Abstract
Around a third of oral bacteria cannot be grown using conventional bacteriological culture media. Community profiling targeting 16S rRNA and shotgun metagenomics methods have proved valuable in revealing the complexity of the oral bacterial community. Studies investigating the role of oral bacteria in health and disease require phenotypic characterizations that are possible only with live cultures. The aim of this study was to develop novel culture media and use an in vitro biofilm model to culture previously uncultured oral bacteria. Subgingival plaque samples collected from subjects with periodontitis were cultured on complex mucin-containing agar plates supplemented with proteose peptone (PPA), beef extract (BEA), or Gelysate (GA) as well as on fastidious anaerobe agar plus 5% horse blood (FAA). In vitro biofilms inoculated with the subgingival plaque samples and proteose peptone broth (PPB) as the growth medium were established using the Calgary biofilm device. Specific PCR primers were designed and validated for the previously uncultivated oral taxa Bacteroidetes bacteria HOT 365 and HOT 281, Lachnospiraceae bacteria HOT 100 and HOT 500, and Clostridiales bacterium HOT 093. All agar media were able to support the growth of 10 reference strains of oral bacteria. One previously uncultivated phylotype, Actinomyces sp. HOT 525, was cultivated on FAA. Of 93 previously uncultivated phylotypes found in the inocula, 26 were detected in in vitro-cultivated biofilms. Lachnospiraceae bacterium HOT 500 was successfully cultured from biofilm material harvested from PPA plates in coculture with Parvimonas micra or Veillonella dispar/parvula after colony hybridization-directed enrichment. The establishment of in vitro biofilms from oral inocula enables the cultivation of previously uncultured oral bacteria and provides source material for isolation in coculture.
INTRODUCTION
The human mouth is colonized by a diverse collection of microorganisms, including fungi, protozoa, viruses, archaea, and bacteria (1). Release 13.2 of the Human Oral Microbiome Database (www.homd.org) lists 707 taxa at the species level, of which 244 have yet to be cultured (2, 3). Oral bacteria are associated with the most common bacterial diseases of humans: dental caries and periodontal disease. In addition, they play a role in systemic disease, both directly by causing infections at other body sites and indirectly, where, for example, periodontal disease is a risk factor for other conditions such as cardiovascular disease and diabetes (4–6). Oral bacteria are also important in health; the presence of the normal microbiota inhibits infection by exogenous organisms. They also play an important role in nitrogen metabolism, whereby oral bacteria produce nitrate reductase to reduce dietary nitrate to nitrite, which is taken up the body and converted to nitric oxide, which is essential for cardiovascular health (7).
The development of culture-independent methods targeting 16S rRNA genes for the characterization of complex bacterial communities has revealed the diversity of the bacterial communities found in the human body (8) and identified species and groups of species that are associated with specific health and disease states such as dental caries (9) and periodontal disease (10). This is clearly valuable information, and associations such as these can be used as biomarkers. However, to elucidate the role that species of interest play in health and disease, further investigation of individual bacterial strains is required, including the identification of virulence factors, susceptibility to antimicrobials, and interactions with the host and other microorganisms. The in vitro culture of representatives of species that have yet to be cultured is therefore a major goal.
There are a number of possible reasons why bacteria cannot be cultured in the laboratory (11). For example, the nutrients required by a strain may not be present in an artificial medium or may not be in a form that can be taken up. It then might be possible to attempt to better mimic in the laboratory the nutrients available in the in vivo environment. Some species of soil bacteria have been isolated by using a combination of nutrient-poor media and extended incubation times (12, 13). Inhibition of growth of some organisms has been shown to result from autoclaving phosphate and agar together during medium preparation (14).
It is also now recognized that bacteria prefer to live naturally in multispecies biofilms and cooperate and communicate with each other. This may involve the regulation of growth rates for the good of the community and the sharing of nutrients or liberation of nutrients from complex substrates. Previously, we successfully cultivated a representative of a previously uncultivated Synergistetes lineage by serial coculture of a subgingival plaque sample while enriching for the target organism by colony hybridization (15). Similarly, Sizova et al. (16) isolated 10 novel strains using three cultivation methods: an in vivo trap to encourage bacterial communication, cultivation of single cells for extended periods of time, and the use of modified media without sugars. Of these three methods, extended incubation of single cells was the most successful (16).
The aim of this study was to evaluate novel culture media and an in vitro biofilm model for their ability to allow growth of previously uncultured oral bacteria in subgingival plaque samples collected from subjects with periodontitis.
MATERIALS AND METHODS
Subjects and sample collection.
Plaque samples were collected from three patients with severe chronic periodontitis who were referred for treatment to the Department of Periodontology at Guy's and St Thomas' NHS Foundation Trust. Ethical approval was granted by South West London REC 3 Research Ethics Proportionate Review Sub-Committee (10/H0803/161), and informed consent was obtained from all study participants. Subjects had at least six teeth with probing depths of ≥6 mm and bone loss, indicating that they were suffering from severe chronic periodontitis and that their subgingival plaque would be likely to harbor a diverse bacterial community. Subjects had not received periodontal or antimicrobial therapy within the previous 6 months, nor did they have any medical conditions known to affect the severity or progression of periodontitis. Subgingival plaque samples were collected with a sterile curette from the advancing front of the lesion, in pockets greater than 8 mm in depth. The samples were placed in 2 ml prereduced periodontitis peg broth (PPB) and taken to the laboratory and processed for culture within 3 min.
Samples were dispersed by vortexing for 30 s and diluted 10-fold in prereduced PPB. An aliquot of 500 μl was retained for bacterial community baseline analysis and the remaining portion plated directly onto selected solid media.
Culture media.
All bacteria were grown in an anaerobic workstation (Don Whitley Scientific Ltd.) at 37°C under anaerobic conditions (80% N2, 10% H2, 10% CO2), unless otherwise stated. Reference strains were cultivated on fastidious anaerobe agar (Lab M, United Kingdom) plus 5% horse blood (FAAB).
To mimic the nutritional environment of the periodontal pocket, a novel set of culture media was devised. The basal liquid medium consisted of Trypticase peptone (0.3%), yeast extract (0.5%), pig gastric mucin (0.25%), KCl (0.25%), hemin (0.001%), menadione (0.0001%), potassium phosphate buffer (0.1 M, pH 8.0), cysteine hydrochloride (0.05%), urea (1 mM), arginine (5 mM), lysine (1 mM), glycine (1 mM), and heat-inactivated horse serum (10%) in 1 liter distilled water. Broth media were prepared by supplementing the base medium with Bacto proteose peptone (PPB), Bacto beef extract (BEB) (BD Biosciences, United Kingdom) or, Gelysate (GB) (BD Biosciences, United Kingdom) (all at 0.07%). Solid versions of the media (PPA, BEA, and GA) were prepared by the addition of granulated agar (15 g; Melford, United Kingdom) to the broth media. The ability of these media to support the growth of oral bacteria was assessed by anaerobic cultivation of the strains listed in Table 1.
TABLE 1.
Growth of reference strains on agar media
| Strain | Growth scorea on: |
|||
|---|---|---|---|---|
| FAA | BEA | GA | PPA | |
| Actinomyces naeslundii ATCC 10301 | ++++ | ++++ | ++++ | +++++ |
| Campylobacter rectus ATCC 33238 | +++ | +++ | +++ | +++ |
| Eikenella corrodens NCTC 10596 | +++ | +++ | +++ | +++ |
| Eubacterium sulci ATCC 35896 | +++ | +++ | ++ | +++ |
| Filifactor alocis ATCC 35896 | +++ | +++ | +++ | +++ |
| Fusobacterium nucleatum ATCC 25586 | +++++ | ++++ | ++++ | ++++ |
| Parvimonas micra ATCC 33270 | ++++ | ++++ | ++++ | +++++ |
| Porphyromonas endodontalis ATCC 35406 | +++++ | ++++ | ++ | +++ |
| Porphyromonas gingivalis ATCC 33277 | +++++ | +++ | ++ | ++ |
| Prevotella intermedia ATCC 25611 | +++++ | +++ | ++ | ++++ |
++, good growth in first streak, sparse over remainder of plate; +++, good growth over whole pate; ++++, heavy growth over whole plate; +++++, confluent, very heavy growth over whole plate.
Subgingival plaque culture.
Tenfold dilutions of the plaque samples were prepared and used to inoculate FAAB, PPA, BEA, and GA plates in triplicate. Plates were incubated for 21 days and photographed after 7 and 14 days. The growth on the 7- and 14-day plates was compared, and colonies seen only at 14 days were subcultured. Additional colonies from the 14-day plate were also subcultured, to a total of 40. The subculture plates were streaked with Staphylococcus aureus NCTC 6715 and Propionibacterium acnes ATCC 6919 at the time of subculture. Isolates were identified by 16S rRNA gene sequence analysis, as described previously (17).
Biofilm culture.
Biofilms were formed by means of the Calgary biofilm device on hydroxyapatite-coated pegs (Innovotech Inc., Edmonton, Alberta, Canada). Pegs were preconditioned in 200 μl heat-inactivated horse serum for 3 h at 37°C under anaerobic conditions. The pegs were then inoculated with subgingival plaque in prereduced PPB. The plates were incubated anaerobically, and the medium was replaced with fresh prereduced PPB every 3.5 days.
Biofilm composition analysis.
Biofilms were harvested every 14 days from 6 pegs, which were snapped off the plate lid. Each peg was placed in 500 μl phosphate-buffered saline (PBS) and the biofilm scraped off using a dental scaler. The resulting 6 suspensions were pooled and resuspended in 500 μl of PBS. To remove free DNA from the samples to ensure that only DNA from intact cells was included in the analysis, the samples were treated with propidium monoazide (PMA) and exposure to light (18): 1.25 μl of 20 mM PMA in water was added and the cells incubated in the dark with occasional shaking for 5 min, and the cell suspension was then placed on ice exposed to a 500-W halogen lamp for 3 min with occasional shaking.
DNA was extracted from the samples by means of the Genelute bacterial genomic DNA kit (Sigma, USA), following the protocol for Gram-positive bacteria. An approximately 500-bp region of the 16S rRNA gene (covering V1 to V3) was PCR amplified from extracted DNA from the three patient samples for 454 sequencing as previously described (19). Sequences were analyzed using the mothur suite mothur v.1.32.1 following the 454 standardized operating procedure. Sequences were clustered into operational taxonomic units (OTUs) at a sequence identity level of 98.5% using an average neighbor algorithm and then classified using a naive Bayesian classifier with the HOMD v 13 reference data set. The Jaccard index and the thetaYC metric were used to generate distance matrices from sequence libraries subsampled to the number of sequences found in the library with the fewest sequences, which were visualized as dendrograms and principal-coordinate analysis (PCoA) plots. Three-dimensional ordination plots were generated in R (r-project.org) using the rgl package.
Design and validation of specific primers for targeted oral taxa.
PCR primers with specificity for Bacteriodetes bacteria HOT 365 and HOT 281, Lachnospiraceae bacteria HOT 100 and HOT 500, and Clostridiales bacterium HOT 093 were designed by manual inspection of 16S rRNA gene alignments (Table 2). Primers were between 17 and 25 nucleotides in length, with a G+C content of between 40 and 50%. They were checked for self-complementarity and validated in silico both by interrogation of the Ribosomal Database Project 16S rRNA database and by BLAST search of the GenBank nucleotide database. Primers were synthesized by MWG Biotech. The primers were further validated by PCR using DNA obtained from P1 baseline sample (Bacteroidetes bacterium HOT 365 and Lachnospiraceae bacterium HOT 500) and P1 170D sample (Clostridiales bacterium HOT 093). PCR was performed using Ready Mix PCR master mixture with 5 pmol of each primer (bacterium-specific primer and 1492R) (Table 2). An initial denaturation was performed at 95°C for 1 min and followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 90 s. A final extension step was performed for 15 min. The amplicons were purified using ExoSAP-IT (Affymetrix), following the manufacturer's instructions, and then sequenced by means of a 3730xl DNA analyzer (Applied Biosystems), using the BigDye Terminator 3.1 cycle sequencing kit (Applied Biosystems) and sequencing primer 519R (20). The sequences were identified by means of the HOMD database identification tool at a sequence identity level of ≥98.5% (2).
TABLE 2.
PCR primers and probes used in the study
| Name | Sequence (5′→3′) | Oligonucleotide type (label) | Specificity | Reference |
|---|---|---|---|---|
| BACT365_281_337F | CGTCGTCTAATATCCCATAACATTG | PCR primer | Bacteroidetes bacteria HOT 365 and HOT 281 | This study |
| LACH500_50F | GCAAGTTGAACGAGAAGTTCC | PCR primer | Lachnospiraceae bacterium HOT 500 | This study |
| CLOST093_312F | CCTCTGTCTACGGGATAACA | PCR primer | Clostridiales bacterium HOT 093 | This study |
| 519R | GWATTACCGCGGCKGCTG | PCR primer | Domain Bacteria | 20 |
| 27FYM | AGAGTTTGATYMTGGCTCAG | PCR primer | Domain Bacteria | 28 |
| Lach500_786 | CTATAGGCCAGCACCTAGC | Probe (Cy5) | Lachnospiraceae bacterium HOT 500 | This study |
| Lach500_1206 | CTTCGCTTCCCTTTGTATCC | Probe (Cy5; DIG) | Lachnospiraceae bacterium HOT 500 | This study |
| Eub338 | GCTGCCTCCCGTAGGAGT | Probe (Cy3) | Domain Bacteria | 29 |
| NonEub | TGAGGATGCCCTCCGTCG | Probe (FITC) | Nonsense probe | 29 |
Detection of targeted bacterial taxa on solid media.
Pooled biofilm samples collected from the pegs were divided into 50-μl aliquots and used to inoculate FAA, FAAB, and PPA plates, which were incubated anaerobically for 7 days. The bacterial growth was harvested by flooding the plate with 2 ml PBS and suspending the colonies with an L-shaped spreader. DNA was extracted from the suspension, and the presence of the target bacteria was detected by PCR using taxon-specific primers (Table 2), as described above.
FISH.
Fluorescent in situ hybridization (FISH) was performed as previously described (21). Microscopic detection of fluorescently labeled cells was performed using a Leica SP2 confocal laser scanning system (Leica Microsystems) fitted with an argon krypton laser (operating at 488 nm), a krypton laser (operating at 568 nm), and a helium-neon laser (operating at 633 nm), using a Leica DMIRE2 inverted microscope with a 60× objective.
Colony hybridization.
Sterile 80-mm-diameter nylon membranes (Amersham Hybond; GE Healthcare) were placed directly onto agar plate cultures. Fifty picograms of digoxigenin (DIG)-labeled pBR328 (DIG nucleic acid detection kit; Roche Diagnostics) was applied to the membrane as positive control. The membrane was baked at 80°C for 40 min, prehybridized in DIG EasyHyb hybridization buffer (Roche Diagnostics) for 1 h at 58°C, and then hybridized with 20 nM DIG-labeled probes at 58°C for 2 h. The membrane was washed twice for 5 min in low-stringency wash buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] plus 0.1% SDS) at room temperature, followed by two 15-min washes in high-stringency wash buffer (0.1× SSC plus 0.1% SDS) at 60°C. The membrane was further washed for 5 min in washing buffer (1× maleic acid buffer plus 0.5% Tween 20; Roche Diagnostics), blocked for 30 min with 1% blocking solution (Roche Diagnostics) in maleic acid buffer, and incubated for 30 min in antibody solution (3 ml anti-DIG–alkaline phosphatase conjugate in 15 ml blocking solution; Roche Diagnostics). After washing twice in washing buffer and equilibrating the membrane with detection buffer (0.1 M Tris-HCl [pH 9.5] plus 0.1 M NaCl [Roche Diagnostics]) for 5 min, DIG/anti-DIG conjugates were detected using nitroblue tetrazolium chloride–5-bromo-4-chloro-3-indolyl-phosphate solution (80 μl in 4 ml detection buffer) (Roche Diagnostics).
Nucleotide sequence accession numbers.
The GenBank accession number for the 16S rRNA gene sequence of strain SP1_1 is KT730048. The sequences obtained from pyrosequencing analysis of the biofilms have been deposited with the NCBI Sequence Read Archive (SRA) under accession no. SRP063656.
RESULTS
Culture on novel agar media.
The ability of the four agar media used in the study, FAA, PPA, BEA, and GA, to support the growth of 10 reference strains of oral bacteria is shown in Table 1. All strains grew on all media with growth scores of at least 2+, and for 9 of the strains, growth on FAA was as strong or stronger than that on the other media. Parvimonas micra ATCC 33270, however, exhibited stronger growth on PPA than on FAA.
Subgingival plaque sample P1 was inoculated onto the four agar media and incubated anaerobically for 21 days, after which slow-growing colonies, including those showing evidence of satelliting, were subcultured and identified with the aim of isolating previous uncultivated phylotypes. The oral taxa identified on the four media are shown in Table 3. Representatives of 38 oral taxa were recovered, including the previously uncultivated phylotype Actinomyces sp. HOT 525 from sample P1.
TABLE 3.
Bacterial taxa isolated from subgingival plaque sample P1 on four anaerobically incubated agar culture media
| Oral taxon | Isolation on: |
|||
|---|---|---|---|---|
| FAA | BEA | PPA | GA | |
| Actinomyces gerencseriae | • | |||
| Actinomyces sp. HOT 525 | • | |||
| Actinomyces sp. HOT 169 | • | |||
| Actinomyces sp. HOT 171 | • | • | • | • |
| Bacteroidaceae bacterium HOT 272 | • | |||
| Campylobacter gracilis | • | • | ||
| Campylobacter showae | • | • | • | |
| Capnocytophaga sp. HOT 326 | • | • | ||
| Capnocytophaga sputigena | • | |||
| Dialister pneumosintes | • | • | • | • |
| Eubacterium infirmum | • | |||
| Eubacterium saphenum | • | • | ||
| Eubacterium sulci | • | |||
| Filifactor alocis | • | • | ||
| Fusobacterium nucleatum subsp. animalis | • | |||
| Fusobacterium nucleatum subsp. nucleatum | • | • | ||
| Mogibacterium diversum | • | |||
| Mogibacterium timidum | • | • | ||
| Parvimonas micra | • | • | • | |
| Peptostreptococcus stomatis | • | • | ||
| Porphyromonas gingivalis | • | • | • | |
| Prevotella intermedia | • | |||
| Prevotella saccharolytica | • | |||
| Prevotella sp. HOT 472 | • | • | ||
| Propionibacterium acnes | • | • | ||
| Pseudoramibacter alactolyticus | • | |||
| Slackia exigua | • | • | • | • |
| Solobacterium moorei | • | |||
| Staphylococcus aureus | • | • | • | |
| Staphylococcus epidermidis | • | |||
| Staphylococcus saprophyticus | • | |||
| Staphylococcus warneri | • | |||
| Streptococcus constellatus | • | |||
| Streptococcus gordonii | • | • | ||
| Streptococcus mitis | • | |||
| Streptococcus sp. HOT 064 | • | |||
| Tannerella forsythia | • | |||
| Veillonella parvula | • | • | ||
Culture-independent analysis of subgingival plaque samples and in vitro biofilms.
The compositions of samples P1, P2, and P3 and biofilms derived from the samples were determined by 16S rRNA gene pyrosequencing. The sequences were identified using the wang method by means of the mothur command classify.seqs with reference to the HOMD v 13 reference data set. Identifications above the bootstrap cutoff of 0.8 are shown in Table S1 in the supplemental material. A diverse bacterial community was seen in all three baseline samples, and a subset of these was seen to constitute the in vitro biofilms.
Previously uncultivated phylotypes were detected in the baseline samples and in the in vitro biofilms, and these are listed in Table 4. Sixty-one such phylotypes were found in the baseline samples, including representatives of the phyla Actinobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Fusobacteria, Proteobacteria, Spirochaetes, Synergistetes, Tenericutes, and “Candidatus Saccharibacteria” (TM7) and division SR1. Sixteen of the uncultured phylotypes were found in the in vitro-cultivated biofilms. It is interesting that some, including Lachnospiraceae bacterium HOT 500, Oribacterium sp. HOT 102, Peptostreptococcaceae bacteria HOT 091 and HOT 103, and Prevotella sp. HOT 304, were detected early in biofilm development and at multiple time points, while the majority were seen in the biofilms only after 105 days of incubation, suggesting that they were present previously but in undetectably low numbers.
TABLE 4.
Previously uncultured phylotypes detected in subgingival plaque samples and in vitro biofilms
| Bacterium | Proportion of microbiota (%) after the indicated time (days) of incubation in sample: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 |
P2 |
P3 |
||||||||||||
| 0a | 15 | 30 | 45 | 60 | 75 | 90 | 105 | 120 | 170 | 0 | 50 | 0 | 50 | |
| Actinomyces sp. HOT 171 | 0.05 | 0.05 | ||||||||||||
| Actinomyces sp. HOT 414 | 0.13 | |||||||||||||
| Actinomyces sp. HOT 449 | 0.04 | |||||||||||||
| Actinomyces sp. HOT 897 | 0.04 | |||||||||||||
| Alloprevotella sp. HOT 308 | 0.04 | |||||||||||||
| Alloprevotella sp. HOT 914 | 0.03 | |||||||||||||
| Bacteroidetes bacterium HOT 280 | 0.54 | 0.03 | 0.34 | |||||||||||
| Bacteroidetes bacterium HOT 505 | 0.1 | |||||||||||||
| Bacteroidetes bacterium HOT 511 | 0.1 | 2.73 | ||||||||||||
| Catonella sp. HOT 451 | 0.05 | |||||||||||||
| Chloroflexi sp. HOT 439 | 0.03 | 0.03 | ||||||||||||
| Clostridiales bacterium HOT 075 | 0.2 | |||||||||||||
| Clostridiales bacterium HOT 085 | 0.08 | |||||||||||||
| Clostridiales bacterium HOT 093 | 0.98 | 0.04 | 0.03 | |||||||||||
| Desulfobulbus sp. HOT 041 | 1.35 | 0.92 | 0.10 | |||||||||||
| Dialister sp. HOT 119 | 0.20 | |||||||||||||
| Erysipelotrichaceae bacterium HOT 904 | 0.13 | 0.03 | ||||||||||||
| Erysipelotrichaceae bacterium HOT 905 | 0.04 | 0.07 | ||||||||||||
| Fretibacterium HOT 359 | 0.1 | 0.11 | 0.17 | 0.30 | ||||||||||
| Fretibacterium HOT 360 | 1.79 | 0.62 | 0.21 | |||||||||||
| Fretibacterium HOT 361 | 1.79 | 0.03 | 0.21 | |||||||||||
| Haemophilus sp. HOT 035 | 0.07 | |||||||||||||
| Johnsonella sp. HOT 166 | 0.27 | 0.10 | ||||||||||||
| Lachnoanaerobaculum sp. HOT 083 | 0.04 | |||||||||||||
| Lachnospiraceae bacterium HOT 080 | 0.03 | |||||||||||||
| Lachnospiraceae bacterium HOT 096 | 0.08 | 0.23 | ||||||||||||
| Lachnospiraceae bacterium HOT 500 | 0.34 | 1.54 | 1.24 | 0.61 | 2.27 | 1.21 | 1.06 | 1.04 | 0.19 | 1.01 | 0.04 | 0.86 | 2.85 | |
| Leptotrichia sp. HOT 215 | 0.03 | |||||||||||||
| Leptotrichia sp. HOT 223 | 0.17 | 0.16 | ||||||||||||
| Leptotrichia sp. HOT 417 | 0.04 | |||||||||||||
| Megasphaera sp. HOT 123 | 0.04 | 0.23 | ||||||||||||
| Megasphaera sp. HOT 841 | 0.03 | |||||||||||||
| Mollicutes bacterium HOT 504 | 0.47 | |||||||||||||
| Mollicutes bacterium HOT 906 | 0.03 | |||||||||||||
| Oribacterium sp. HOT 102 | 0.13 | 0.03 | 0.11 | 0.56 | 0.39 | 0.51 | ||||||||
| Peptostreptococcaceae bacterium HOT 081 | 0.03 | 0.24 | ||||||||||||
| Peptostreptococcaceae bacterium HOT 091 | 0.22 | 0.08 | 0.08 | 0.96 | 0.51 | |||||||||
| Peptostreptococcaceae bacterium HOT 103 | 0.08 | 0.11 | 0.36 | 0.26 | 0.05 | 0.68 | 0.17 | 0.36 | ||||||
| Peptostreptococcaceae bacterium HOT 495 | 0.08 | |||||||||||||
| Prevotella sp. HOT 292 | 0.13 | 0.03 | ||||||||||||
| Prevotella sp. HOT 300 | 1.17 | |||||||||||||
| Prevotella sp. HOT 301 | 0.08 | 0.03 | ||||||||||||
| Prevotella sp. HOT 304 | 8.99 | 3.69 | 1.89 | 1.50 | 3.37 | 4.46 | 4.00 | 5.05 | 0.03 | 0.21 | 0.03 | 2.63 | ||
| Prevotella sp. HOT 376 | 0.07 | |||||||||||||
| Prevotella sp. HOT 526 | 0.30 | 0.05 | 0.59 | 0.10 | ||||||||||
| Selenomonas sp. HOT 126 | 0.27 | |||||||||||||
| SR1 bacterium HOT 345 | 0.10 | |||||||||||||
| Streptococcus sp. HOT 058 | 0.03 | 0.03 | 0.05 | |||||||||||
| Tannerella sp. HOT 286 | 0.36 | |||||||||||||
| Tannerella sp. HOT 808 | 0.04 | 0.75 | ||||||||||||
| Saccharibacteria bacterium HOT 346 | 0.07 | 0.25 | 0.29 | |||||||||||
| Saccharibacteria bacterium HOT 349 | 0.03 | 0.08 | ||||||||||||
| Saccharibacteria bacterium HOT 356 | 2.52 | 3.94 | ||||||||||||
| Treponema sp. HOT 238 | 0.03 | |||||||||||||
| Treponema sp. HOT 254 | 0.07 | |||||||||||||
| Treponema sp. HOT 234 | 0.14 | |||||||||||||
| Treponema sp. HOT 237 | 0.24 | |||||||||||||
| Treponema sp. HOT 258 | 0.03 | |||||||||||||
| Treponema sp. HOT 262 | 0.46 | |||||||||||||
| Treponema sp. HOT 270 | 0.07 | 0.92 | ||||||||||||
| Veillonellaceae sp. HOT 132 | 0.10 | |||||||||||||
Baseline samples.
The mothur wang classification method assigns sequences to reference taxa by sequence identity and applies a confidence threshold on the basis of bootstrapping. Sequences are not classified either if they do not match sequences in the database or if they cannot be unambiguously identified. This can be problematic for genera such as Streptococcus where many valid species have highly similar 16S rRNA sequences. The sequences that were not classified in the primary analysis were therefore reanalyzed by BLAST comparison with the HOMD extended set and GenBank nucleotide databases. An additional 32 previously uncultivated phylotypes were thus identified and are shown in Table 5. Ten of these were detected in the in vitro biofilms. In addition, 9 groups of sequences with less than 98.5% sequence identity to human oral taxa in the HOMD extended data set were identified. These are listed in Table 6. Seven of the nine taxa were found only in baseline samples, but two were detected in the in vitro biofilms.
TABLE 5.
Identification of unclassified sequences detected in subgingival plaque samples and in vitro biofilms
| Bacterium | Proportion of microbiota (%) after the indicated time (days) of incubation in sample: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 |
P2 |
P3 |
||||||||||||
| 0a | 15 | 30 | 45 | 60 | 75 | 90 | 105 | 120 | 170 | 0 | 50 | 0 | 50 | |
| Bacteroidetes bacterium HOT 365 | 0.51 | 0.08 | ||||||||||||
| Bacteroidetes bacterium HOT F70 | 0.03 | 0.04 | ||||||||||||
| Burkholderiales bacterium HOT A57 | 0.04 | |||||||||||||
| Comamonadaceae bacterium HOT F47 | 0.03 | 0.03 | ||||||||||||
| Fusobacterium HOT C10 | 0.68 | |||||||||||||
| Fusobacterium HOT H27 | 0.04 | |||||||||||||
| Lachnospiraceae bacterium HOT G33 | 0.27 | |||||||||||||
| Leptotrichia sp. HOT 417 | 0.38 | 1.92 | ||||||||||||
| Leptotrichia sp. HOT B57 | 0.57 | 2.47 | ||||||||||||
| Peptococcus sp. HOT D92 | 0.10 | |||||||||||||
| Peptostreptococcaceae bacterium HOT 369 | 0.03 | |||||||||||||
| Porphyromonas sp. HOT B94 | 0.14 | |||||||||||||
| Prevotella sp. HOT 300 | 0.21 | |||||||||||||
| Prevotella sp. HOT 396 | 0.51 | |||||||||||||
| Propionivibrio sp. HOT C33 | 0.03 | |||||||||||||
| Selenomonas sp. HOT 126 | 0.03 | 0.36 | ||||||||||||
| Selenomonas sp. HOT B30 | 0.21 | |||||||||||||
| Selenomonas sp. HOT E44 | 0.17 | 0.08 | ||||||||||||
| Selenomonas sp. HOT E83 | 0.42 | |||||||||||||
| Selenomonas sp. HOT F29 | 0.14 | 0.10 | 0.04 | 0.71 | ||||||||||
| Selenomonas sp. HOT G51 | 0.05 | |||||||||||||
| Selenomonas sp. HOT G67 | 0.14 | |||||||||||||
| Streptococcus sp. HOT 064 | 0.03 | |||||||||||||
| Streptococcus sp. HOT 431 | 0.10 | |||||||||||||
| Streptococcus sp. HOT C56 | 0.03 | |||||||||||||
| Streptococcus sp. HOT C65 | 0.03 | |||||||||||||
| Fretibacterium HOT 360 | 0.10 | 0.04 | 0.03 | |||||||||||
| Fretibacterium HOT 362 | 0.07 | |||||||||||||
| Fretibacterium HOT 453 | 0.17 | 0.13 | ||||||||||||
| Veillonellaceae bacterium HOT 135 | 0.03 | 0.04 | ||||||||||||
| Terrahaemophilus sp. HOT G25 | 0.08 | |||||||||||||
Baseline samples.
TABLE 6.
Novel human oral taxa identified in the study
| Bacterium | HOMD HOT no. | Proportion of microbiota (%) after the indicated time (days) of incubation in sample: |
||||
|---|---|---|---|---|---|---|
| P1 |
P2 (0) | P3 (0) | ||||
| 0 | 120 | 170 | ||||
| Bacteroidetes [G-5] H1 | H71 | 2.16 | ||||
| Peptostreptococcus [XI][G-7] H2 | 922 | 0.08 | ||||
| Peptostreptococcus [XI][G-5] H3 | 923 | 0.29 | ||||
| Acholeplasmatales H5 | H72 | 0.91 | ||||
| Stomatobaculum H6 | 924 | 0.03 | ||||
| Leptotrichia H7 | 925 | 0.03 | 0.08 | |||
| Prevotella H8 | 926 | 0.03 | ||||
| Fretibacterium H9 | H73 | 0.20 | ||||
Culture of a previously uncultivated oral taxon.
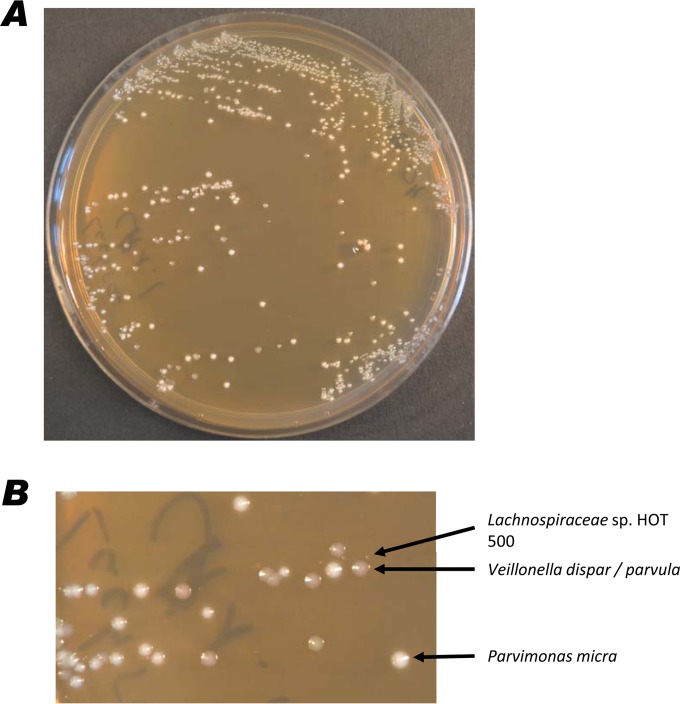
Taxa representing three previously uncultured lineages were chosen as targets for further investigation. The choice was made based on their frequency of detection in the biofilms generated from subgingival plaque samples and previous detection in the studies used to construct the HOMD database (3). The target taxa were Bacteroidetes bacteria HOT 365 and HOT 281, Lachnospiraceae bacterium HOT 500, and Clostridiales bacterium 093. The PCR primers shown in Table 2 were used to amplify PCR products from the in vitro biofilms derived from the subgingival plaque samples and were confirmed to have the appropriate specificity by sequencing of the products obtained. Table S1 in the supplemental material reveals that Lachnospiraceae bacterium HOT 500 was found in the biofilms derived from samples P1, P2, and P3 at all time points. Lachnospiraceae bacterium HOT 500 and Bacteroidetes bacterium HOT 365 were detected in the biofilms from P1 (34 weeks) and P2 (30 weeks) and on anaerobically incubated PPA plates inoculated with biofilm material after 6 days of incubation. Colony hybridization of these PPA plates with the Lach500_1206 DIG-labeled probe (Table 2) revealed areas of positive hybridization, and bacterial growth from these regions was subcultured onto fresh PPA plates. After 5 days of incubation, three colony types were observed on the plate, one of which appeared to be exhibiting satellite growth around the other types, with small colonies up to 3 mm away from the other colony types (Fig. 1).
FIG 1.
Culture of Lachnospiraceae sp. HOT 500 on a PPA agar plate from an inoculum enriched by colony hybridization. (A) Whole plate; (B) part of plate showing colonies of Lachnospiraceae sp. HOT 500, Parvimonas micra, and Veillonella dispar/parvula.
The colony types exhibiting independent growth were identified by 16S rRNA gene sequence analysis as Parvimonas micra and Veillonella dispar/parvula. The satellite colonies, designated isolate SP1_1, displayed >98.5% 16S rRNA gene sequence identity to Lachnospiraceae bacterium HOT 500. SP1_1 was not capable of independent growth but was able to grow in coculture with Parvimonas micra or Veillonella dispar/parvula, as well as Fusobacterium nucleatum subsp. nucleatum, Staphylococcus epidermidis, and Propionibacterium acnes. SP1_1 was able to grow in coculture on all agar media used in the study, but growth was typically more abundant on PPA medium.
DISCUSSION
This study has confirmed that subgingival plaque includes a high proportion of previously uncultivated oral taxa. The first strategy used to cultivate previously uncultivated organisms was to design novel agar culture media in an attempt to better mimic the nutrients available in the subgingival environment. Proteose peptone was chosen as the primary nitrogen source for the base medium because the wide range of peptide sizes makes it suitable for the culture of fastidious bacteria. Mucin was included since it is the principal glycoprotein in human saliva and its addition to culture media in vitro has resulted in increased bacterial growth overall and an increased proportion of anaerobes, including fastidious organisms such as spirochetes (22). Gelysate, a pancreatic digest of gelatin, was added to one version of the medium because gelatin is extracted from collagen, one of the major connective tissue components of the periodontal tissues. Beef extract includes a mixture of peptides, amino acids, nucleotide fractions, organic acids, minerals, and vitamins likely to benefit the growth of the target group of organisms. The results obtained from extended anaerobic incubation of the range of novel agar media were rather disappointing in that only one previously uncultured oral taxon was recovered, and this belonged to the genus Actinomyces, a genus whose members are not normally regarded as difficult to culture and which may therefore represent a chance finding.
In contrast, the establishment of an in vitro biofilm derived from the samples by means of the Calgary biofilm device resulted in a species-rich oral bacterial community, which included a number of previously uncultivated oral bacterial taxa. This can be regarded as a first step to in vitro isolation. The biofilm culture was then available as a source of material for further investigation and attempts to recover previously uncultivated taxa, which proved successful for Lachnospiraceae bacterium HOT 500. This model has been shown to generate reproducible complex oral biofilms (23), which will be valuable for future work to use modified and supplemented growth media and conditions and inocula from different oral disease states in order to isolate more novel oral bacterial taxa.
The growth of previously uncultivated species in coculture with other bacteria confirms the hypothesis that oral bacteria have evolved as a mixed community and that many members of the community depend on the presence of other bacteria for growth, presumably by sharing nutrients or because of a requirement for intercellular signaling to control growth (11). The recent successful isolation of a member of the phylum TM7 by He et al. (24) has shown that the relationship between oral bacteria can be more intimate, with the demonstration that the TM7 organism isolated has an intracellular lifestyle, growing within the cells of another species.
The importance of developing methods to isolate previously uncultivated bacteria has been recently emphasized by the isolation of a strain of a new bacterial species, “Eleftheria terrae,” that produces a novel antimicrobial, teixobactin, with clinically relevant activity (25). The strain was isolated by means of the iChip (26), whereby soil samples were diluted so that a single bacterial cell was present in channels of the device, which was then covered by a semipermeable membrane and placed in soil so that growth factors produced by the soil community could access the culture chamber. A similar coculture approach using soft agar and a membrane filter has been successful in isolating novel human gut bacteria (27). Further developments of these and related methods will be of value in further increasing the proportion of the oral bacterial biome that can be cultured in vitro.
Supplementary Material
ACKNOWLEDGMENTS
This work has been supported in part by grant DE016937 from the National Institute of Dental and Craniofacial Research.
We thank V. Booth and R. Palmer for collecting the subgingival plaque samples used in this study. We thank J. Kistler for assistance with the pyrosequencing analyses. We thank S. Vartoukian for helpful discussions and comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02156-15.
REFERENCES
- 1.Wade WG. 2013. Characterisation of the human oral microbiome. J Oral Biosci 55:143–148. doi: 10.1016/j.job.2013.06.001. [DOI] [Google Scholar]
- 2.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Southerland JH, Moss K, Taylor GW, Beck JD, Pankow J, Gangula PR, Offenbacher S. 2012. Periodontitis and diabetes associations with measures of atherosclerosis and CHD. Atherosclerosis 222:196–201. doi: 10.1016/j.atherosclerosis.2012.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Chee B, Park B, Bartold PM. 2013. Periodontitis and type II diabetes: a two-way relationship. Int J Evid Based Healthc 11:317–329. doi: 10.1111/1744-1609.12038. [DOI] [PubMed] [Google Scholar]
- 6.Beck JD, Offenbacher S. 2005. Systemic effects of periodontitis: epidemiology of periodontal disease and cardiovascular disease. J Periodontol 76:2089–2100. doi: 10.1902/jop.2005.76.11-S.2089. [DOI] [PubMed] [Google Scholar]
- 7.Kapil V, Haydar SM, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. 2013. Physiological role for nitrate-reducing oral bacteria in blood pressure control. Free Radic Biol Med 55:93–100. doi: 10.1016/j.freeradbiomed.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanasi E, Dewhirst FE, Chalmers NI, Kent R Jr, Moore A, Hughes CV, Pradhan N, Loo CY, Tanner AC. 2010. Clonal analysis of the microbiota of severe early childhood caries. Caries Res 44:485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol 44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vartoukian SR, Palmer RM, Wade WG. 2010. Strategies for culture of ‘unculturable’ bacteria. FEMS Microbiol Lett 309:1–7. doi: 10.1111/j.1574-6968.2010.02000.x. [DOI] [PubMed] [Google Scholar]
- 12.Davis KE, Joseph SJ, Janssen PH. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl Environ Microbiol 71:826–834. doi: 10.1128/AEM.71.2.826-834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joseph SJ, Hugenholtz P, Sangwan P, Osborne CA, Janssen PH. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl Environ Microbiol 69:7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka T, Kawasaki K, Daimon S, Kitagawa W, Yamamoto K, Tamaki H, Tanaka M, Nakatsu CH, Kamagata Y. 2014. A hidden pitfall in the preparation of agar media undermines microorganism cultivability. Appl Environ Microbiol 80:7659–7666. doi: 10.1128/AEM.02741-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vartoukian SR, Palmer RM, Wade WG. 2010. Cultivation of a Synergistetes strain representing a previously uncultivated lineage. Environ Microbiol 12:916–928. doi: 10.1111/j.1462-2920.2009.02135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sizova MV, Hohmann T, Hazen A, Paster BJ, Halem SR, Murphy CM, Panikov NS, Epstein SS. 2012. New approaches for isolation of previously uncultivated oral bacteria. Appl Environ Microbiol 78:194–203. doi: 10.1128/AEM.06813-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munson MA, Pitt Ford T, Chong B, Weightman AJ, Wade WG. 2002. Molecular and cultural analysis of the microflora associated with endodontic infections. J Dent Res 81:761–766. doi: 10.1177/154405910208101108. [DOI] [PubMed] [Google Scholar]
- 18.Nocker A, Mazza A, Masson L, Camper AK, Brousseau R. 2009. Selective detection of live bacteria combining propidium monoazide sample treatment with microarray technology. J Microbiol Methods 76:253–261. doi: 10.1016/j.mimet.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Kistler JO, Booth V, Bradshaw DJ, Wade WG. 2013. Bacterial community development in experimental gingivitis. PLoS One 8:e71227. doi: 10.1371/journal.pone.0071227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 21.Vartoukian SR, Palmer RM, Wade WG. 2009. Diversity and morphology of members of the phylum “synergistetes” in periodontal health and disease. Appl Environ Microbiol 75:3777–3786. doi: 10.1128/AEM.02763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glenister DA, Salamon KE, Smith K, Beighton D, Keevil CW. 1988. Enhanced growth of complex communities of dental plaque bacteria in mucin-limited continuous culture. Microb Ecol Health Dis 1:31–38. doi: 10.3109/08910608809140176. [DOI] [Google Scholar]
- 23.Kistler JO, Pesaro M, Wade WG. 2015. Development and pyrosequencing analysis of an in-vitro oral biofilm model. BMC Microbiol 15:24. doi: 10.1186/s12866-015-0364-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A 112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schaberle TF, Hughes DE, Epstein S, Jones M, Lazarides L, Steadman VA, Cohen DR, Felix CR, Fetterman KA, Millett WP, Nitti AG, Zullo AM, Chen C, Lewis K. 2015. A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi: 10.1038/nature14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nichols D, Cahoon N, Trakhtenberg EM, Pham L, Mehta A, Belanger A, Kanigan T, Lewis K, Epstein SS. 2010. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol 76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, Benno Y. 2015. Application of a single-colony coculture technique to the isolation of hitherto unculturable gut bacteria. Microbiol Immunol 59:63–70. doi: 10.1111/1348-0421.12220. [DOI] [PubMed] [Google Scholar]
- 28.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.