Abstract
The nitrilase-mediated pathway has significant advantages in the production of optically pure aromatic α-hydroxy carboxylic acids. However, low enantioselectivity and activity are observed on hydrolyzing o-chloromandelonitrile to produce optically pure (R)-o-chloromandelic acid. In the present study, a protein engineering approach was successfully used to enhance the performance of nitrilase obtained from Burkholderia cenocepacia strain J2315 (BCJ2315) in hydrolyzing o-chloromandelonitrile. Four hot spots (T49, I113, Y199, and T310) responsible for the enantioselectivity and activity of BCJ2315 were identified by random mutagenesis. An effective double mutant (I113M/Y199G [encoding the replacement of I with M at position 113 and Y with G at position 199]), which demonstrated remarkably enhanced enantioselectivity (99.1% enantiomeric excess [ee] compared to 89.2% ee for the wild type) and relative activity (360% of the wild type), was created by two rounds of site saturation mutagenesis, first at each of the four hot spots and subsequently at position 199 for combination with the selected beneficial mutation I113M. Notably, this mutant also demonstrated dramatically enhanced enantioselectivity and activity toward other mandelonitrile derivatives and, thus, broadened the substrate scope of this nitrilase. Using an ethyl acetate-water (1:9) biphasic system, o-chloromandelonitrile (500 mM) was completely hydrolyzed in 3 h by this mutant with a small amount of biocatalyst (10 g/liter wet cells), resulting in a high concentration of (R)-o-chloromandelic acid with 98.7% ee, to our knowledge the highest ever reported. This result highlights a promising method for industrial production of optically pure (R)-o-chloromandelic acid. Insight into the source of enantioselectivity and activity was gained by homology modeling and molecular docking experiments.
INTRODUCTION
Optically pure aromatic α-hydroxy carboxylic acids, such as mandelic acid and its derivatives, are widely used as intermediates and resolving agents for the production of many pharmaceutical and agricultural products (1, 2). Among them, (R)-o-chloromandelic acid is one of the most preferred chiral building blocks for industrial synthesis of the antithrombotic agent (S)-clopidogrel, commercialized under the brand name Plavix (3, 4).
Considering the great importance of (R)-o-chloromandelic acid in pharmaceuticals and the ever-growing demand for green catalytic processes, tremendous efforts have been made to establish enantioselective routes of production (1). Among them, the nitrilase-mediated pathway is a simple and practical approach for the commercial production of (R)-o-chloromandelic acid and its derivatives because it does not involve a cofactor, uses a less-expensive starting material, and theoretically yields 100% of the desired product.
However, few nitrilases have been identified with high enantioselectivity and activity toward o-chloromandelonitrile (2, 3, 5–7). This severely reduces the practical applications of nitrilase-mediated production of optically pure (R)-o-chloromandelic acid. A nitrilase with high enantioselectivity and specific activity is crucial to fit the manufacturing process and reduce industrial production costs. These requirements may be met by discovering new enzymes or by applying protein engineering to enhance the performance of the existing nitrilases, and convincing successes have been achieved by both methods (8).
In our previous study, we identified and characterized a novel arylacetonitrilase, BCJ2315, from Burkholderia cenocepacia strain J2315 (9). BCJ2315 demonstrated high activity toward o-chloromandelonitrile and tolerated up to 50 mM o-chloromandelonitrile, though the o-chloromandelonitrile was highly toxic to the enzyme, demonstrating great potential for production of (R)-o-chloromandelic acid under high o-chloromandelonitrile concentrations (4). Unfortunately, BCJ2315 showed relatively low enantioselectivity toward o-chloromandelonitrile, providing the desired product with only 89.2% enantiomeric excess (ee), which reduces its value for industrial production of (R)-o-chloromandelic acid (4).
In the present study, we aspired to improve the fitness of BCJ2315 for the hydrolysis of o-chloromandelonitrile to efficiently produce (R)-o-chloromandelic acid. A protein engineering approach was successfully used to improve the enantioselectivity and activity of BCJ2315 by changing the enzyme structure, and a biphasic system was applied to produce optically pure (R)-o-chloromandelic acid in high concentrations by alleviating the inhibition of the toxic o-chloromandelonitrile. Homology modeling and molecular docking experiments were performed to gain insights into the binding mode of the substrates and the active sites.
MATERIALS AND METHODS
Materials.
B. cenocepacia J2315 was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Escherichia coli strain M15 and the plasmid pQE30 were used to express the nitrilase BCJ2315 (GenBank accession number WP_012492804). Mandelonitrile, mandelic acid, and their derivatives were ordered from Trademax Pharmaceuticals & Chemicals Co., Ltd. (Shanghai, China). The other chemicals used were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Random mutagenesis and library construction.
The expression plasmid pQE30/BCJ2315 containing the wild-type (WT) nitrilase gene from B. cenocepacia J2315 was used as the template for error-prone PCR (epPCR). Error-prone PCR was performed by using an instant error-prone PCR kit according to the manufacturer's instructions (Tiandz, Beijing, China) using primers 5′-ATCACGGATCCATGACCATCAATC-3′ and 5′-GCTAATTAAGCTTAAGCGGGTGTG-3′. The amplified DNA fragments were digested with restriction enzymes (BamHI/HindIII) targeting sites incorporated into the primers, and the resulting fragments were ligated into pQE30 digested with the same restriction enzymes. The ligation mixture was then transformed into E. coli M15.
Single colonies were picked and seeded into square 96-well plates containing ZYM-5052 autoinducing medium (10) and final concentrations of 100 mg/liter ampicillin and 50 mg/liter kanamycin. The plates were cultured at 30°C with shaking at 150 rpm for 36 h. The induced cells were harvested by centrifugation at 4°C for 10 min and washed twice with normal saline. The pellet obtained was resuspended in phosphate buffer (pH 8.0) and incubated at 30°C for 10 min before the addition of o-chloromandelonitrile (10 mM) to initiate the reaction. The reaction was performed in a rotary shaker (200 rpm) at 30°C for 12 h and then quenched with 1/10 (vol/vol) 2 M HCl. The reaction mixture was centrifuged at 10,000 × g for 10 min, and the resulting supernatant was used for analysis. Mutant activity was first assayed by the Berthelot assay to eliminate inactive mutants (11), and the enantioselectivity of active mutants was analyzed by high-performance liquid chromatography (HPLC).
Site-directed mutagenesis.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer's instructions. Primers containing the appropriate base changes are listed in Table S1 in the supplemental material.
Site saturation mutagenesis.
Site saturation mutagenesis was performed by introducing degenerate oligonucleotides (NNK) at the selected hot spots (sites 49, 113, 199, and 310) using the QuikChange site-directed mutagenesis kit as described by Zheng et al. (12).
Expression and purification of WT and engineered enzymes.
BCJ2315 and the mutants were expressed as His-tagged proteins in E. coli M15 and purified using Ni-nitrilotriacetic acid (NTA) Superflow columns (Qiagen, Hilden, Germany) as described previously (9).
Culture of the I113M/Y199G mutant.
Fermentation of the I113M/Y199G (MG) mutant (encoding the replacement of I with M at position 113 and Y with G at position 199) was performed as described previously (13) with modified fermentation medium (10 g/liter peptone, 5 g/liter yeast extract, 5 g/liter NaCl, 2 g/liter MgSO4, and 8 g/liter glycerol) and supplementary medium (10 g/liter peptone, 5 g/liter yeast extract, 5 g/liter NaCl, 2 g/liter MgSO4, and 20 g/liter glycerol).
Nitrilase activity of the WT and mutants.
The standard reaction was carried out at 30°C in a 1-ml reaction mixture containing 100 μmol sodium phosphate (pH 8.0), 10 μmol o-chloromandelonitrile (200 mM stock solution in methanol), and 1 mg dry cell weight (DCW)/ml culture or 10 μg purified nitrilase. Aliquots (100 μl) were withdrawn at different time intervals, and the reaction was quenched with 10 μl 2 M HCl. The cells (or nitrilases) were removed by centrifugation, and the supernatant was used to determine the activity by HPLC as described in “Analytical methods” below. One unit of enzyme activity was defined as the amount of cells (or enzyme) that produced 1 μmol o-chloromandelic acid per min under the standard assay conditions. All the experiments were performed in triplicate. The means and standard deviations were calculated from the results of three independent experiments.
Enantioselective hydrolysis of mandelonitrile and its derivatives with BCJ2315 and the MG mutant in an aqueous monophasic system.
The hydrolysis activities of purified BCJ2315 and the MG mutant toward 10 mM concentrations of different mandelonitrile derivatives (200 mM stock solution in methanol) were compared under standard conditions. The activity was determined by reversed-phase HPLC as described below in “Analytical methods” after reacting for 10 min. The ee values were measured by chiral HPLC when the substrate was exhausted. All of the experiments were performed in triplicate. The mean values and standard deviations were calculated from the results of three independent experiments.
Structure and binding conformation determination.
The three-dimensional homology model of BCJ2315 was constructed using MODELER in Discovery Studio (DS) 4.1 (Accelrys Software, San Diego, CA, USA). Five nitrilase crystal structures with the highest homology to BCJ2315 were chosen as the templates: Nit6803 (PDB accession number 3WUY, 57% similarity and 36% identity) from Synechocystis sp. strain PCC6803 (14), PaNit (PDB accession number 3IVZ, 42% similarity and 25% identity) from Pyrococcus abyssi (15), hypothetical protein Ph0642 (PDB accession number 1J31, 41% similarity and 25% identity) from Pyrococcus horikoshii (16), mouse nitrilase 2 (PDB accession number 2W1V, 41% similarity and 23% identity) (17), and Nit3 (PDB accession number 1F89, 38% similarity and 25% identity) from Saccharomyces cerevisiae (18). The structure ranking was based on the probability density function (PDF) total energy and evaluated by Verify Protein (Profiles-3D) and Ramachandran plots in DS 4.1. According to these validation results, the best homology model was chosen for further refinement. The structure model of BCJ2315 that was generated was improved further by refining the loop conformations based on the compatibility of the amino acid sequences with known PDB structures using the Protein Health module in Discovery Studio. The refined model was used to perform docking studies. Both enantiomers of o-chloromandelonitrile were docked into the models of WT and mutant BCJ2315 using C-DOCKER in DS 4.1 (19). The active sites were defined as the amino acid residues enclosed within a sphere with an 8-Å radius centered on the catalytic triad. The substrate orientation that yielded the lowest interaction energy was chosen for further analysis. All structure figures were prepared with Chimera (http://www.cgl.ucsf.edu/chimera).
Production of (R)-o-chloromandelic acid by the MG mutant in biphasic medium.
The enantioselective hydrolysis of o-chloromandelonitrile in biphasic medium was conducted in a 250-ml controlled stirred tank reactor. The reaction temperature was maintained at 30°C by a thermostat-controlled water bath, and the pH was maintained by a pH/oxidation reduction potential (ORP) controller. The biphasic reaction mixture (100 ml) contained 1 g wet cells suspended in phosphate buffer (100 mM, pH 8.0), 0.5 M o-chloromandelonitrile, and 10% (vol/vol) ethyl acetate. Samples (100 μl) were withdrawn periodically and diluted with an equal volume of methanol. The extents of conversion and enantioselectivity were determined by HPLC analysis. When the biotransformation was completed, the product was isolated as described by Xue et al. (7).
Analytical methods.
The quantitative analysis of mandelonitrile, mandelic acid, and their derivatives was performed using HPLC with a Zorbax SB-Aq column (4.6 mm by 250 mm and 5-μm particle size) (Agilent Technologies, Ltd., Carlsbad, CA, USA) at a flow rate of 0.8 ml/min with an eluting solvent system of phosphoric acid (0.1%, vol/vol) and methanol (40:60, vol/vol). The absorbance at 210 nm (A210) was measured.
The optical purity of mandelic acid and other derivatives was determined by HPLC analysis of the enantiomers. The A210 was measured. Chiral analytical conditions for the products are provided in Table S2 in the supplemental material.
RESULTS
Screening mutants with improved enantioselectivity from the epPCR library.
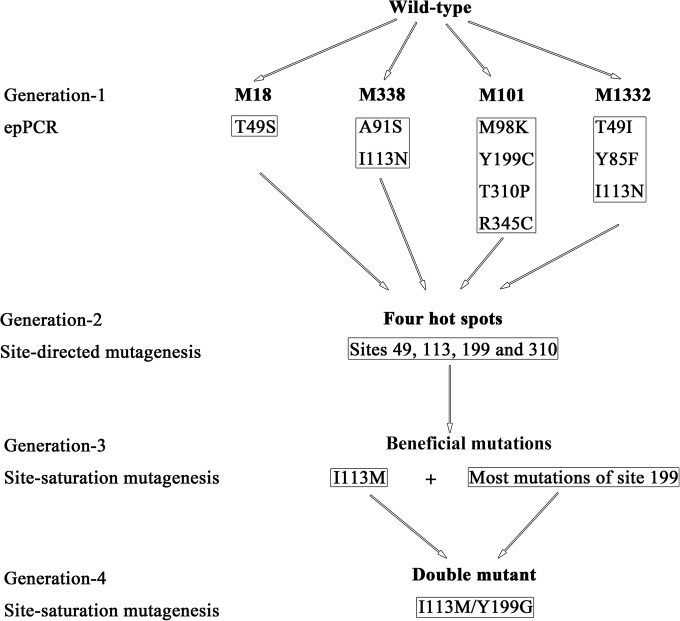
The process of protein engineering of BCJ315 is depicted in Fig. 1. Initially, random mutagenesis was performed to identify beneficial mutants with enhanced enantioselectivity. The gene encoding the nitrilase BCJ2315 was randomly mutated by error-prone PCR using an instant error-prone PCR kit. The amplified nitrilase gene was ligated with the linearized expression vector pQE30 and then transformed into E. coli M15 to generate a library of mutated nitrilases. About 6,000 of the 8,000 clones screened for hydrolysis of o-chloromandelonitrile using whole cells in 96-well plates showed nitrilase activity as determined by the Berthelot assay to measure the release of ammonia and were further analyzed for enantioselectivity by direct HPLC analysis. Four BCJ2315 mutants with enhanced enantioselectivity (ee > 95%) were selected. For convenience, the nitrilase BCJ2315 was designated as the WT. The four selected BCJ2315 mutants were purified, and their enantioselectivities and relative activities were determined as described in Materials and Methods (see Table S3 in the supplemental material for a summary). All four BCJ2315 mutants showed enhanced enantioselectivity, with ee values ranging from 95.9 to 99.3%, compared with 89.2% for the WT. The relative activities of the four mutants were also compared with that of the WT. However, all four mutants exhibited decreased relative activity. The highest relative activity was 87% for the M18 mutant.
FIG 1.
Scheme of the protein engineering of nitrilase BCJ2315. Initially, random mutagenesis by error-prone PCR (epPCR) yielded four BCJ2315 mutants with enhanced enantioselectivity but decreased activity containing 10 different amino acid substitutions. Site-directed mutagenesis was conducted, and four hot spots (sites 49, 113, 199, and 310) responsible for enantioselectivity and activity of BCJ2315 were characterized. Subsequently, two rounds of site saturation mutagenesis were performed, first at each of the four hot spots and subsequently at position 199 for combination with the selected beneficial mutation I113M, and a I113M/Y199G double mutant was generated that showed pronounced enhancements in enantioselectivity (99.1% ee) and relative activity (360%) compared to those of the WT.
Characterization of selected mutants.
DNA sequencing identified nine mutations, comprising eight amino acid substitutions in the four mutants. Each mutant presented one to four amino acid substitutions. M18 and M1332 presented different amino acid substitutions at the same site, position 49, which replaced the threonine there with a serine and an isoleucine, respectively. M338 and M1332 presented the same mutation (I113N).
Site-directed mutagenesis was performed to determine which of the eight mutation positions obtained were responsible for the enantioselectivity and activity of BCJ2315. Nine mutations were generated individually, and the resulting mutants were purified to determine their enantioselectivities and activities. The results are summarized in Table S3 in the supplemental material. Five mutations (T49S, T49I, I113N, Y199C, and T310P) contributed to the enhanced enantioselectivity of BCJ2315, with ee values ranging from 94.3 to 98.4%. The highest enantioselectivity was observed with I113N. Additionally, the mutations decreased the relative activity of BCJ2315, except for the Y199C mutant, whose activity was more than double that of the WT. The remaining four mutations were silent mutations and had no influence on the enantioselectivity and activity of BCJ2315.
Saturation mutagenesis at selected hot spots.
To obtain a mutant with high enantioselectivity and specific activity, we performed saturation mutagenesis at each of the four selected sites (T49, I113, Y199, and T310). The results are summarized in Table S4 in the supplemental material. Mutants with ee values greater than 95% and improved activity were chosen for further study. Under these criteria, no beneficial amino acid substitutions were obtained at sites T49 and T310 (see Table S4). Only the I113M mutation at site 113 presented a high enantioselectivity (97.2%) and a relatively high activity of 138% compared with that of the WT. In contrast, most of the Y199 mutations exhibited improved enantioselectivity and relative activity. The three best mutations (Y199A, Y199E, and Y199G) exhibited ee values of 95.6%, 95.1%, and 96.7% and relative activities of 273%, 191%, and 306%, respectively. Among them, the mutation Y199G showed the highest enantioselectivity and relative activity.
Combination of mutations.
Since beneficial mutations often have an additive effect, combinations of mutations are often used to improve enzyme performance (20). Therefore, we used the I113M mutant as a template and conducted saturation mutagenesis at position 199 to generate double mutants that further improved the enantioselectivity and activity of BCJ2315.
The results of the combinatorial mutagenesis are summarized in Table S5 in the supplemental material. Notably, most of the mutations demonstrated increased enantioselectivity and relative activity compared with those of the I113M mutant, demonstrating the synergistic effects of beneficial mutations. All mutations demonstrated high enantioselectivity, with ee values ranging from 97.2 to 98.7%. The highest enantioselectivity (98.7% ee) and relative activity (376%) were observed with the I113M/Y199G (MG) double mutant. Therefore, we chose the MG mutant for further studies.
Enantioselective hydrolysis of mandelonitrile and its derivatives with BCJ2315 and the MG mutant in an aqueous monophasic system.
The WT and MG mutant enzymes were purified, and their relative activities and enantioselectivities were compared. The purified MG mutant showed 3.6-fold greater activity than the WT for hydrolysis of o-chloromandelonitrile (Table 1), which was similar to the 3.76-fold increase for the whole-cell system (see Table S5 in the supplemental material). The enantioselectivity was increased from 89.2% to 99.1% with the purified MG mutant.
TABLE 1.
Enantioselective hydrolysis of mandelonitrile and its derivatives with WT nitrilase and I113M/Y199G mutant
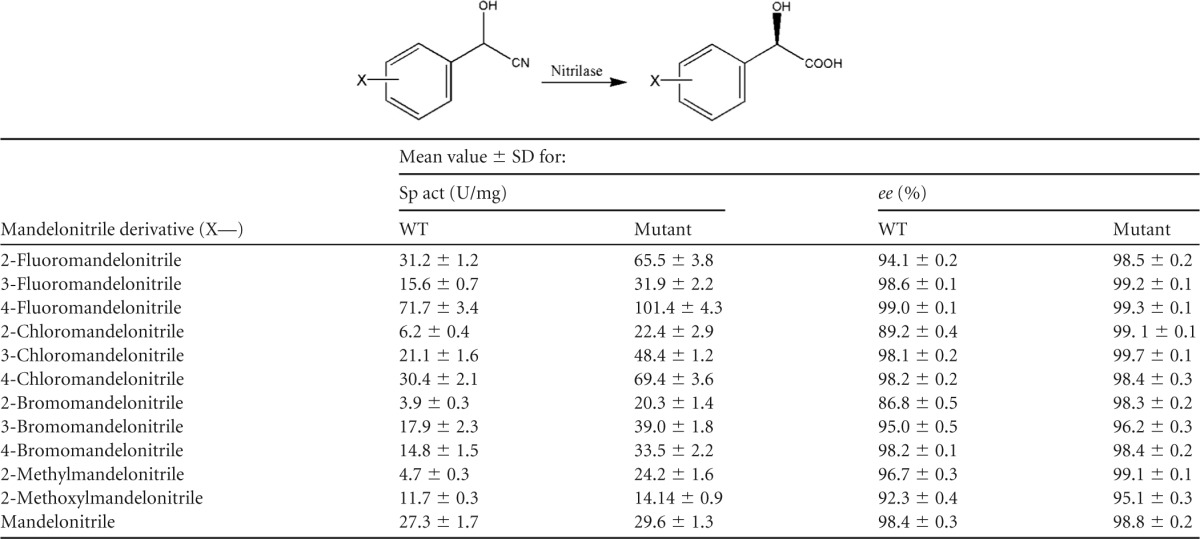
To explore the substrate scope, reactions with mandelonitrile and its derivatives were conducted with the WT and MG mutant. The results are shown in Table 1. The WT had broad substrate acceptance of mandelonitrile and its derivatives and showed good enantioselectivity toward most of the assayed substrates, except the ortho-substituted mandelonitrile. This may be attributed to the stronger hindrance effect in the ortho position than in the meta and para positions. Surprisingly, the MG mutant demonstrated enhanced activity and enantioselectivity toward all of the substituted mandelonitriles tested, especially the ortho-substituted substrates (o-fluoro, o-chloro, and o-bromo). Moreover, no amide was observed in any of the substrates tested. All of the ee values for carboxylic acids produced by the MG mutant were in an acceptable range (>95%), which is particularly noteworthy for the more sterically encumbered ortho-substituted derivatives. Additionally, the specific activity was dramatically improved toward all of the substrates tested with the MG mutant. The greatest improvement was observed with o-bromomandelonitrile (5.1-fold greater than that of the WT).
Production of (R)-o-chloromandelic acid with the MG mutant in biphasic medium.
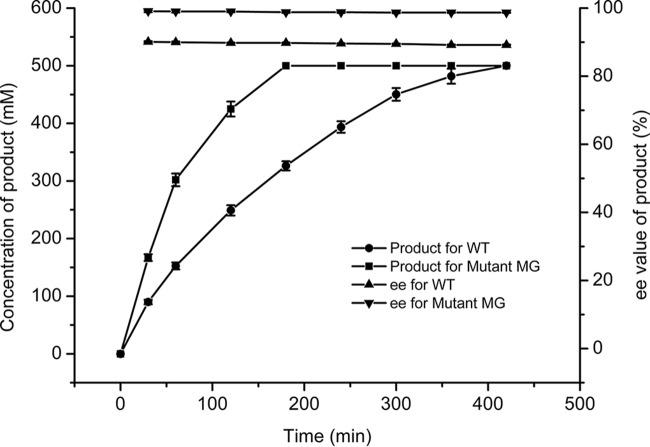
To explore the potential synthetic value of the MG mutant, the whole-cell biocatalyst was used to produce (R)-o-chloromandelic acid at a high concentration in an ethyl acetate-water biphasic system. Hydrolysis of 500 mM o-chloromandelonitrile by MG mutant and WT whole cells was carried out in an ethyl acetate-water (1:9, vol/vol) system. The time course of the reaction is shown in Fig. 2. The WT could completely hydrolyze 500 mM o-chloromandelonitrile in 7 h, providing a productivity of 319.8 g liter−1 day–1 with a product ee value of 89.2%. The mutant MG exhibited enhanced activity, completely hydrolyzing the substrate in 3 h with an ee value of 98.7%. This is similar to the ee value observed in the aqueous monophasic system, in contrast to the negative effect of the biphasic system on enantioselectivity often observed with nitrilases (13), probably due to the low proportion of organic solvent used herein (10%, vol/vol). Product production for the MG mutant was dramatically improved to 746.36 g liter−1 day–1. After crystallization in toluene, the product was obtained with a total isolation yield of 91% by the MG mutant, with an ee of 99.6%.
FIG 2.
Time course of hydrolyzing o-chloromandelonitrile mediated by the WT enzyme (BCJ2315) and the MG (I113M/Y199G) double mutant in an ethyl acetate-water (1:9, vol/vol) system. The reaction mixture contained 10 g/liter wet cells, 0.5 M o-chloromandelonitrile, 10% (vol/vol) ethyl acetate, and 90% sodium phosphate buffer (pH 8.0).
DISCUSSION
(R)-o-Chloromandelic acid is a key intermediate for the synthesis of clopidogrel, a platelet aggregation inhibitor with high market occupancy that can reduce the risk of cardiovascular events in patients with acute coronary syndromes. An advanced method for obtaining (R)-o-chloromandelic acid would be the direct hydrolysis of racemic o-chloromandelonitrile. Surprisingly, though hundreds of nitrilases have been discovered, there are few reports on nitrilase-mediated pathways, mainly due to the lack of nitrilases that are truly enantioselective toward ortho-substituted mandelonitrile. To solve this problem, we used a protein engineering approach on an existing nitrilase, BCJ2315, and successfully generated a mutant (MG) with enhanced enantioselectivity (99.1% ee, compared with 89.2% ee for the WT) and relative activity (360% of the WT). It is noteworthy that this mutant also demonstrated dramatically enhanced enantioselectivity and activity toward other mandelonitrile derivatives, which greatly extended its scope of application.
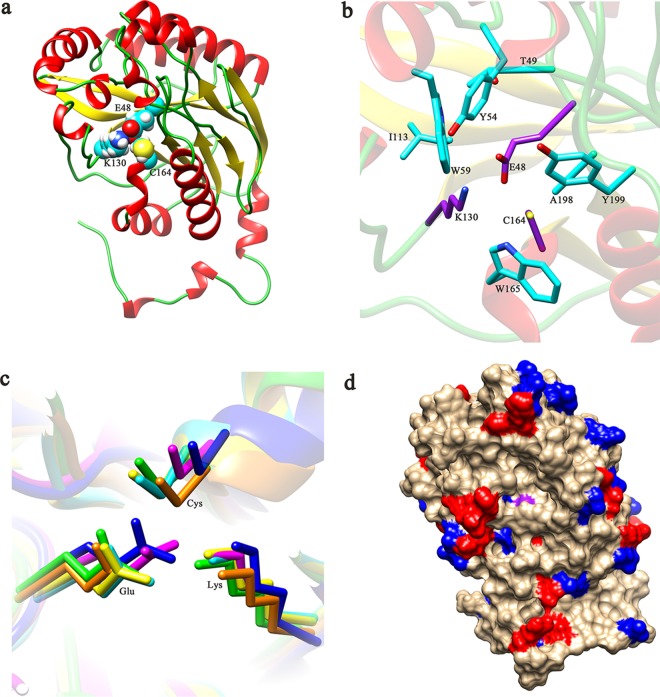
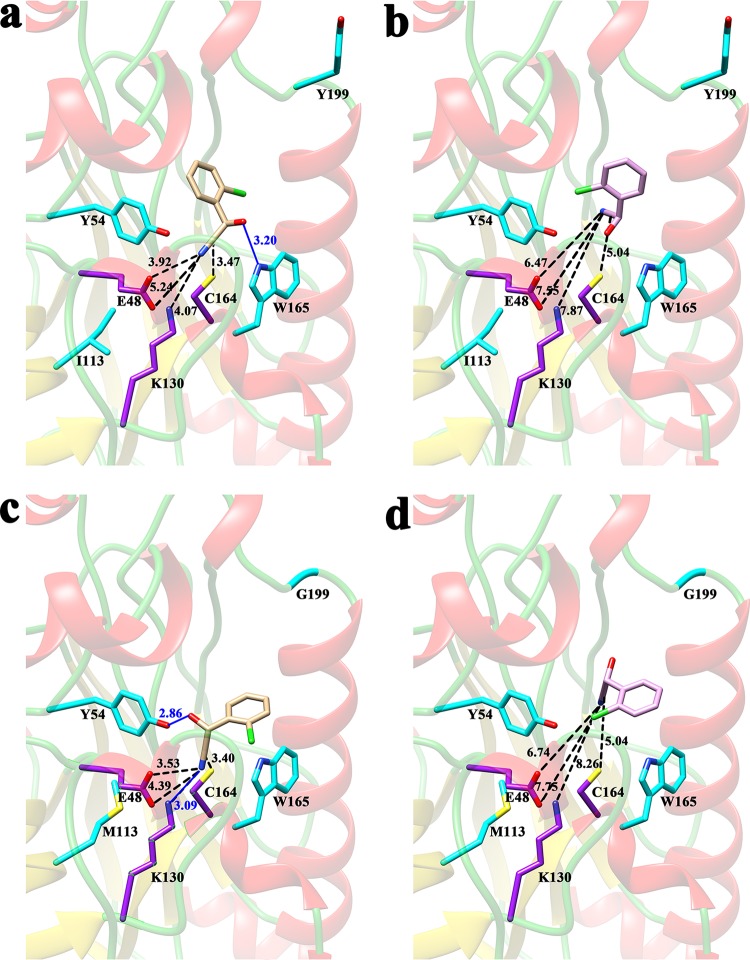
Homology modeling and molecular docking were performed to gain insights into the structure and binding conformation of BCJ2315 and its substrates. The three-dimensional homology model of BCJ2315 (Fig. 3) was obtained by comparative modeling using Accelrys Discovery Studio 4.1. The extended C terminus (residues 310 to 349) of BCJ2315 was not modeled due to the lack of a template. The model was further checked by PROCHECK, and the Ramachandran plot shows that 91.0% of residues are in the most favored regions, 7.8% are in additional allowed regions, 0.7% are in the generously allowed regions, and 0.4% (E83) are in the disallowed regions. Further analysis suggests that E83 is in the disallowed regions located far away from the active sites of nitrilase. This indicates that the backbone dihedral angles psi and phi in the BCJ2315 model were reasonably accurate. The conserved catalytic triad C164-E48-K130 is labeled in the structure (Fig. 3a and b) and presented an orientation and location similar to those of the templates after BCJ2315 was superimposed with the five homologous models (Fig. 3c), thus also demonstrating a proper model conformation. A cavity was predicted to be the binding pocket, with C164 forming the floor of the cavity (Fig. 3d). This was further supported, as the substrate was precisely positioned into this cavity by molecular docking (Fig. 4). Twenty-five residues, including the catalytic triad of C164-E48-K130, i.e., Y54, L56, W59, T134, E137, W165, W188, P189, S190, F191, S192, A197, A198, G202, P203, N206, L246, Q247, A248, G249, and G250, were predicted to be within 5 Å of the substrate, forming the substrate binding pocket.
FIG 3.
Structure of BCJ2315. (a) Structure of BCJ2315 created using DS 4.1. The α-helices, β-sheets, and loops are shown in red, yellow, and green, respectively. The atomic structures of the catalytic residues, E48, K130, and C164, are represented by spheres: the carbon, hydrogen, sulfur, oxygen, and nitrogen atoms are shown in cyan, white, yellow, red, and blue, respectively. (b) Close-up view of the catalytic triad, with carbon atoms in purple, as well as other critical residues, with carbon atoms in cyan. (c) Superimposition of the nitrilase catalytic triads. Orange, 1F89 (PDB); yellow, 1J31 (PDB); green, 2W1V (PDB); cyan, 3IVZ (PDB); blue, 3WUY (PDB); magenta, BCJ2315 (this study). (d) Solvent-accessible surface of BCJ2315. Positive and negative residues are shown in blue and red, respectively. The nucleophile C164 is shown in purple.
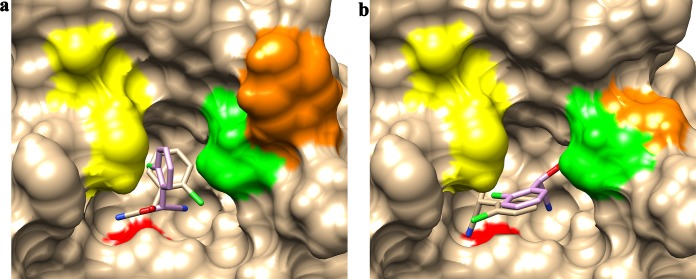
FIG 4.
Binding modes of (R)-/(S)-o-chloromandelonitrile in the binding pockets of the WT enzyme (a) and the MG mutant (b). The nucleophile C164 is shown in red on the bottom of the binding pocket. Residues W59, A198, and Y199, which are located at the entrance of the binding pocket, are shown in yellow, green, and orange, respectively. (R)- and (S)-o-chloromandelonitrile are shown as stick figures with carbon atoms in tan and lilac, respectively.
The conserved catalytic triad C164-E48-K130 plays an important role in the proposed nitrilase reaction mechanism (8, 21, 22). Diffusion of o-chloromandelonitrile into the pocket of BCJ2315 allows interactions with substrate binding residues that position it in a correct orientation with respect to C164-E48-K130. Hydrolysis of the substrate is initiated by nucleophilic attack on the cyano carbon atom by the S atom of C164 and followed by protonation of the nitrogen to form a covalent thioimidate intermediate. Subsequently, K130 activates a water molecule for further attack on the nitrile carbon to form a tetrahedral intermediate, which may be stabilized by the positive charge of K130. This tetrahedral intermediate eventually breaks down to produce the carboxylic acid and release the nitrilase. E48 acts as a general base to increase the nucleophilicity of the cysteine and participate in proton transfer to form ammonia (8, 21, 22). The first step of nucleophilic attack that is mediated by C164 is crucial for determining the hydrolysis efficiency. A sufficiently small through-space distance (d) value between the nucleophilic sulfur of C164 and the cyano carbon atom of the substrate could correspond to a near-attack conformer, as discussed previously for nitrilase and other enzyme-catalyzed reactions (14, 23), and this distance should be shorter in the case of the preferred enantiomer.
To gain insight into the binding conformation of BCJ2315 and its substrates, a docking study was performed using CDOCKER, and the best docking pose with the lowest binding energy was chosen for further analysis. Figure 4 shows the orientations of the cyano groups on (R)- and (S)-o-chloromandelonitrile docked in the binding pockets of the WT and the MG mutant, based on predictions from the molecular docking analysis. For the preferred (R)-enantiomer, the cyano group on the substrate was modeled to point directly toward the nucleophilic C164 of the catalytic triad. In contrast, in the disfavored (S)-enantiomer, the cyano group points away from C164, which is expected to be an unproductive orientation. The different substrate orientation toward the C164 of the catalytic triad may cause different catalytic efficiencies, thus establishing BCJ2315's enantiopreference.
The differences in the orientations of the (R)- and (S)-enantiomers in the predicted binding pocket were further clarified by the distance (d) between the nucleophilic S atom of C164 and the cyano carbon atom of the nitrile, based on the prediction of molecular docking analysis (Fig. 5). The d values of the preferred (R)-o-chloromandelonitrile (dR) were 3.47 Å and 3.40 Å for the WT and the MG mutant (Fig. 5a and c), respectively, but the disfavored (S)-o-chloromandelonitrile was positioned much further away, with d values (dS) of 4.13 Å and 5.04 Å, respectively (Fig. 5b and d). The increased distance may prevent efficient nucleophilic attack and, thus, could explain the preference for the (R)-enantiomer. The Δd (dS − dR) value increased from 0.66 Å to 1.64Å going from the WT to the MG mutant, which could give an explanation of the enhanced performance of the MG mutant. The distances between the N atom of the cyano group and either E48 or K130 were shortened for the (R)-enantiomer and increased for the (S)-enantiomer in the MG mutant model relative to the distances in the WT. This may lead to efficient interaction between the substrate and E48-K130 and stabilize the tetrahedral transition state. Thus, structural changes in the binding pocket imposed by the evolutionary process are predicted to make nucleophilic attack much more difficult with the disfavored enantiomer due to the longer distance between the cyano carbon and the S atom of C164. These observations could roughly explain the enhanced enantioselectivity and relative activity of the MG mutant compared with those of the WT.
FIG 5.
Docking poses of (R)-o-chloromandelonitrile in the binding pockets of the WT enzyme (a) and the MG mutant (c) and of (S)-o-chloromandelonitrile in the binding pockets of the WT (b) and the MG mutant (d). The catalytic triad (C164-E48-K130) is shown with carbon atoms in purple, as well as other critical residues, with carbon atoms in cyan. Distances between (R)-o-chloromandelonitrile or (S)-o-chloromandelonitrile and the catalytic triads are shown by dashed black lines, and H bonds are shown by solid blue lines. (R)- and (S)-o-chloromandelonitrile are shown in stick figures, with carbon atoms in tan and lilac, respectively. The chlorine, sulfur, oxygen, and nitrogen atoms are shown in green, yellow, red and blue, respectively. All of the atoms of G199 are shown, as it does not have a side chain.
The hydrogen bonds in the models were analyzed carefully since they play significant roles in substrate orientation and enzyme-substrate intermediate complex formation (19). In the WT-substrate model, no hydrogen bonding was predicted between (S)-o-chloromandelonitrile and the enzyme (Fig. 5b). For the preferred (R)-enantiomer, the hydroxyl oxygen of the nitrile was modeled to form a hydrogen bond with the N atom in the ring of W165, which may help orient the substrate in a proper situation (Fig. 5a). In the MG mutant-substrate model also, no hydrogen bonding was predicted between (S)-o-chloromandelonitrile and the enzyme. For the preferred (R)-enantiomer, although the hydrogen bonds predicted in the WT-substrate model disappeared, two useful hydrogen bonds were predicted to enhance the interactions of the enzyme and the substrate (Fig. 5c). The newly formed hydrogen bond between K130 of the catalytic triad and the (R)-enantiomer was important for the formation of the covalent substrate-enzyme complex and was helpful to form a tetrahedral intermediate. Y54 formed a strong predicted hydrogen bonding interaction with the (R)-enantiomer (2.86 Å), which may help position the substrate in a more suitable orientation than does the hydrogen bond (3.20 Å) formed between W165 and the (R)-enantiomer in the WT-substrate model (Fig. 5a and c). The different interactions between the enzyme and the two enantiomers of the nitrile caused by the hydrogen bonds would be an import factor in determining the enantioselectivity of BCJ2315, and also, the different interactions caused by hydrogen bonds could explain the enhanced performance of the MG mutant compared with that of the WT.
Almost all replacements of the tyrosine (Y199) residue resulted in increased enantioselectivity and relative activity (see Table S4 in the supplemental material). Y199 was modeled to lie in the surface loop of the enzyme and far from the catalytic triad (>10 Å) and the substrate (>5 Å); thus, it does not directly participate in the catalytic reaction (Fig. 3b). It is well known that most active nitrilases form spiral structures which consist of 4 to 22 subunits. This process could influence the activity and stability of the nitrilases. Based on multiple-alignment analysis of BCJ2315 sequences with those of 7 microbial nitrilases and 11 members with available structural information for the nitrilase superfamily reported by Thuku et al., Y199 does not take part in the spiral structure formation of BCJ2315, as it is not involved in the interaction areas named A, C, D, and F surfaces which hold the subunits together (22; data not shown). The structure model obtained suggests that Y199 may be located at the entrance of the binding pocket (Fig. 4) and may be involved in a steric clash with the substrate, as the bulky side chain of the tyrosine is expected to sterically clash with the relatively bulky chlorine atom to hinder efficient substrate diffusion into the active site. Replacements of Y199 with smaller amino acids would therefore be beneficial by reducing this clash. This model is consistent with our experimental results (see Table S4), which showed higher activities for smaller residues at position 199, such as Y199G, Y199A, and Y199S, demonstrating relative activities of 306%, 273%, and 324%, respectively, compared with the WT. In contrast, decreased activity was observed for bulky side chains at position 199, such as Y199W, Y199F, and Y199H, which demonstrated relative activities of 121%, 105%, and 114%, respectively, compared with the WT. Based on this assumption, W59 and A198, which are modeled as localized around the entrance of the binding pocket (Fig. 4), may also influence the enantioselectivity and specific activity by steric hindrance, providing additional hot spots for further reengineering of BCJ2315. Engineering the residues in the binding pocket is also an attractive approach for modification of BCJ2315's enantioselectivity and specific activity by controlling the substrate-accessible volume within the active site (24).
While our study was in process, another study reported successful improvement of the performance of nitrilase NitA for hydrolyzing o-chloromandelonitrile (7). Based on molecular docking analysis of the binding mode of (R)-o-chloromandelonitrile into NitA, four potential active-site residues (Y52, T132, F189, and S190) in close proximity to the substrate were chosen, and two rounds of site saturation mutagenesis were conducted to obtain an excellent double mutant, T132A/F189T, with 4.37-fold-higher specific activity (7.39 U/mg) toward o-chloromandelonitrile than WT NitA. Furthermore, the enantioselectivity was improved from 17.34 to >200 to give a product with high enantiopurity (7). In this study, we identified four hot spots responsible for the enantioselectivity and activity of nitrilase BCJ2315 by random mutagenesis targeting the entire protein. Compared with the above-mentioned residues, which are in close proximity to the substrate, the four hot spots herein were more distant (>5 Å) from the substrate, based on the docking models. Random mutagenesis seeks to mutate all amino acids equally and would create a great number of distant mutations that are also critical for the improvement of catalytic properties, extending the possibility of creating beneficial mutants.
Due to the great importance of (R)-o-chloromandelic acid, several nitrilase-mediated pathways to produce (R)-o-chloromandelic acid at high concentrations have been developed. Five of these processes are compared in Table 2. The highest product concentration (500 mM) was observed with the engineered MG mutant nitrilase in this study, slightly higher than that of the engineered T132A/F189T nitrilase mutant (450 mM) (7). The highest volumetric productivity (g liter−1 day−1) and catalyst productivity (g/g DCW) were also achieved by the MG mutant in this study. Moreover, catalyst loading for the MG mutant was extremely low (3.85 g DCW/liter, equal to 10 g/liter wet cells), which is nearly 1 order of magnitude lower than those of the other three nitrilases listed in Table 2, demonstrating highly efficient production of (R)-o-chloromandelic acid by the mutant MG.
TABLE 2.
Comparison of data for the production of (R)-o-chloromandelic acid mediated by nitrilases from different sources
| Nitrilase sourcea | Catalyst loading (g DCW/liter) | Substrate concn; process type | Yield (%) | Product concn (mM) | Product ee (%) | Volumetric productivity (g/liter/day) | Catalyst productivity (g/g DCW) | Reference or source |
|---|---|---|---|---|---|---|---|---|
| Labrenzia aggregata | 52 | 300 mM; 10% (vol/vol) toluene | 94.5 | 276 | 96.3 | 154.4 | 0.99 | 3 |
| Arthrobacter sp. strain F-73b | 58 | 4 × 100 mM; fed batch, 20% (vol/vol) ethyl acetate | 92 | 400 | 98.5 | 199 | 1.29 | 5 |
| Metagenomic library 2A6c | 30 | 5 × 100 mM; 20% (vol/vol) toluene | 87 | 450 | 99 | 671.76 | 2.8 | 7 |
| Burkholderia cenocepacia J2315 | 3.85 | 494 mM; extended fed batch | —d | 415 | 97.6 | 232.3 | 20.11 | 4 |
| Burkholderia cenocepacia J2315 I113M/Y199G mutant | 3.85 | 500 mM; 10% (vol/vol) ethyl acetate | 91 | 500 | 98.7 | 746.36 | 24.24 | This study |
Recombinant E. coli strains containing the respective nitrilase genes were used here.
The wild strain of Arthrobacter sp. strain F-73 was used as the biocatalyst here.
The nitrilase mutant was used here.
—, data not available.
In conclusion, a nitrilase double mutant, denoted MG (I113M/Y199G), that demonstrated dramatically enhanced enantioselectivity and relative activity toward o-chloromandelonitrile and other mandelonitrile derivatives was successfully generated using a protein engineering strategy. The MG mutant demonstrated potential in the industrial production of optically pure (R)-o-chloromandelic acid due to its high enantioselectivity, high product concentration, high volumetric productivity, and high catalyst productivity. The binding modes of the substrates and the active sites were constructed by homology modeling and molecular docking, providing structural insights into the differences observed in the activities and enantioselectivities of the WT and the MG mutant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 21406068/B060804), China Postdoctoral Science Foundation-funded project (grant no. 2014M560308), National Major Science and Technology Projects of China (grant no. 2012ZX09304009), and National Basic Research Program of China (grant no. 2012CB721103).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02688-15.
REFERENCES
- 1.Groger H. 2001. Enzymatic routes to enantiomerically pure aromatic alpha-hydroxy carboxylic acids: a further example for the diversity of biocatalysis. Adv Synth Catal 343:547–558. doi:. [DOI] [Google Scholar]
- 2.DeSantis G, Zhu Z, Greenberg WA, Wong K, Chaplin J, Hanson SR, Farwell B, Nicholson LW, Rand CL, Weiner DP, Robertson DE, Burk MJ. 2002. An enzyme library approach to biocatalysis: development of nitrilases for enantioselective production of carboxylic acid derivatives. J Am Chem Soc 124:9024–9025. doi: 10.1021/ja0259842. [DOI] [PubMed] [Google Scholar]
- 3.Zhang CS, Zhang ZJ, Li CX, Yu HL, Zheng GW, Xu JH. 2012. Efficient production of (R)-o-chloromandelic acid by deracemization of o-chloromandelonitrile with a new nitrilase mined from Labrenzia aggregata. Appl Microbiol Biotechnol 95:91–99. doi: 10.1007/s00253-012-3993-4. [DOI] [PubMed] [Google Scholar]
- 4.Wang HL, Sun HH, Gao WY, Wei DZ. 2014. Efficient production of (R)-o-chloromandelic acid by recombinant Escherichia coli cells harboring nitrilase from Burkholderia cenocepacia J2315. Org Process Res Dev 18:767–773. doi: 10.1021/op400174a. [DOI] [Google Scholar]
- 5.Nagasawa T, Yoshida T, Matsuyama A. August 2006. Method for producing the carboxylic acid. Japanese patent JP2006223246.
- 6.Schreiner U, Hecher B, Obrowsky S, Waich K, Klempier N, Steinkellner G, Gruber K, Rozzell JD, Glieder A, Winkler M. 2010. Directed evolution of Alcaligenes faecalis nitrilase. Enzyme Microb Technol 47:140–146. doi: 10.1016/j.enzmictec.2010.05.012. [DOI] [Google Scholar]
- 7.Xue YP, Shi CC, Xu Z, Jiao B, Liu ZQ, Huang JF, Zheng YG, Shen YC. 2015. Design of nitrilases with superior activity and enantioselectivity towards sterically hindered nitrile by protein engineering. Adv Synth Catal 357:1741–1750. doi: 10.1002/adsc.201500039. [DOI] [Google Scholar]
- 8.Martinkova L, Kren V. 2010. Biotransformations with nitrilases. Curr Opin Chem Biol 14:130–137. doi: 10.1016/j.cbpa.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Wang HL, Sun HH, Wei DZ. 2013. Discovery and characterization of a highly efficient enantioselective mandelonitrile hydrolase from Burkholderia cenocepacia J2315 by phylogeny-based enzymatic substrate specificity prediction. BMC Biotechnol 13:14. doi: 10.1186/1472-6750-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Studier FW. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Weatherburn MW. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 12.Zheng L, Baumann U, Reymond JL. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ni K, Wang H, Zhao L, Zhang M, Zhang S, Ren Y, Wei D. 2013. Efficient production of (R)-(−)-mandelic acid in biphasic system by immobilized recombinant E. coli. J Biotechnol 167:433–440. doi: 10.1016/j.jbiotec.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Zhang LJ, Yin B, Wang C, Jiang SQ, Wang HL, Yuan YA, Wei DZ. 2014. Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Synechocystis sp. PCC6803. J Struct Biol 188:93–101. doi: 10.1016/j.jsb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Raczynska JE, Vorgias CE, Antranikian G, Rypniewski W. 2011. Crystallographic analysis of a thermoactive nitrilase. J Struct Biol 173:294–302. doi: 10.1016/j.jsb.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Sakai N, Tajika Y, Yao M, Watanabe N, Tanaka I. 2004. Crystal structure of hypothetical protein PH0642 from Pyrococcus horikoshii at 1.6Å resolution. Proteins 57:869–873. doi: 10.1002/prot.20259. [DOI] [PubMed] [Google Scholar]
- 17.Barglow KT, Saikatendu KS, Bracey MH, Huey R, Morris GM, Olson AJ, Stevens RC, Cravatt BF. 2008. Functional proteomic and structural insights into molecular recognition in the nitrilase family enzymes. Biochemistry 47:13514–13523. doi: 10.1021/bi801786y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumaran D, Eswaramoorthy S, Gerchman SE, Kycia H, Studier FW, Swaminathan S. 2003. Crystal structure of a putative CN hydrolase from yeast. Proteins 52:283–291. doi: 10.1002/prot.10417. [DOI] [PubMed] [Google Scholar]
- 19.Song DW, Zhu SZ, Li XZ, Zheng GJ. 2014. Homology modeling and docking studies of BjGL, a novel (+) gamma-lactamase from Bradyrhizobium japonicum. J Mol Graph Model 47:1–7. doi: 10.1016/j.jmgm.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Fujii R, Nakagawa Y, Hiratake J, Sogabe A, Sakata K. 2005. Directed evolution of Pseudomonas aeruginosa lipase for improved amide-hydrolyzing activity. Protein Eng Des Sel 18:93–101. doi: 10.1093/protein/gzi001. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes BCM, Mateo C, Kiziak C, Chmura A, Wacker J, van Rantwijk F, Stolz A, Sheldon RA. 2006. Nitrile hydratase activity of a recombinant nitrilase. Adv Synth Catal 348:2597–2603. doi: 10.1002/adsc.200600269. [DOI] [Google Scholar]
- 22.Thuku RN, Brady D, Benedik MJ, Sewell BT. 2009. Microbial nitrilases: versatile, spiral forming, industrial enzymes. J Appl Microbiol 106:703–727. doi: 10.1111/j.1365-2672.2008.03941.x. [DOI] [PubMed] [Google Scholar]
- 23.Reetz MT, Bocola M, Wang LW, Sanchis J, Cronin A, Arand M, Zou J, Archelas A, Bottalla AL, Naworyta A, Mowbray SL. 2009. Directed evolution of an enantioselective epoxide hydrolase: uncovering the source of enantioselectivity at each evolutionary stage. J Am Chem Soc 131:7334–7343. doi: 10.1021/ja809673d. [DOI] [PubMed] [Google Scholar]
- 24.Lafaquiere V, Barbe S, Puech-Guenot S, Guieysse D, Cortes J, Monsan P, Simeon T, Andre I, Remaud-Simeon M. 2009. Control of lipase enantioselectivity by engineering the substrate binding site and access channel. Chembiochem 10:2760–2771. doi: 10.1002/cbic.200900439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.