Pierce) and separated on a 10% SDS-polyacrylamide gel, followed by transfer onto a nitrocellulose membrane. Western blotting assays were conducted using a NifH antiserum (a kind gift of L. Huergo) at a 1:5,000 dilution, essentially as described previously (11).
RESULTS
Nutrient scavenging is a hallmark of clumping cells.
To characterize the physiology of clumping cells, we used two chemotaxis mutant strains, BS106 (ΔcheB1 ΔcheR1) and AB104 (ΔcheY1), which, respectively, do not clump and clump significantly more than the wild-type strain when grown under elevated-aeration conditions conducive to clumping (Tables 3 and 4). Under these conditions, there is no detectable change in the growth rate of these strains (7). We then used shotgun proteomics to compare the physiology of these two strains with that of the wild type, when growing under clumping conditions.
TABLE 3.
Proteins upregulated during growth under clumping conditions in BS106 and AB104 relative to their levels of regulation in the wild-type strain
| Functional group, locus tag | NCBI GI accession no. | Product | Fold change in expressiona |
|
|---|---|---|---|---|
| BS106 | AB104 | |||
| Amino acid metabolism | ||||
| AZOBR_p110151 | 392377302 | Dihydroxyacid dehydratase, IlvD | 2.00 | |
| AZOBR_p1110080 | 392378209 | Bifunctional (acetylornithine and succinyldiaminopimelate aminotransferase, ArgD) | 2.06 | |
| AZOBR_p1120047 | 392378339 | 4-Aminobutyrate transaminase, GabT | 2.99 | |
| AZOBR_120050 | 392382411 | Cysteine synthase, CysK2 | 4.13 | |
| AZOBR_100230 | 392382078 | Nitrogen regulatory protein P-II, GlnB | 9.54 | |
| Carbohydrate metabolism | ||||
| AZOBR_10049 | 392380562 | Acetyl-CoA synthetase, AcsA | 2.46 | |
| AZOBR_p60069 | 392380456 | Phosphomannomutase, ExoC | 2.68 | |
| AZOBR_p280010 | 392384431 | Acetoacetyl-CoA reductase, PhbB | 3.25 | |
| AZOBR_p110080 | 392377233 | Glycogen phosphorylase, GlgP | 3.54 | |
| AZOBR_p440019 | 392379933 | Acetoacetyl-CoA reductase, PhbB | 3.95 | |
| AZOBR_70042 | 39238167 | Granule-associated protein (phasin) | 6.36 | |
| AZOBR_p110146 | 392377297 | Granule-associated protein (phasin) | 2.61 | 3.20 |
| AZOBR_p420023 | 392379838 | Pyruvate formate lyase 1, PflB | 12.13 | |
| Energy metabolism | ||||
| AZOBR_p110118 | 392377271 | Cytochrome c, CycA | 2.20 | |
| AZOBR_p140082 | 392377731 | Electron transfer flavoprotein alpha subunit, EtfA | 2.30 | |
| AZOBR_10053 | 392380566 | Putative enzyme of heme biosynthesis (HemY-like) | 3.03 | |
| Lipid metabolism, AZOBR_110041 | 392382289 | Enoyl-(acyl-carrier-protein) reductase (NADH), FabI1 | 4.12 | |
| Metabolism of cofactors, vitamins, AZOBR_140242 | 392382705 | Riboflavin synthase, alpha subunit, RibC | 6.62 | |
| Nucleotide metabolism, AZOBR_p130122 | 392377557 | IMP dehydrogenase, GuaB | 2.20 | 2.26 |
| Cell division, AZOBR_p220010 | 392383868 | DNA-binding protein HU, Hup | 2.41 | 3.03 |
| Translation | ||||
| AZOBR_160012 | 392383025 | 50S ribosomal subunit protein L11, RplK | 2.07 | |
| AZOBR_110102 | 392382348 | 50S ribosomal protein L9, RplI | 2.45 | |
| AZOBR_10172 | 392380677 | Putative tRNA/rRNA methyltransferase | 2.66 | |
| AZOBR_150245 | 392383000 | 50S ribosomal subunit protein L24, RplX | 2.85 | |
| AZOBR_70161 | 392381783 | 50S ribosomal protein L36, RpmJ | 2.89 | |
| AZOBR_150249 | 392383004 | 50S ribosomal subunit protein L16, RplP | 3.18 | |
| AZOBR_150246 | 392383001 | 50S ribosomal subunit protein L14, RplN | 4.80 | |
| Membrane transport | ||||
| AZOBR_p1120016 | 392378310 | d-Methionine ABC transporter protein, periplasmic binding protein, MetQ2 | 2.05 | |
| AZOBR_p470016 | 392380221 | Putative ABC transporter (substrate-binding protein) | 2.58 | |
| AZOBR_100136 | 392381988 | Putative ABC transporter, periplasmic ligand-binding component | 4.24 | |
| AZOBR_p440178 | 392380086 | Putative RND efflux transporter, MFP subunit | 18.01 | |
| Motility and signal transduction | ||||
| AZOBR_p1160045 | 392378761 | Flagellin | 2.83 | |
| AZOBR_180037 | 392383138 | Methyl-accepting chemotaxis protein | 7.95 | |
| Protein turnover | ||||
| AZOBR_100239 | 392382087 | ATP-dependent protease Lon | 2.11 | |
| AZOBR_20015 | 392381054 | Small subunit of chaperonin GroESL, GroES | 2.14 | |
| Cell envelope biogenesis, AZOBR_140018 | 392382489 | Outer membrane lipoprotein of the Pal-Tol system, Omp | 3.42 | |
| Protein secretion | ||||
| AZOBR_p1140133 | 392378687 | Type VI secretion protein | 2.86 | |
| AZOBR_180276 | 392383362 | Conserved virulence factor protein | 7.06 | |
| Unknown function | ||||
| AZOBR_200174 | 392383617 | Hypothetical protein | 2.37 | |
| AZOBR_100025 | 392381887 | Hypothetical protein | 3.06 | |
| AZOBR_100035 | 392381895 | Hypothetical protein | 3.93 | |
| AZOBR_p140066 | 392377715 | Hypothetical protein | 4.66 | |
| AZOBR_10054 | 392380567 | Hypothetical protein | 4.80 | |
| AZOBR_p440099 | 392380009 | Conserved protein of unknown function | 4.98 | |
| AZOBR_180136 | 392383231 | Hypothetical protein | 5.13 | |
| AZOBR_p130059 | 392377496 | Hypothetical protein | 7.75 | |
| AZOBR_p1110075 | 392378204 | Hypothetical protein | 8.04 | 18.87 |
| AZOBR_10026 | 392380539 | Conserved exported protein of unknown function | 65.91 | 50.64 |
The fold change in expression was determined as described in the Materials and Methods section.
TABLE 4.
Proteins downregulated during growth under clumping conditions in strains BS106 and AB104 relative to their levels of regulation in the wild-type strain
| Functional group, locus tag | NCBI GI accession no. | Product | Fold change in expressiona |
|
|---|---|---|---|---|
| BS106 | AB104 | |||
| Amino acid metabolism | ||||
| AZOBR_110031 | 392382279 | Urease, gamma subunit | 5.36 | |
| AZOBR_p1110049 | 392378180 | Aspartokinase | 2.70 | |
| Carbohydrate metabolism | ||||
| AZOBR_40292 | 392381393 | Dihydrolipoyl dehydrogenase flavoprotein | 2.63 | |
| AZOBR_100325 | 392382171 | Enolase (2-phosphoglycerate dehydratase) | 2.63 | |
| AZOBR_40299 | 392381400 | Transaldolase | 3.52 | |
| Energy metabolism | ||||
| AZOBR_100248 | 392382096 | NADH-quinone oxidoreductase, subunit F, NuoF | 3.51 | |
| AZOBR_70054 | 392381682 | Formate dehydrogenase, A subunit | 17.34 | 3.05 |
| Nucleotide metabolism, AZOBR_10040 | 392380553 | Bifunctional PurH | 2.11 | |
| Motility and signal transduction | ||||
| AZOBR_10322 | 392380823 | Putative transcriptional regulator, HTH-XRE family | 3.44 | 15.78 |
| AZOBR_p410056 | 392379767 | Flagellin, Laf1 | 6.91 | 22.56 |
| Translation | ||||
| AZOBR_150112 | 392382875 | Putative endoribonuclease L-PSP | 2.25 | |
| AZOBR_40325 | 392381424 | 50S ribosomal subunit protein L27 | 2.97 | |
| AZOBR_150240 | 392382997 | 50S ribosomal subunit protein L6 | 3.45 | |
| AZOBR_p170010 | 392377861 | Tyrosine tRNA synthetase | 7.43 | |
| Membrane transport | ||||
| AZOBR_p220082 | 392383940 | Glycine betaine/proline transport system substrate-binding protein | 3.19 | |
| AZOBR_120040 | 392382401 | Amino acid ABC transporter, substrate-binding component | 3.40 | |
| Protein turnover | ||||
| AZOBR_10528 | 392381029 | Putative intracellular protease | 2.11 | 6.54 |
| AZOBR_10367 | 392380868 | ATP-dependent protease, component of the HslUV proteasome | 7.91 | |
| Oxidative stress, AZOBR_p120093 | 392377402 | Superoxide dismutase, iron/manganese cofactor | 2.95 | |
| Unknown function, AZOBR_40088 | 392381199 | Hypothetical protein | 7.78 | |
The fold change in expression was determined as described in the Materials and Methods section.
The A. brasilense Sp7 control yielded 1,366 protein identifications, while mutant strains BS106 and AB104 gave 1,355 and 1,280 identifications, respectively. The three strains had 921 common proteins, and the reproducibility between technical replicates was 63% for AB104 culture samples and 71% for both BS106 and wild-type culture samples. The AB104 and BS106 cultures had 1,003 common protein identifications, giving a good representation of the proteins required for basic metabolic functions in both mutant cultures. Patterns of up- and downregulation relative to the regulation in the wild type (Tables 3 and 4) were very different for the nonclumping BS106 strain and the hyperclumping AB104 strain, indicating that each mutant strain has a unique physiological response under clumping conditions, as expected. No protein identified in AB104 was inversely regulated in BS106, even though these strains have opposite clumping. This suggests that the differentially expressed proteins (relative to their expression in the wild type) detected in both AB104 and BS106 are not directly associated with clumping (Tables 3 and 4). Therefore, differences in the patterns of protein expression among the wild type, AB104, and BS106 likely reflect physiological changes experienced by clumping, hyperclumping, and nonclumping cells grown under elevated-aeration conditions.
The proteins uniquely up- or downregulated in BS106 could represent proteins with functions related to the compensatory response caused by its inability to clump rather than clumping per se. Proteins downregulated only in the BS106 strain may also represent proteins with functions whose expression is not needed by clumping cells (Table 4). However, comparison of the up- and downregulated proteins in clumping wild-type cells with those in the hyperclumping AB104 strain is expected to reflect proteins responsible for physiological traits specific to clumping cells. The largest class of up- and downregulated proteins identified in AB104 corresponds to proteins for which the function is unknown. Of the proteins uniquely upregulated in AB104 relative to their regulation in the wild type, those corresponding to amino acid metabolism, metabolism of cofactors and vitamins, motility and signal transduction, protein turnover (chaperones), and protein secretion represented the most significant sets, since they were specific to clumping cells, with no change in expression being detected in the nonclumping BS106 cells (Table 3). Several key enzymes in amino acid biosynthesis were significantly overexpressed in the clumping mutant AB104 relative to their levels of expression in the wild type (Table 3). A putative urease and aspartokinase were also specifically downregulated in the hyperclumping AB104 (Table 4). The increase in amino acid biosynthesis coincident with a decreased expression of proteins implicated in amino acid utilization during catabolism is consistent with clumping cells adopting a metabolism aimed at conserving amino acids. This assumption was further supported by the observation that the expression of a tyrosine tRNA synthetase was significantly lower in hyperclumping cells than in the wild type, suggesting that these cells experience some amino acid limitation. The increased expression of a PII GlnB protein in hyperclumping AB104 cells is consistent with this hypothesis, since the expression of glnB increases in A. brasilense under nitrogen-limiting conditions (12). As clumping increased, the expression of proteins implicated in the accumulation of carbon storage materials (granules of polyhydroxybutyrate [PHB], glycogen, acetyl coenzyme A [CoA] synthetase, phasins; Table 3) also increased. This elevated carbon storage activity as clumping increased in AB104 was concomitant with a reduction in the expression of several proteins involved in glycolysis (dihydrolipoyl dehydrogenase flavoprotein, enolase) and the pentose phosphate pathway (transaldolase) (Table 4). A shift in metabolism in clumping cells was also evidenced by the decreased expression in a major enzyme of the oxidative electron transport chain (NuoF), the reduced expression of a superoxide dismutase (Table 4), and the elevated expression of different enzymes involved in electron transfer and energy metabolism and in the synthesis of major cofactors to assist with these activities (Table 3). The increased expression of a Lon protease and GroES subunit of the GroESL chaperone in the hyperclumping AB104 relative to that in wild-type cells also shows that these cells experience stress, but it also supports the hypothesis that clumping is associated with a nutrient-scavenging strategy. A major reorganization of the cell's metabolism during clumping is also suggested by the up- and downregulation of several transporters as clumping increases (Table 3). The elevated levels of expression of a protein belonging to the type VI secretion system and of an outer membrane protein in the hyperclumping strain are consistent with increased cell-to-cell contacts and cell envelope remodeling, respectively, as clumping progresses (Table 3). The increased levels of expression of a chemotaxis receptor and an outer membrane lipoprotein in the hyperclumping AB104 relative to those in the wild-type strain further suggest that cells experience their surroundings increasingly differently, again linking this behavior to changes in metabolism (Tables 3 and 4). A significant increase in the expression of several proteins that comprise the large subunit of the ribosome exclusively in hyperclumping AB104 cells is also consistent with major changes in the cells' physiology (Table 3). While most large ribosomal subunit proteins detected to be differentially expressed between the mutants and the wild-type strain had elevated levels of expression in the hyperclumping strain, two 50S ribosomal proteins were downregulated in the AB104 mutant relative to their levels of expression in the wild-type strain. These different changes in the expression patterns of several proteins that comprise the large subunit of the ribosome may indicate specific responses of the translation machinery to the metabolic stress experienced by the cells.
Intracellular nitrogen status triggers a transition from clumping to flocculation.
Clumping is required for flocculation, and both behaviors are induced under conditions of metabolism imbalance triggered by elevated aeration and, for flocculation only, limitations in the availability of a nitrogen source. The data presented above suggest that clumping cells are scavenging nitrogen compounds and accumulating polyhydroxybutyrate granules. To further test this hypothesis, we analyzed clumping and flocculation in several mutant strains affected in carbon or nitrogen metabolism (Table 1).
A. brasilense wild-type cells grown under conditions of flocculation (elevated aeration and limited nitrogen nutrition) clumped at about 9 h postinoculation and flocculated at about 24 h postinoculation (Fig. 1). A mutation in the poly-beta-hydroxybutyrate polymerase (encoded by phbC), involved in PHB granule synthesis, led to significant defects in the ability of cells to clump, while the effect on flocculation was minimal. Mutants defective in genes involved in nitrogen sensing and metabolism (amtB, ntrC, glnB, and glnZ) were also tested. The amtB and ntrC mutants had a minor clumping and flocculation phenotype, while the glnB and glnZ mutants were impaired in clumping and severely affected in flocculation. These data suggest that nitrogen limitation is a trigger signal for flocculation. It follows that provision of a source of combined nitrogen to flocculated cells should restore vegetative growth. We tested this possibility by supplementing the cultures of flocculated wild-type A. brasilense cells with either a preferred carbon source (malate) or different sources of nitrogen (Fig. 2). The addition of ammonium chloride and, to a lesser extent, glutamate, which also corresponds to the reduced assimilated form of ammonium, but no other treatment (including nitrate) was sufficient to rescue flocculation. These results support the hypothesis that flocculation is induced by fixed nitrogen (in the form of ammonium) starvation.
FIG 1.
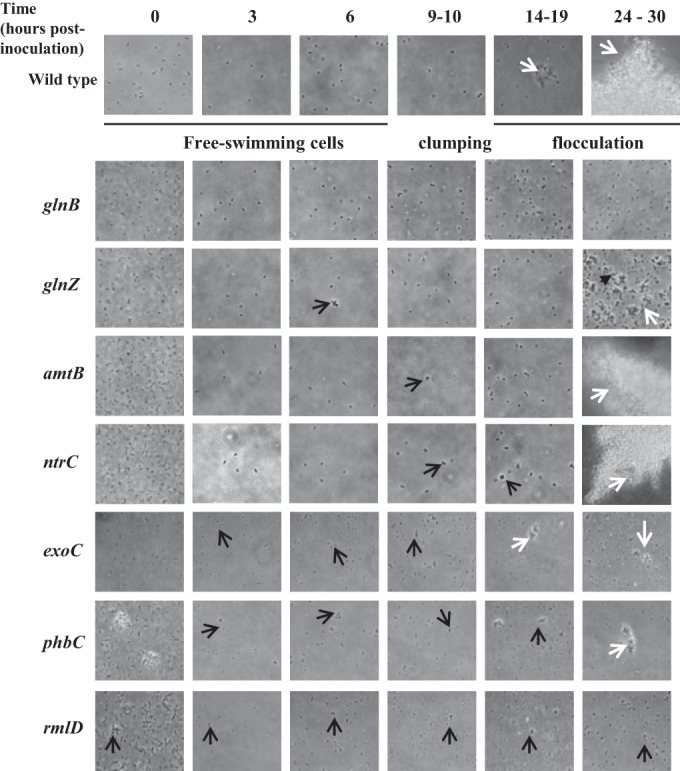
Effect of mutations in genes for cell envelope proteins and carbon and nitrogen metabolism on clumping and flocculation. Representative images of cells at the different times indicated at the top are shown. Black arrows, clumps (transient clusters of motile cells); white arrows, flocs (stable aggregates of nonmotile cells).
FIG 2.
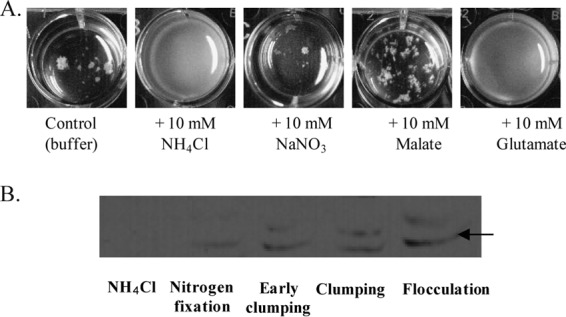
Evidence for different metabolisms in clumping and flocculated cells. (A) Rescue of flocculation by the exogenous addition of a carbon or nitrogen source. At 24 h, a flocculated culture of the wild-type strain was supplemented with the indicated compound and allowed to grow for an additional 12 h. At 24 h, the initial flocculated culture is similar to that shown for the control. The white aggregates seen in the wild-type control cultures are flocs of cells. (B) Western blot using a crude anti-NifH antiserum against whole protein extracts of the wild-type strain grown under the conditions indicated. A band at about 42 kDa (arrow), corresponding to the NifH subunit, could be detected in cultures grown under nitrogen fixation or flocculation conditions but not when ammonium chloride was present in the culture. A cross-reacting band, the nature of which is unknown, is also seen in cultures grown under clumping and flocculation conditions. Early clumping, cells incubated under flocculation conditions for 3 h postinoculation; Clumping, cells incubated under flocculation conditions for 9 h postinoculation; Flocculation, wild-type cells grown under flocculation conditions and collected at 24 h postinoculation.
Proteomics data suggested that as clumping increases, cells experience their surroundings differently, which could possibly be related to the increased cell-to-cell contacts and cell envelope remodeling. We thought to test this hypothesis by analyzing clumping and flocculation in cell envelope mutant strains (lacking the rmlD or the exoC gene). Both mutants analyzed were found to clump precociously and to be significantly impaired in flocculation, supporting the notion that cell envelope remodeling is required for clumping and flocculation (Fig. 1).
Proteomics also suggested that cells experience a different environment as clumping increases. We hypothesized that oxygen diffusion may be more limited in clumping cells. We reasoned that if this hypothesis is correct, then both clumping and flocculated cells should express functions that are typically tightly repressed by the presence of oxygen. We tested this hypothesis by determining, using an anti-NifH antiserum (gift of Luciano F. Huergo, Universidade Federal do Paraná, Brazil), whether the nitrogenase enzyme, for which expression is tightly controlled by oxygen and ammonium availability, was expressed by wild-type cells grown to clumping or to an early flocculation stage (Fig. 2). Note that we used cells that flocculated early after inoculation (about 24 h) because the prolongation of growth under these conditions led to the production of dormant cells and cysts that were very difficult to lyse (data not shown). Consistent with our hypothesis, a faint band corresponding to NifH was seen under conditions of nitrogen fixation, clumping, and early flocculation (Fig. 2). These results suggest that intracellular conditions during clumping and early flocculation are sufficient to induce the expression of an oxygen-sensitive enzyme, such as the nitrogenase.
Screening of a transposon mutant library for additional genes implicated in cell envelope remodeling.
The data presented above linked clumping behavior with nutrient scavenging for adaptation of cells to metabolically limiting elevated-aeration conditions. To identify additional functions modulating clumping and flocculation, we screened a transposon mutant library for defects in flocculation. We enriched for mutants that were impaired in the extent or timing of flocculation and then selected for those with defects in cell envelope polysaccharides, which are detected by the binding of dyes specific for diverse EPS and are associated with flocculation ability (33). This approach yielded a set of 32 mutants which were able to grow on a nitrogen-free medium and which did not display a growth defect in the flocculation medium (Table 5). Some of these mutants did not flocculate at all, while others had only a reduced ability to flocculate. Some, but not all mutants, were also impaired in the timing of clumping.
TABLE 5.
Transposon mutants and their clumping and flocculation phenotypes
| No. of mutants isolated | GI (GenBank) accession no. | Predicted function | Functional category | Clumping time (h postinoculation) | Flocculationa | Functional complementationb |
|---|---|---|---|---|---|---|
| 3 | gi|13105000 (YP_005031193.1) | Putative alpha-amylase (alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase) | Cell envelope metabolism | 3 | Absent | Yes |
| 7 | gi|392380387 (YP_004987578.1) | Putative glycosyltransferase, family 2, putative dolichyl-phosphate beta-d-mannosyltransferase | Cell envelope metabolism | 6 | Absent | Partial (stable clumps but no flocculation) |
| 2 | gi|392383473 (YP_005032670.1) | Putative sugar phosphatase, HAD family | Cell envelope metabolism | 6 | Reduced | Yes |
| 6 | gi|392383472 (YP_005032669.1) | Putative glycosyltransferase, group 1 | Cell envelope metabolism | 9 | Reduced | Yes |
| 1 | gi|392380402 (YP_004987559.1) | NoeL/GDP mannose dehydratase | Cell envelope metabolism | 9 | Reduced | Yes |
| 1 | gi|392382484 (YP_005031681.1) | Acyl-CoA thioester hydrolase, YbgC-like | Cell envelope metabolism | 6 | Absent | Yes |
| 2 | gi|392380513 (YP_005029834.1) | Putative magnesium/cobalt transporter | Membrane transport | 6 | Severely reduced | Yes |
| 1 | gi|392379600 (YP_004986759.1) | TonB-dependent siderophore receptor | Membrane transport | 3 | Absent | Yes |
| 6 | gi|56875477 (HE577327) | Intergenic region, 7 bp at 5′ side (molybdenum cofactor biosynthesis protein MoaA) and 30 bp at 3′ side ([4Fe-4S] ferredoxin) | Intergenic | 9 | Absent | Not determinedc |
| 1 | gi|612168671 (EZQ05301.1) | CheA3 (ACF pathway) | Signal transduction | 9 | Absent | Yes |
Flocculation relative to that of the wild-type strain at 24 h postinoculation, determined as described in the Materials and Methods section. Absent, no flocculation; Reduced, a flocculation level less than 50% of that of the wild-type strain; Severely reduced, a flocculation level less than 5% of that of the wild-type strain.
For functional complementation, the clumping and flocculation phenotype of a transposon mutant strain carrying an empty vector or carrying the parental gene expressed from the constitutive nptII promoter present on the vector as described in the Materials and Methods section.
The transposon insertion did not lead to a loss of function but caused a downstream gene to be constitutively expressed, as shown in Fig. S1 in the supplemental material.
Transposon mutations that cause defects both in the time to clumping and in the ability of cells to flocculate include insertions located within genes for a putative alpha-amylase (alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase), a putative glycosyltransferase (putative dolichyl-phosphate beta-d-mannosyltransferase), a sugar phosphatase of the HAD family, and an acyl-CoA thioester hydrolase encoded by a gene similar to the E. coli ybgC gene (Table 5). The last transposon is inserted in a ybgC-like gene located upstream of a cluster of genes coding for homologs of TolAQR, similar to the genomic organization in E. coli. All the genes affected by the transposon insertions described above are likely impaired in the production of proteins involved in early cell envelope structure/function, since they affect both clumping and flocculation. A similar inference can be made regarding the transposon insertions located within the gene for a homolog of a TonB receptor, as well as within the genes for a magnesium/cobalt transporter.
Transposon insertions that abolish or reduce flocculation without any detectable effect on clumping are probably located in genes with roles related to the transition of clumping to flocculation and, thus, in later stages of adaptation to the metabolic stress caused by exposure to persistent elevated-aeration conditions. These include insertions in the intergenic region between the gene for a putative molybdenum cofactor biosynthesis protein (MoaA) and the gene for a [4Fe-4S] ferredoxin (Table 5). The latter insertion causes the moaA gene to be constitutively expressed in the transposon mutant under flocculation conditions, while it is turned off in the wild type under these conditions (see Fig. S1 in the supplemental material). The constitutive expression of moaA in the mutant is likely caused by the activity of a promoter located within Tn5, as shown in other studies (e.g., see reference 33). An insertion within the gene for a chemotaxis histidine kinase homolog encoded by a gene within a locus coding for an ACF-like chemotaxis signal transduction system also causes a defect in flocculation.
Additional transposon insertions which cause specific defects in flocculation but not clumping were located in genes encoding a putative glycosyltransferase, a GDP-mannose dehydratase previously implicated in EPS biosynthesis in A. brasilense and named NoeL (4, 5) (Table 5). These mutations likely affect EPS production and, thus, cell envelope remodeling at later stages of adaptation to the oxygen stress experienced by the cells.
All mutations but one (a putative dolichyl-phosphate beta-d-mannosyltransferase) could be functionally rescued by expressing the parental genes from a constitutive promoter on a low-copy-number plasmid (Table 4). These results indicate that the transposon insertions are nonpolar and that they directly cause the defects in clumping and/or flocculation that are observed. Stable clumps, in which cells loose motility (see Fig. S2 in the supplemental material), could be observed by expressing the parental gene in the strain in which the transposon is inserted into a putative mannosyltransferase gene, but flocculation could not be restored. There are two possible explanations for this failure to complement. First, the expression of the mannosyltransferase may be more transient and at levels significantly different from those available from the constitutive promoter. Second, it may be that the role of the mannosyltransferase in flocculation is indirect and that an additional mutation, probably selected by the mutant enrichment method used, acting alone or in combination with the defect in the mannosyltransferase is responsible for the flocculation defect. The results thus suggest that this putative enzyme may contribute to the transition from dynamic clumps to stable clumps, but its exact role in flocculation should be confirmed in the future.
DISCUSSION
Elevated oxygen concentrations impede the oxidative metabolism of A. brasilense, for which energy metabolism is best under microaerophilic conditions (4, 5, 7, 8, 11). Cell-to-cell clumping is a dynamic and transient behavioral response of motile A. brasilense cells to the metabolic stress caused by elevated aeration (8, 14, 16). This behavior is initiated by a transient decrease in swimming speed, and the cells resume free swimming if conditions improve (7, 14, 16). In contrast, clumping cells become irreversibly adherent to one another if conditions worsen and may eventually flocculate if nitrogen is limiting (7, 8). Here, we provide evidence that clumping cells adjust their metabolism and respond to the imposed stress by scavenging both carbon and nitrogen nutrients. The proteomic data indicate that clumping cells adopt a nutrient-scavenging strategy and prepare for further nutritional stresses by, for example, increasing carbon storage and conserving amino acids to cope with elevated-aeration conditions; the characterization of several mutants affected in nitrogen and carbon metabolism validated this assumption. In this respect, the physiology of clumping cells resembles that of nondifferentiating bacteria exposed to transient starvation conditions and includes a scavenging metabolism to prepare cells for further environmental stress (1). Metabolic scavenging during clumping primes the cells for flocculation: clumping cells accumulate carbon in the form of PHB granules and secrete some in the form of EPS. EPS secretion is expected to modify the properties of cell surfaces, which our mutagenesis analysis, biased for the identification of cell envelope mutants, confirmed is critical for flocculation. Clumping cells also scavenge nitrogen compounds. Our data indicate that clumping cells ready their physiology for subsequent nitrogen limitation by preparing for nitrogen fixation. In contrast to clumping, flocculation is a process triggered by metabolic stress, and it involves significant morphological changes, including a loss of motility, increased accumulation of PHB granules, and increased EPS production, representing a reorganization of the cell's physiology more profound than that caused by clumping (8, 9, 13, 20, 34–36). Numerous studies have linked flocculation with resistance to various stresses, since mutations that abolish or reduce flocculation also impair stress endurance (20, 37–40). The data obtained in the present study provide a physiological rationale for the increased stress resistance of flocculated cells and highlight a role for clumping as a transitional and preparatory behavior for resistance to metabolic stress.
Both clumping and flocculation are characterized by changes in the adhesive properties of cell surfaces, including changes in the nature of the EPS produced (7, 10, 35, 36). The results obtained here are consistent with these previous findings. The transposon screen for clumping and flocculation defects identified enzymes involved in cell surface biosynthesis and remodeling, suggesting that cell surface remodeling is a strategy to cope with metabolic stress caused by elevated aeration. Similar changes in cell surface properties to cope with oxidative stress have previously been reported in A. brasilense (40).
The genes identified by the transposon screen used in this study may be indirectly related only to clumping and flocculation; i.e., the genes may primarily affect other functions that mediate nutrient stress and, as a result, promote flocculation. In fact, many of the mutations identified in this screen likely affect general cellular functions (e.g., alpha-amylase, TonB receptor, the YbgC homolog), even if they do not affect growth under the clumping and flocculation conditions used here. We hypothesize that the mutations in these functions alter the cells' ability to cope with oxygen stress, hence resulting in modified clumping and flocculation behaviors. Similarly, several other studies have linked general cellular functions with flocculation in Azospirillum (37–41).
Our results indicate that clumping and flocculating cells are protected from the persistent oxygen stress since these cells are able to maintain a microaerophilic intracellular environment compatible with the expression of an oxygen-sensitive enzyme, the nitrogenase. While there is some level of nitrogenase expression in clumping and early-flocculated cells, we have no evidence that the cells are able to fix nitrogen under these conditions. The upregulation of GlnB and the accumulation of PHB granules in hyperclumping cells detected by proteomics are consistent with these cells experiencing nitrogen limitation. The ability to rescue flocculation by the exogenous addition of ammonium or glutamate and the flocculation defect caused by mutations inactivating PII-like proteins (GlnB and GlnZ), which monitor the intracellular nitrogen status (42), further support this hypothesis. PHB accumulation was previously linked to flocculation (37, 43), and evidence indicates that PHB metabolism is regulated, in part, by GlnB, GlnZ, and NtrC (44, 45). Therefore, clumping and flocculating cells appear to adapt their physiology to maintain a potential for nitrogen fixation under otherwise inhibitory conditions. Further evidence supporting this contention was obtained in a recent study by Hou et al. (41) which identified similar stress endurance functions, including those implicated in PHB and nitrogen metabolism, nitrogen fixation, and oxygen tolerance, to be contributors to flocculation in A. brasilense. Interestingly, some A. brasilense mutants, including one with a mutation affecting an orphan response regulator, FlcA (13), and another with a mutation affecting a putative universal stress protein (encoded by orf280) (39), do not flocculate but were reported to possess an enhanced nitrogen fixation ability (17, 18). We have previously shown that the flcA mutant clumps but does not flocculate, which causes the culture to become filled with clumping cells over time (8). Nitrogen fixation in the orf280 mutant strain was induced under slightly elevated oxygen concentrations which were not compatible with nitrogen fixation in the wild-type strain (18), but the clumping phenotype of this mutant is not known. It is possible that the enhanced nitrogen fixation abilities of the flcA mutant and perhaps also the orf280 mutant are a direct result of their ability to clump.
We identified a chemotaxis histidine kinase CheA-like homolog to be a potential regulator of the transition between clumping and flocculation. The gene for this CheA-like homolog is located within a putative chemotaxis operon predicted to control cellular functions other than flagellum-based directed motility (28). Interestingly, this CheA-like protein is homologous to the CheA3 protein of the closely related organism Rhodospirillum centenum. In R. centenum, CheA3 is also encoded by a gene within an ACF-like operon which was shown to control cyst production (46). The transposon mutant with a mutation within the cheA-like gene in A. brasilense did not flocculate and was not expected to produce cysts since flocculation is required for cyst development (9). Similarly, an R. centenum mutant lacking CheA3 does not produce cysts, but the underlying mechanism is not known. The lack of flocculation in the transposon mutant identified here suggests that the ACF-like CheA functions in a pathway that affects cell envelope remodeling.
The exact mechanism by which metabolic stress is sensed by A. brasilense to trigger clumping and flocculation is yet to be elucidated. Previous studies have implicated cyclic GMP (cGMP) signaling in cyst development in R. centenum and A. brasilense (47), consistent with the identification of an R. centenum ACF-type CheA homolog that is involved in flocculation in A. brasilense. Further, clumping in A. brasilense is regulated by the ability of a chemotaxis receptor protein (Tlp1) to bind cyclic di-GMP (c-di-GMP) (14). Both c-GMP and c-di-GMP thus represent candidate molecules for signaling metabolic stress during clumping and flocculation. This hypothesis is also supported by the observation that a mutant with a mutation in a phosphodiesterase (chsA::Tn5) produces elevated levels of intracellular c-di-GMP and clumps abundantly (14). We have no evidence that clumping or flocculation is mediated by the ubiquitous starvation-signaling pp(p)Gpp molecule. However, it is reasonable to expect the pp(p)Gpp alarmone to be implicated in the transition from clumping to flocculation, given the role of flocculation in stress endurance (12) and the established function of the pp(p)Gpp alarmone in the adaptation of bacteria to nutritional stress (48). The observation of the upregulation of several, but not all, 50S ribosomal proteins during clumping and the downregulation of a few is relevant in this respect. Specifically, hyperclumping cells displayed elevated levels of expression of the L11 ribosomal protein, which interacts with the RelA enzyme on stalled ribosomes, triggering alarmone synthesis under conditions of nutrient limitation (49, 50). Similarly, L24 is present at the ribosome E site, where it interacts with chaperones, which are typically overexpressed during stress, including here in clumping cells (49). The pattern of regulation seen in clumping cells may thus reflect a specific adaptive response of the translational machinery to the nutritional stress experienced by the cell.
Together, the results obtained here reveal an ordered set of physiological adaptations that couple a transient behavior with a long-term adaptive response: metabolic scavenging during clumping primes the cells for survival in response to additional stress, including a readiness for nitrogen fixation, ultimately permitting long-term persistence in the presence of metabolic stress by flocculation.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Vanderleyden, C. Elmerich, Y. Okon, and L. Huergo for sharing strains and materials, as well as Kellie Cox, Brandon Seaver, and Ryan Hammond for help with characterization of some of the transposon mutations and Manesh Shah for proteome data analysis.
This work was supported by NSF awards MCB-0919819 and MCB-1330344 and by the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract DE-AC05-00OR22725.
Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02782-15.
REFERENCES
- 1.Kjelleberg S, Albertson N, Flärdh K, Holmquist L, Jouper-Jaan Å, Marouga R, Östling J, Svenblad B, Weichart D. 1993. How do non-differentiating bacteria adapt to starvation? Antonie Van Leeuwenhoek 63:333–341. doi: 10.1007/BF00871228. [DOI] [PubMed] [Google Scholar]
- 2.He K, Bauer CE. 2014. Chemosensory signaling systems that control bacterial survival. Trends Microbiol 22:389–398. doi: 10.1016/j.tim.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karatan E, Watnick P. 2009. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev 73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okon Y, Houchins JP, Albrecht SL, Burris RH. 1977. Growth of Spirillum lipoferum at constant partial pressures of oxygen, and the properties of its nitrogenase in cell-free extracts. J Gen Microbiol 98:87–93. doi: 10.1099/00221287-98-1-87. [DOI] [PubMed] [Google Scholar]
- 5.Zhulin IB, Bespalov VA, Johnson MS, Taylor BL. 1996. Oxygen taxis and proton motive force in Azospirillum brasilense. J Bacteriol 178:5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandre G, Greer SE, Zhulin IB. 2000. Energy taxis is the dominant behavior in Azospirillum brasilense. J Bacteriol 182:6042–6048. doi: 10.1128/JB.182.21.6042-6048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bible AN, Stephens BB, Ortega DR, Xie Z, Alexandre G. 2008. Function of a chemotaxis-like signal transduction pathway in modulating motility, cell clumping, and cell length in the alphaproteobacterium Azospirillum brasilense. J Bacteriol 190:6365–6375. doi: 10.1128/JB.00734-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bible A, Russell MH, Alexandre G. 2012. The Azospirillum brasilense Che1 chemotaxis pathway controls swimming velocity, which affects transient cell-to-cell clumping. J Bacteriol 194:3343–3355. doi: 10.1128/JB.00310-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadasivan L, Neyra CA. 1985. Flocculation in Azospirillum brasilense and Azospirillum lipoferum: exopolysaccharides and cyst formation. J Bacteriol 163:716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards AN, Siuti P, Bible AN, Alexandre G, Retterer ST, Doktycz MJ, Morrell-Falvey JL. 2011. Characterization of cell surface and extracellular matrix remodeling of Azospirillum brasilense chemotaxis-like 1 signal transduction pathway mutants by atomic force microscopy. FEMS Microbiol Lett 314:131–139. doi: 10.1111/j.1574-6968.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z, Ulrich LE, Zhulin IB, Alexandre G. 2010. PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis. Proc Natl Acad Sci U S A 107:2235–2240. doi: 10.1073/pnas.0910055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burdman S, Okon Y, Jurkevitch E. 2000. Surface characteristics of Azospirillum brasilense in relation to cell aggregation and attachment to plant roots. Crit Rev Microbiol 26:91–110. doi: 10.1080/10408410091154200. [DOI] [PubMed] [Google Scholar]
- 13.Pereg-Gerk L, Paquelin A, Gounon P, Kennedy IR, Elmerich C. 1998. A transcriptional regulator of the LuxR-UhpA family, FlcA, controls flocculation and wheat root surface colonization by Azospirillum brasilense Sp7. Mol Plant Microbe Interact 11:177–187. doi: 10.1094/MPMI.1998.11.3.177. [DOI] [PubMed] [Google Scholar]
- 14.Russell MH, Bible AN, Fang X, Gooding JR, Campagna SR, Gomelsky M, Alexandre G. 2013. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway. mBio 4:e00001-13. doi: 10.1128/mBio.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siuti P, Green C, Edwards AN, Doktycz MJ, Alexandre G. 2011. The chemotaxis-like Che1 pathway has an indirect role in adhesive cell properties of Azospirillum brasilense. FEMS Microbiol Lett 323:105–112. doi: 10.1111/j.1574-6968.2011.02366.x. [DOI] [PubMed] [Google Scholar]
- 16.Qi X, Nellas RB, Byrn MW, Russell MH, Bible AN, Alexandre G, Shen T. 2013. Swimming motility plays a key role in the stochastic dynamics of cell clumping. Phys Biol 10:026005. doi: 10.1088/1478-3975/10/2/026005. [DOI] [PubMed] [Google Scholar]
- 17.Katupitiya S, Millet J, Vesk M, Viccars L, Zeman A, Lidong Z, Elmerich C, Kennedy IR. 1995. A mutant of Azospirillum brasilense Sp7 impaired in flocculation with a modified colonization pattern and superior nitrogen fixation in association with wheat. Appl Environ Microbiol 61:1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Revers LF, Passaglia LM, Marchal K, Frazzon J, Blaha CG, Vanderleyden J, Schrank IS. 2000. Characterization of an Azospirillum brasilense Tn5 mutant with enhanced N2 fixation: the effect of ORF280 on nifH expression. FEMS Microbiol Lett 183:23–29. doi: 10.1111/j.1574-6968.2000.tb08928.x. [DOI] [PubMed] [Google Scholar]
- 19.Pereg Gerk L, Gilchrist K, Kennedy IR. 2000. Mutants with enhanced nitrogenase activity in hydroponic Azospirillum brasilense-wheat associations. Appl Environ Microbiol 66:2175–2184. doi: 10.1128/AEM.66.5.2175-2184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michiels K, Verreth C, Vanderleyden J. 1990. Azospirillum lipoferum and Azospirillum brasilense surface polysaccharide mutants that are affected in flocculation. J Appl Bacteriol 69:705–711. doi: 10.1111/j.1365-2672.1990.tb01567.x. [DOI] [Google Scholar]
- 21.Del Gallo M, Negi M, Neyra CA. 1989. Calcofluor- and lectin-binding exocellular polysaccharides of Azospirillum brasilense and Azospirillum lipoferum. J Bacteriol 171:3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 23.Burdman S, Jurkevitch E, Schwartsburd B, Hampel M, Okon Y. 1998. Aggregation in Azospirillum brasilense: effects of chemical and physical factors and involvement of extracellular components. Microbiology 144(Pt 7):1989–1999. [DOI] [PubMed] [Google Scholar]
- 24.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 25.Wolters DA, Washburn MP, Yates JR. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem 73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 26.McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR. 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom 219:245–251. doi: 10.1016/S1387-3806(02)00563-8. [DOI] [Google Scholar]
- 27.Eng JK, McCormack AL, Yates JR III. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Wisniewski-Dye F, Borziak K, Khalsa-Moyers G, Alexandre G, Sukharnikov LO, Wuichet K, Hurst GB, McDonald WH, Robertson JS, Barbe V, Calteau A, Rouy Z, Mangenot S, Prigent-Combaret C, Normand P, Boyer M, Siguier P, Dessaux Y, Elmerich C, Condemine G, Krishnen G, Kennedy I, Paterson AH, Gonzalez V, Mavingui P, Zhulin IB. 2011. Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet 7:e1002430. doi: 10.1371/journal.pgen.1002430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabb DL, McDonald WH, Yates JR III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. 2006. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A 103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Wen ZH, Washburn MP, Florens L. 2010. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal Chem 82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57:289–300. [Google Scholar]
- 33.Berg DE, Weiss A, Crossland L. 1980. Polarity of Tn5 insertion mutations in Escherichia coli. J Bacteriol 142:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valverde A, Okon Y, Burdman S. 2006. cDNA-AFLP reveals differentially expressed genes related to cell aggregation of Azospirillum brasilense. FEMS Microbiol Lett 265:186–194. doi: 10.1111/j.1574-6968.2006.00482.x. [DOI] [PubMed] [Google Scholar]
- 35.Bahat-Samet E, Castro-Sowinski S, Okon Y. 2004. Arabinose content of extracellular polysaccharide plays a role in cell aggregation of Azospirillum brasilense. FEMS Microbiol Lett 237:195–203. doi: 10.1111/j.1574-6968.2004.tb09696.x. [DOI] [PubMed] [Google Scholar]
- 36.Lerner A, Castro-Sowinski S, Valverde A, Lerner H, Dror R, Okon Y, Burdman S. 2009. The Azospirillum brasilense Sp7 noeJ and noeL genes are involved in extracellular polysaccharide biosynthesis. Microbiology 155:4058–4068. doi: 10.1099/mic.0.031807-0. [DOI] [PubMed] [Google Scholar]
- 37.Kadouri D, Jurkevitch E, Okon Y. 2003. Involvement of the reserve material poly-beta-hydroxybutyrate in Azospirillum brasilense stress endurance and root colonization. Appl Environ Microbiol 69:3244–3250. doi: 10.1128/AEM.69.6.3244-3250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lerner A, Castro-Sowinski S, Lerner H, Okon Y, Burdman S. 2009. Glycogen phosphorylase is involved in stress endurance and biofilm formation in Azospirillum brasilense Sp7. FEMS Microbiol Lett 300:75–82. doi: 10.1111/j.1574-6968.2009.01773.x. [DOI] [PubMed] [Google Scholar]
- 39.Galindo Blaha CA, Schrank IS. 2003. An Azospirillum brasilense Tn5 mutant with modified stress response and impaired in flocculation. Antonie Van Leeuwenhoek 83:35–43. doi: 10.1023/A:1022937828186. [DOI] [PubMed] [Google Scholar]
- 40.Wasim M, Bible AN, Xie Z, Alexandre G. 2009. Alkyl hydroperoxide reductase has a role in oxidative stress resistance and in modulating changes in cell-surface properties in Azospirillum brasilense Sp245. Microbiology 155:1192–1202. doi: 10.1099/mic.0.022541-0. [DOI] [PubMed] [Google Scholar]
- 41.Hou X, McMillan M, Coumans JVF, Poljak A, Raftery MJ, Pereg L. 2014. Cellular responses during morphological transformation in Azospirillum brasilense and its flcA knockout mutant. PLoS One 9:e114435. doi: 10.1371/journal.pone.0114435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Zamaroczy M. 1998. Structural homologues PII and Pz of Azospirillum brasilense provide intracellular signalling for selective regulation of various nitrogen-dependent functions. Mol Microbiol 29:449–463. doi: 10.1046/j.1365-2958.1998.00938.x. [DOI] [PubMed] [Google Scholar]
- 43.Kadouri D, Burdman S, Jurkevitch E, Okon Y. 2002. Identification and isolation of genes involved in poly(beta-hydroxybutyrate) biosynthesis in Azospirillum brasilense and characterization of a phbC mutant. Appl Environ Microbiol 68:2943–2949. doi: 10.1128/AEM.68.6.2943-2949.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun J, Peng X, Van Impe J, Vanderleyden J. 2000. The ntrB and ntrC genes are involved in the regulation of poly-3-hydroxybutyrate biosynthesis by ammonia in Azospirillum brasilense Sp7. Appl Environ Microbiol 66:113–117. doi: 10.1128/AEM.66.1.113-117.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun J, Van Dommelen A, Van Impe J, Vanderleyden J. 2002. Involvement of glnB, glnZ, and glnD genes in the regulation of poly-3-hydroxybutyrate biosynthesis by ammonia in Azospirillum brasilense Sp7. Appl Environ Microbiol 68:985–988. doi: 10.1128/AEM.68.2.985-988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berleman JE, Bauer CE. 2005. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol Microbiol 56:1457–1466. doi: 10.1111/j.1365-2958.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- 47.Marden JN, Dong Q, Roychowdhury S, Berleman JE, Bauer CE. 2011. Cyclic GMP controls Rhodospirillum centenum cyst development. Mol Microbiol 79:600–615. doi: 10.1111/j.1365-2958.2010.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 49.Brodersen DE, Nissen P. 2005. The social life of ribosomal proteins. FEBS J 272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 50.Jenvert RM, Schiavone LH. 2007. The flexible N-terminal domain of ribosomal protein L11 from Escherichia coli is necessary for the activation of stringent factor. J Mol Biol 365:764–772. doi: 10.1016/j.jmb.2006.10.065. [DOI] [PubMed] [Google Scholar]
- 51.Michiels K, Vanderleyden J, Van Gool A, Signer E. 1988. Isolation and characterization of Azospirillum brasilense loci that correct Rhizobium meliloti exoB and exoC mutations. J Bacteriol 170:5401–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephens BB, Loar SN, Alexandre G. 2006. Role of CheB and CheR in the complex chemotactic and aerotactic pathway of Azospirillum brasilense. J Bacteriol 188:4759–4768. doi: 10.1128/JB.00267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jofre E, Lagares A, Mori G. 2004. Disruption of dTDP-rhamnose biosynthesis modifies lipopolysaccharide core, exopolysaccharide production, and root colonization in Azospirillum brasilense. FEMS Microbiol Lett 231:267–275. doi: 10.1016/S0378-1097(04)00003-5. [DOI] [PubMed] [Google Scholar]
- 54.Van Dommelen A, Keijers V, Vanderleyden J, de Zamaroczy M. 1998. (Methyl)ammonium transport in the nitrogen-fixing bacterium Azospirillum brasilense. J Bacteriol 180:2652–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang YY, Arsene F, Elmerich C. 1993. Characterization of the ntrBC genes of Azospirillum brasilense Sp7: their involvement in the regulation of nitrogenase synthesis and activity. Mol Gen Genet 240:188–196. doi: 10.1007/BF00277056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


