Abstract
Bacterial microcompartments (BMCs) are proteinaceous organelles encapsulating enzymes that catalyze sequential reactions of metabolic pathways. BMCs are phylogenetically widespread; however, only a few BMCs have been experimentally characterized. Among them are the carboxysomes and the propanediol- and ethanolamine-utilizing microcompartments, which play diverse metabolic and ecological roles. The substrate of a BMC is defined by its signature enzyme. In catabolic BMCs, this enzyme typically generates an aldehyde. Recently, it was shown that the most prevalent signature enzymes encoded by BMC loci are glycyl radical enzymes, yet little is known about the function of these BMCs. Here we characterize the glycyl radical enzyme-associated microcompartment (GRM) loci using a combination of bioinformatic analyses and active-site and structural modeling to show that the GRMs comprise five subtypes. We predict distinct functions for the GRMs, including the degradation of choline, propanediol, and fuculose phosphate. This is the first family of BMCs for which identification of the signature enzyme is insufficient for predicting function. The distinct GRM functions are also reflected in differences in shell composition and apparently different assembly pathways. The GRMs are the counterparts of the vitamin B12-dependent propanediol- and ethanolamine-utilizing BMCs, which are frequently associated with virulence. This study provides a comprehensive foundation for experimental investigations of the diverse roles of GRMs. Understanding this plasticity of function within a single BMC family, including characterization of differences in permeability and assembly, can inform approaches to BMC bioengineering and the design of therapeutics.
INTRODUCTION
In the last decade, an accumulation of countervailing evidence has overturned the long-held verdict that bacteria are primitive organisms lacking intracellular organization. In addition to membrane-bound organelles like the magnetosomes of magnetotactic bacteria (1, 2) or the anammoxosomes found in some members of the Planctomycetes, bacteria also form protein-based organelles known as bacterial microcompartments (BMCs) (3–5). The defining feature of all BMCs is a shell composed of homologous proteins containing the Pfam00936 domain. A protein with a single copy of the Pfam00936 domain that forms hexamers is referred to as BMC-H; a second type of shell protein consists of a fusion of two Pfam00936 domains and forms pseudohexameric trimers and is referred to as BMC-T. The hexamers and pseudohexamers are thought to tile into the facets of the shell (3, 6). A third type of shell protein containing the Pfam03319 domain forms pentamers that are assumed to cap the vertices of an apparently icosahedral shell and is referred to as BMC-P (3, 7). The BMC-H, BMC-T, and BMC-P oligomers are typically perforated by pores, through which metabolite exchange with the cytosol is assumed to be mediated. Residues flanking the pores are generally polar and well conserved among orthologs, and mutation of the residues can lead to growth defects, leakage of intermediates, or impaired uptake of substrates (8, 9), while others are tolerated (10). Comparable to a lipid bilayer, the protein shell is predicted to act as a semipermeable membrane that isolates the BMC lumen from the cytosol. Notably, this permeability is opposite in selectivity to that of a lipid bilayer; it allows the passage of charged and polar molecules, whereas uncharged and nonpolar compounds do not freely pass.
While the protein shell defines the organelle and is conserved among all BMCs, the enzymes associated with BMCs vary; as a result, BMCs are functionally diverse. The first BMC identified, the carboxysome, encapsulates ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (11, 12) together with a carbonic anhydrase (13, 14) to enhance the efficiency of CO2 fixation in cyanobacteria and some chemo- and photoautotrophs. In contrast, the five other types of BMCs that have been experimentally characterized to date are catabolic. These BMCs function to degrade various carbon compounds and are referred to as propanediol-utilizing (PDU) and ethanolamine-utilizing (EUT) BMCs (3, 15–17), ethanol-utilizing BMCs (18, 19), and two distinct types of BMCs that degrade fucose (20, 21).
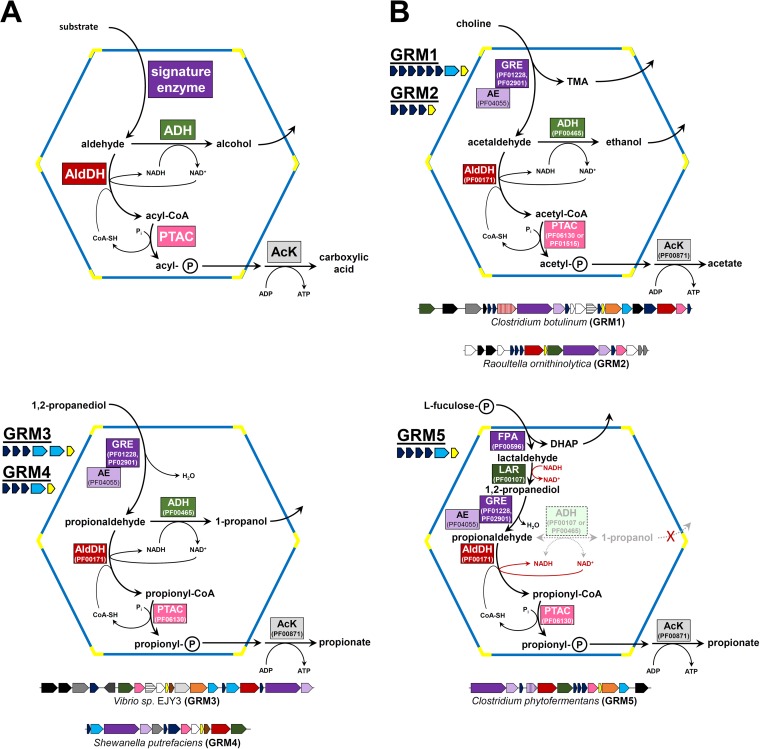
A common feature of all known catabolic BMCs, referred to as metabolosomes (22), is the formation of a volatile and/or reactive aldehyde (Fig. 1) that is subsequently either reduced to an alcohol by an alcohol dehydrogenase or oxidized to an acyl coenzyme A (CoA) thioester by an acylating aldehyde dehydrogenase. The CoA thioester is then used to produce ATP (Fig. 1) via reactions catalyzed by a phosphotransacylase and an acyl kinase. The alcohol dehydrogenase and the phosphotransacylase are important for the regeneration of NAD+ and CoA within the BMC lumen (23–25) (Fig. 1). These cofactors are presumably too large to cross the shell; thus, BMCs maintain private cofactor pools insulated from the cytosol. Collectively, the aldehyde dehydrogenase, the alcohol dehydrogenase, and the phosphotransacylase constitute the core enzymes conserved across the majority of catabolic BMCs (5, 26). The aldehyde-generating enzyme, which differs among metabolosome types, is considered the signature enzyme (5, 26) because it defines the substrate of the BMC (e.g., 1,2-propanediol dehydratase for PDU BMCs, ethanolamine ammonia-lyase for EUT BMCs, a sugar phosphate aldolase for the BMCs involved in fucose degradation).
FIG 1.
Schematics of the functions of metabolosomes. (A) Generalized functional scheme for metabolism by experimentally characterized catabolic bacterial microcompartments. The signature enzyme generates a toxic and/or volatile aldehyde, which is successively metabolized by an acylating aldehyde dehydrogenase (AldDH), a phosphotransacylase (PTAC), and an acyl kinase (AcK). The cosubstrates CoA and NAD+ are regenerated by the PTAC and an alcohol dehydrogenase (ADH) within the lumen of the compartment. (B) Schematics of the proposed functions of the different GRE-associated microcompartments (GRMs). The GREs in GRM1 and GRM2 are predicted to be choline TMA-lyases or ethanolamine ammonia-lyases, whereas the GREs in GRM3 to GRM5 are proposed to function as 1,2-propanediol dehydratases. GRM5 BMC loci also encode a fuculose phosphate aldolase (FPA) and a putative lactaldehyde reductase (LAR). The function of the lactaldehyde reductase would negate the necessity for a propanol dehydrogenase. Representative gene clusters for each type of locus are depicted below the schematic with the corresponding coloring: purple, signature enzymes; purple hatched, FPA; red, AldDH; red hatched, inactive (dud) AldDH; green, ADH; pink, PTAC; light gray, AcK; orange, PduS (PF13375, PF13534, PF01512, PF10531); gray hatched, EutJ (PF11104); brown, DUF336 (PF03928); dark gray, putative transporters (various Pfams); black, putative regulators (various Pfams); white, other ancillary genes. The average number and types of BMC shell proteins of the different GRMs are also indicated; dark blue, BMC-H; light blue, BMC-T; yellow, BMC-P.
Recently, a comprehensive study (5) identified and categorized a surprisingly large number of different types of BMCs on the basis of genomic analyses and genome locus composition. Interestingly, the most abundant single class of BMCs as defined by the signature enzyme is almost entirely uncharacterized. These BMC loci contain genes encoding a glycyl radical enzyme (GRE) and its associated activating enzyme (AE). The glycyl radical enzyme family, which utilizes radicals of the amino acids glycine and cysteine during catalysis, is considered to be evolutionarily ancient (27); members have a vast array of different substrate and reaction specificities. Examples of experimentally characterized GREs are pyruvate formate-lyase (28), anaerobic ribonucleotide reductase (29), benzylsuccinate synthase (30), glycerol dehydratase (31), 4-hydroxyphenylacetate decarboxylase (32), and choline trimethylamine (TMA)-lyase (33). GREs are synthesized in a catalytically inactive form that requires activation by an iron-sulfur cluster-containing enzyme, which catalyzes the reductive cleavage of S-adenosylmethionine (SAM), producing a transient radical intermediate (34). This radical intermediate abstracts a hydrogen from a strictly conserved glycine residue (35), generating the active form of the GRE. Subsequently, the radical is transferred to a conserved cysteine residue in the active site, which then takes part in the radical reaction with the substrate of the GRE. The active form of a GRE is extremely oxygen sensitive; exposure to oxygen leads to inactivation by cleavage of the polypeptide chain at the position of the radical (36). Encapsulation in a BMC may protect the enzyme from molecular oxygen; this would parallel the function of the carboxysome shell, which provides a diffusion barrier to CO2 (37) and, presumably, O2.
Genome loci encoding GRE-associated microcompartments (GRMs) are prevalent and phylogenetically diverse, being found in the Actinobacteria, Firmicutes, Alphaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria (5). The GRE of a GRM locus in a clostridial species was suggested to act as a 1,2-propanediol dehydratase, analogous to the PDU BMC (20). A GRE associated with another type of GRM locus has been shown to catalyze a choline trimethylamine-lyase reaction (33, 38). This hints at the functional diversity of GRMs. In contrast to the PDU and EUT metabolosome loci for which identification of the signature enzyme (e.g., ethanolamine ammonia-lyase) is sufficient to predict function, for GRM loci the identification of the GRE is only a first step to predicting function. This is due to the range of their prospective substrates.
Here we present a comprehensive bioinformatic characterization of the GRM loci. By sequence, structural, and phylogenetic comparison of the associated core and ancillary enzymes and analysis of the shell properties, we show that the GRM loci not only are the most prevalent BMC loci (5) but also encode the most functionally diverse family of metabolosomes known. These data enable prediction of three distinct functions for GRM loci and hint that many are involved in pathogenesis. Moreover, we found evidence for distinctive differences in how the GRM subtypes assemble. Finally, our data indicate how the GRMs may have evolved, likely descending from PDU BMCs. Collectively, these results provide a basis for experimental efforts to characterize this diverse family of BMCs that appear to play key roles in organismal fitness in ecophysiologically important microbes. Moreover, it expands our understanding of the structural and functional diversity of BMCs.
MATERIALS AND METHODS
The GRM locus database was extracted from the work of Axen et al. (5). Multiple-sequence alignments were performed with the MUSCLE program (39) using standard parameters, and alignments were visualized using the JalView program (40).
The evolutionary histories of the proteins and enzymes were inferred using the maximum likelihood method based on the Le and Gascuel model (41). All positions containing gaps were eliminated. Evolutionary analyses were conducted in the MEGA (version 6) program (42) with 100 bootstraps. Only for the phylogenetic analyses of BMC-H sequences, all positions in the alignments with less than 95% site coverage were eliminated, allowing fewer than 5% alignment gaps at any position.
Homology modeling and secondary structure predictions were performed by using the Phyre2 online server (43), RaptorX web server (44), and SWISS-MODEL server (45). The models were analyzed using the PyMOL (https://www.pymol.org) and UCSF Chimera (46) programs. Homology modeling for surface electrostatics analysis was performed using the SWISS-MODEL server. Multiple templates were used, and only models with the highest global model quality estimation values and the highest values from the QMEAN server (47) were used for subsequent surface electrostatics analysis. The surface electrostatics were generated using the PDB2PQR program (48), the APBS program (49), and PyMOL. Pore diameters were analyzed using the Caver (http://caver.cz) plug-in for PyMOL.
RESULTS
Analysis of signature enzymes.
The different types of GRE-associated BMCs are referred to as GRM1 to GRM5 (Fig. 1B) according to the classification of Axen et al. (5), which grouped loci on the basis of the encoded Pfam domains and calculation of locus similarity networks. We compared the primary structures of the enzymes, created homology models based on known crystal structures, and constructed phylogenetic trees. We hypothesized that enzymes would phylogenetically cluster together according to their function (i.e., substrate and reaction specificities), likely corresponding to the BMC locus types, as opposed to the species phylogeny of the host organism.
(i) GREs.
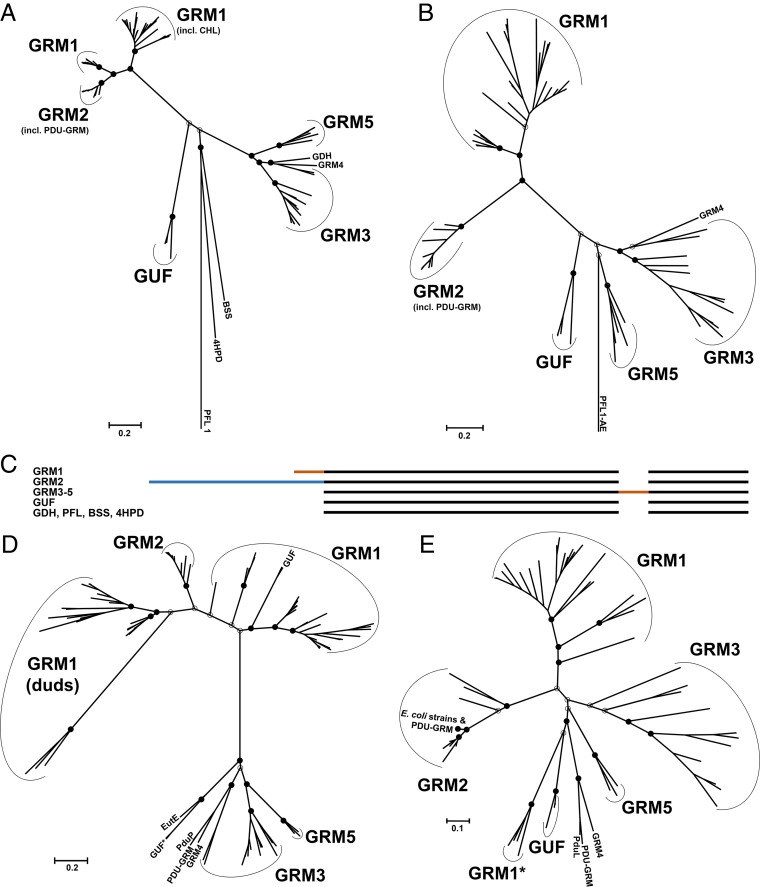
A list of all analyzed BMC-associated glycyl radical enzymes (GREs) (with GenBank accession numbers) is provided in Table S1 in the supplemental material. A phylogenetic tree of the BMC-associated GREs (Fig. 2A) shows that they fall into distinct groups that are, for the most part, congruent with the Pfam domain-based taxonomy of the BMC loci (5). The GRM1 GREs include choline trimethylamine-lyase (5, 33, 38), which forms acetaldehyde in a reaction similar to that of the ethanolamine ammonia-lyase of EUT BMCs. The GREs of GRM1 fall into two distinct clades in the phylogenetic analysis (Fig. 2A). The major clade comprises GREs from the Firmicutes, Deltaproteobacteria, and Actinobacteria. The smaller clade is monophyletic and contains representatives only from the family of Peptococcaceae, which also belong to the Firmicutes. Representatives of both phylogenetic clades exhibit well-conserved N-terminal extensions of about 60 amino acids that share features of encapsulation peptides (EPs) and their linkers (see the supplemental material). EPs have been shown to interact with shell proteins in experimentally characterized BMCs (50–52) and may also play a role in the nucleation of encapsulated enzymes (53).
FIG 2.
Phylogenetic trees of GRM-associated enzymes. Maximum likelihood trees are drawn to scale, and branch lengths are based on the number of substitutions per site. Bootstrap values for important nodes are represented as filled circles (above 50%) and empty circles (below 50%). (A) GRE tree generated from 83 amino acid sequences. The sequences used for calibration were those of biochemically characterized GREs not associated with BMCs: vitamin B12-independent glycerol dehydratase (GDH) from Clostridium butyricum, the benzylsuccinate synthase (BSS) from Thauera aromatica, the 4-hydroxyphenylacetate decarboxylase (4HPD) from Clostridium difficile, and pyruvate formate-lyase type 1 (PFL1) from E. coli. PDU-GRM, the locus in Escherichia fergusonii which appears to be a fusion of the PDU BMC and GRM loci; GUF, GREs of unknown function; CHL, choline TMA-lyase. (B) Tree of the activating enzymes inferred from 79 amino acid sequences. The activating enzyme of the pyruvate formate-lyase type 1 from E. coli (PFL1-AE) was used as an outgroup. (C) Cartoon representation of the multiple-sequence alignment of the GREs. Orange segments, extensions/insertions that likely constitute EPs with linkers; blue segment, the N-terminal extension of the GREs of the GRM2 BMCs that appears to be a partial domain duplication. (D) Tree of the acylating aldehyde dehydrogenases (AldDHs) inferred from 111 amino acid sequences. The AldDH homologs from the conventional ethanolamine-utilizing BMC (EutE) and propanediol-utilizing BMC (PduP) from Salmonella enterica were included in the analysis. GRM1 (duds), presumably inactive AldDH homologs; GUF*, AldDHs from D. psychrophila. (E) Tree of PduL-like phosphotransacylase homologs inferred from 74 amino acid sequences. The phosphotransacylase (PduL) from the experimentally characterized PDU BMC of S. enterica was included for comparison. GRM1*, the PduL homologs of Desulfovibrio spp.
A few GREs (from Desulfitobacterium dehalogenans, Desulfitobacterium hafniense DCB-2 and Y51, and Desulfotalea psychrophila) that were previously classified as members of the GRM1 locus family apparently lack EPs. These GREs cluster separately in the phylogenetic tree and appear to be more closely related to nonencapsulated GREs (Fig. 2A); these are designated GREs of unknown function (GUFs). Homology modeling of the GUF structure results in an active-site architecture very different from that of the other types of GRM GREs (see Fig. S1 in the supplemental material). Although the genes encoding the GUFs are located in the vicinity of BMC gene clusters (5), they are oriented in the opposite direction from the BMC loci (except in D. psychrophila). Notably, the genomes of all of the organisms that harbor the GUF loci also contain a second BMC locus with GRE orthologs that cluster with GRM1-type GREs (Table 1). The lack of an EP and the inverted gene orientation indicate that the GUFs are likely not part of a BMC. For more detailed information on the GUF loci, see the supplemental material.
TABLE 1.
Organisms whose genomes encode GRMs and additional BMC loci
| Organism | GRM locus type(s) | Additional BMC locus type(s)a |
|---|---|---|
| Alkaliphilus metalliredigens QYMF | GRM1 | BUF |
| Alkaliphilus oremlandii OhILAs | GRM1 | EUT2, BUF |
| Clostridium botulinum A3 strain Loch Maree | GRM1 | Satellite |
| Clostridium botulinum B strain Eklund 17B | GRM1 | MUF |
| Clostridium botulinum B1 strain Okra | GRM1 | Satellite |
| Clostridium botulinum E3 strain Alaska E43 | GRM1 | MUF |
| Clostridium botulinum F strain Langeland | GRM1 | Satellite |
| Clostridium ljungdahlii DSM 13528 | GRM1, GRM3 | Satellite, satellite-like |
| Clostridium phytofermentans ISDg | GRM1, GRM5 | EUT2 |
| Clostridium saccharolyticum WM1 | GRM1 | EUT2, PVM-like |
| Clostridium tetani E88 | GRM1 | EUT2 |
| Desulfitobacterium dehalogenans ATCC 51507 | GRM1, GUF | |
| Desulfitobacterium hafniense DCB-2 | GRM1, GUF | EUT3 |
| Desulfitobacterium hafniense Y51 | GRM1, GUF | EUT3 |
| Desulfosporosinus meridiei DSM 13257 | GRM1 | EUT2 (2×) |
| Desulfosporosinus orientis DSM 765 | GRM1 | EUT2, EUT other |
| Desulfotalea psychrophila LSv54 | GRM1, GUF | |
| Desulfotomaculum reducens MI-1 | GRM1 | EUT2 |
| Escherichia coli 536 | GRM2 | EUT1 |
| Escherichia coli APEC O1 | GRM3 | EUT1 |
| Escherichia coli CFT073 | GRM3 | EUT1 |
| Escherichia coli ED1a | GRM3 | EUT1 |
| Escherichia coli IAI39 | GRM2 | EUT1 |
| Escherichia coli O7:K1 strain CE10 | GRM2 | EUT1 |
| Escherichia coli S88 | GRM3 | EUT1 |
| Escherichia coli UTI89 | GRM2 | EUT1 |
| Escherichia fergusonii ATCC 35469 | PDU/GRM2-fusion | EUT1 |
| Klebsiella oxytoca E718 | GRM2 | EUT1, PDU1 |
| Klebsiella oxytoca KCTC 1686 | GRM2 | EUT1, PDU1 |
| Klebsiella pneumoniae 342 | GRM2 | EUT1, PDU1 |
| Klebsiella variicola At-22 | GRM2 | EUT1, PDU1 |
| Raoultella ornithinolytica B6 | GRM2, GRM3 | |
| Ruminococcus obeum A2-162 | GRM5 | Satellite, satellite-like |
| Ruminococcus sp. SR1/5 | GRM5 | Satellite, satellite-like |
| Ruminococcus torques L2-14 | GRM5 | Satellite, satellite-like |
The BMC locus nomenclature is that described by Axen et al. (5). BUF, BMC of unknown function; MUF, metabolosome of unknown function; PVM, Planctomycetes and Verrucomicrobia microcompartment.
The GREs of the GRM2 loci likewise cluster as a separate clade (Fig. 2A). A distinctive feature of the GRM2-associated GREs is an ∼350-amino-acid N-terminal extension (Fig. 2C; see also Fig. S2 in the supplemental material) that is the result of an apparent duplication of ∼1 kb of the 5′ end of the original gene. The primary structure of the extension domain is ∼28% identical to the main domain of the GRE. Because the N-terminal extension does not include the glycine or the cysteine residues involved in the radical catalysis, it presumably is not enzymatically active (54). A role for this partial domain in the assembly of the BMC core is discussed below.
The GREs of GRM3, GRM4, and GRM5 form clades that cluster together in the phylogenetic tree (Fig. 2A). There is no evidence of an N- or C-terminal EP for these GREs; instead, they contain a feature distinguishing them from all other GREs: an insertion of about 50 to 60 amino acids between the structural domains (Fig. 2C) that may represent a novel interdomain EP (see the supplemental material). Notably, the non-BMC-associated vitamin B12-independent glycerol dehydratase (GDH) clusters together with the GRM3, GRM4, and GRM5 GREs in the phylogenetic tree (Fig. 2A) and does not have an insertion (Fig. 2C). This supports the hypothesis that the insertion is a specific feature of these BMC-associated GREs, likely functioning as an EP.
The GRE of a GRM5 locus was previously predicted to function as 1,2-propanediol dehydratase (20). Our analysis supports this prediction, and the close phylogenetic relationship among the GREs of the GRM3, GRM4, and GRM5 loci implies that they all have the same function. Furthermore, the canonical (nonencapsulated) GDH is nested within the greater GRM3, GRM4, and GRM5 clade and shares ∼60% amino acid sequence identity with these GREs. The close physicochemical resemblance of their presumed substrate, 1,2-propanediol, to glycerol supports the functioning of these GREs as propanediol dehydratases. Furthermore, homology modeling of their structures shows that they have nearly identical active-site architectures (see Fig. S1A in the supplemental material). There are two conserved substitutions: a serine and a tyrosine residue present in the GDH are replaced in the proposed propanediol dehydratases by a valine and a phenylalanine, respectively. The valine side chain would form a close, unfavorable contact to the one primary hydroxyl group of glycerol (see Fig. S1A in the supplemental material) that is not present in 1,2-propanediol. In addition, both the valine and the phenylalanine would provide a more hydrophobic environment, accommodating the terminal methyl group of 1,2-propanediol. Therefore, the GRM3, GRM4, and GRM5 GREs likely represent vitamin B12-independent alternatives to the propanediol dehydrates of the canonical PDU BMC.
Three GREs that were previously classified as members of GRM3-type loci (5) cluster together with enzymes from the GRM1 type (Clostridium ljungdahlii, Desulfosporosinus meridiei, Desulfosporosinus orientis). These GREs also do not contain the internal putative EP that is found in GRM3 GREs. They do, however, possess the N-terminal EP typical of GRM1 GRE homologs. This is one of the few examples of incongruence between the GRM classification based on the locus composition of the encoded Pfam domains (5) and the phylogenetic relationship among specific constituent proteins. For further information on the reclassified loci, see the supplemental material.
(ii) Glycyl radical AEs.
The phylogenetic tree of the activating enzymes (AEs) associated with the different GRMs is congruent with the GRE phylogeny (Fig. 2A and B). These observations indicate a coevolution of the AEs and GREs; the AEs may be specific in their ability to activate the colocalized GRE.
Comparison of the primary structures of the BMC-associated AEs reveals that all but the GRM5 AEs contain one or two additional [4Fe-4S] cluster motifs, in addition to the catalytically active iron-sulfur cluster. These extra motifs are found within an insertion of ∼60 amino acids downstream of the catalytic cluster (see Fig. S3 in the supplemental material). However, the primary structures of the insertions differ even among AEs of the same GRM type (see Fig. S3 in the supplemental material). The presence of additional Fe-S clusters in some non-BMC-related AEs has been noted previously (27, 55). Recently, it was proposed that these additional Fe-S clusters are involved in the generation of a more long-lived glycyl radical and are not required for electron transfer to the catalytic Fe-S cluster responsible for SAM cleavage (56). None of the BMC-associated AEs appear to contain an EP; they are presumably recruited to the BMC by the interaction with their cognate GREs.
(iii) Fuculose phosphate aldolases and lactaldehyde reductases of the GRM5 loci.
The GRM5 loci are unique in encoding an additional signature enzyme—a class II aldolase—that was proposed to cleave l-fuculose phosphate and l-rhamnulose phosphate into dihydroxyacetone phosphate and lactaldehyde (20). The latter is believed to be reduced to 1,2-propanediol, the substrate for the GRE-type propanediol dehydratase (20). The GRM5 fuculose phosphate aldolase (FPA) possesses a canonical C-terminal EP, implying that the FPA localizes in the interior of the GRM5 BMC. Another gene typically found only in GRM5 loci encodes a putative zinc-containing alcohol dehydrogenase (comprising the Pfam00107 and Pfam08240 domains), in addition to the iron-containing ADH (see Table S2 in the supplemental material). These zinc-containing enzymes perhaps serve as lactaldehyde reductases, a reaction that is required only in the GRM5 BMCs (Fig. 1B). This could also explain why two of the GRM5 loci do not contain the iron-containing ADH required for NAD+ regeneration, a function that would be fulfilled by the lactaldehyde reductase. The inclusion of FPA and the putative lactaldehyde reductase is the major difference between the GRM5 and GRM3 loci (5) and represents the progressive incorporation of the remainder of the pathway-relevant upstream enzymes into the organelle.
Core enzyme analysis supports the GRM phylogeny.
Apart from the signature enzymes, the GREs, and their respective AEs, the analysis of the three common metabolosome core enzymes, namely, aldehyde dehydrogenase, phosphotransacylase, and alcohol dehydrogenase (5), provides more evidence for divergent GRM functions.
(i) Acylating AldDHs.
A phylogenetic tree of the GRM-associated acylating aldehyde dehydrogenases (AldDHs) also exhibits distinct branching patterns (Fig. 2D) that concur with the previous classification of the GRM loci (5). A comparison of their primary structures reveals that AldDHs from GRM1 and GRM2 possess C-terminal EPs. Interestingly, there are two AldDH isoforms encoded by the majority (31 of 34) of GRM1 loci analyzed (Fig. 2D). However, in one of the two paralogs, a catalytically important cysteine residue in the active site is replaced by either a proline or a serine (5), presumably rendering the enzyme inactive (57, 58); these are labeled “duds” in the phylogenetic tree (Fig. 2D). In contrast, the GRM3 to GRM5 loci encode only one AldDH homolog, which lacks the C-terminal EP found in the AldDHs of the GRM1 and GRM2 loci. Instead, they contain N-terminal EPs comprising up to two amphipathic helices (see the supplemental material).
(ii) PTACs.
The PduL (Pfam06130) and PTA_PTB (Pfam01515) homologs are two evolutionarily distinct phosphotransacylases (PTACs) associated with catabolic BMCs (59). All but four of the GRM loci encode a PduL homolog as their core PTAC. One of these four loci, the GUF locus in D. psychrophila, encodes a PTA_PTB-like PTAC; these are typically associated with only a subset of EUT BMC loci (5). PduL homologs contain an N-terminal EP for encapsulation within the BMC (25). Phylogenetically, the PduL homologs from the same locus type tend to group together and form clades (Fig. 2E). However, only PduLs from the GRM2 and GRM3 loci form true monophyletic clades, supporting their classification as distinct locus types. All PTACs of the GRM1 loci except for those of the deltaproteobacterial Desulfovibrio genera also cluster together in one large clade in the phylogenetic tree (Fig. 2E). The branching distance may indicate that the PduL homolog of the deltaproteobacterial Desulfovibrio genera was acquired by a relatively recent horizontal gene transfer event.
(iii) Iron-containing ADHs.
In the metabolosome lumen, iron-containing alcohol dehydrogenases (ADHs; Pfam00465) are believed to be essential for NADH recycling to NAD+ (24), which is a cofactor required for the acylating aldehyde dehydrogenases (Fig. 1). The ADHs are very similar among all types of GRM loci, and only a few loci lack these genes (see Table S2 in the supplemental material). Only the ADHs from the GRM2 loci form a monophyletic clade, possibly because these loci are confined to the gammaproteobacteria (see Fig. S4 and Data Set S1 in the supplemental material). None of the ADHs appear to have EPs; they are likely to be encapsulated through interactions with other lumen proteins (26, 53). For example, PduQ, the ADH of the PDU BMC, physically interacts with the AldDH PduP (23), which contains an EP.
Ancillary enzymes/proteins of GRM loci.
Interestingly, a putative pduS gene can be found in roughly half of the GRM1 loci, a minority of the GRM3 loci, and in all GRM5 loci. PduS is a flavoprotein which acts as a cobalamin reductase in the maturation and repair of the vitamin B12 cofactor of the canonical propanediol dehydratase in PDU BMCs. Since no vitamin B12-dependent enzymes are encoded in GRM loci, the role of the pduS gene product is enigmatic; it may have been co-opted for a new function. We suggest that PduS serves as an electron shuttle (using the flavin cofactor), providing electrons to the AE. Consistent with this hypothesis, pduS genes always co-occur with pduT-like genes, whose products presumably serve as electron conduits across the shell (60). All GRM-associated PduS homologs contain conserved amino acid sequence motifs that have been proposed to be involved in the binding of NADH, flavin mononucleotide, and two [4Fe-4S] clusters (60, 61) and thus appear to be redox active. Notably, an alternative flavoprotein gene is found in 12 out of the 19 GRM3 loci. Interestingly, in all but one of the GRM3 loci, genes encoding the alternative flavoproteins and PduS homologs are mutually exclusive (see Data Set S1 in the supplemental material). Moreover, it was recently noted that flavoproteins are widespread among diverse BMC loci, yet their functions are unknown (5, 26). Other occurrences of genes encoding additional ancillary BMC enzymes and proteins are summarized in the supplemental material.
The shell composition of GRM locus types is consistent with distinctive functions.
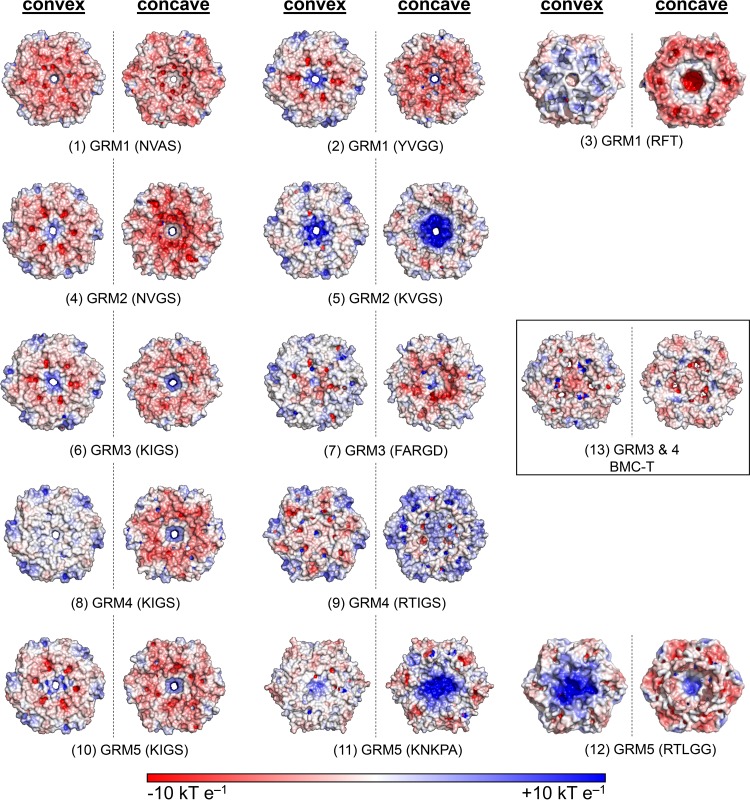
Given the proposed differences in the substrate specificities of their signature enzymes, GRM1 to GRM5 BMCs presumably require different substrates and products to cross their respective shells (Fig. 1; Table 2). These differences should be reflected in the permeability properties of their shell proteins. We compared the shell compositions of the GRM loci by extracting the primary structures of all BMC-H, BMC-T, and BMC-P proteins from each GRM locus, constructed multiple-sequence alignments and phylogenetic trees (see Fig. S5 in the supplemental material), and built homology models.
TABLE 2.
GRM locus shell protein oligomer properties based on homology modelinga
| Locus | Predicted function | Metabolites predicted to cross the shell | Shell protein type | Pore motif residues (diam [Å]) | Pore properties | Model in Fig. 3 | Closest homolog characterizedb (% identity) |
|---|---|---|---|---|---|---|---|
| GRM1 | Choline degradation | Choline, trimethylamine, ethanol, acetyl-phosphate | BMC-H | Y-V-G-G (4.7) | Positive | 2 | PduJ (76) |
| BMC-H | G-X-C-P-Q | Occluded (Fe-S cluster) | NAc | GrpU (>50d) | |||
| BMC-H | R-F-T/S | Occluded, possibly gated negatively charged β-barrel | 3 | PduU (61) | |||
| BMC-H | N-I/V-A/G-S (4.5) | Polar, neutral/negative | 1 | PduA (56) | |||
| BMC-T | S/P-T/V-C-P | Occluded (Fe-S cluster) | NA | PduT (40) | |||
| GRM2 | Choline degradation | Choline, trimethylamine, ethanol, acetyl-phosphate | BMC-H | K-V-G-S (4.5) | Positive | 5 | PduA (51) |
| BMC-H | N-V/I-G-S (4.3) | Polar, neutral/negative | 4 | PduJ (66) | |||
| BMC-T | NA | NA | NA | NA | |||
| GRM3 | Propanediol degradation | Propanediol, propanol, propionyl-phosphate | BMC-H | K-I-G-S (4.3) | Positive | 6 | PduA (85) |
| BMC-H | G-X-C-P-Q | Occluded (Fe-S cluster) | NA | GrpU (>50d) | |||
| BMC-H | L/F-T/A-K/R-G-G/D | Occluded | 7 | None | |||
| BMC-T | G-A/G-G-H (3.7) | Negative (3 pores) | 13 | PduB (67) | |||
| GRM4 | Propanediol degradation | Propanediol, propanol, propionyl-phosphate | BMC-H | K-I-G-S (4.7) | Positive | 8 | PduJ (82) |
| BMC-H | R-T-I-G-S | Occluded | 9 | PduK (49) | |||
| BMC-T | G-A-G-H | Negative (3 pores) | 13 | PduB (77) | |||
| GRM5 | Fuculose- and/or rhamnulose-1-phosphate degradation via propanediol | Fuculose- and/or rhamnulose-1-phosphate, DHAP,e (propanol),f propionyl-phosphate | BMC-H | R/K-N-K-P-A | Occludedg | 11 | CcmK1/2 (35)g |
| BMC-H | K-I-G-S (4.4) | Positive | 10 | PduA (79) | |||
| BMC-H | R-K-L-G-G | Occluded | 12 | PduJ (40) | |||
| BMC-T | T-V-C-P | Occluded (Fe-S cluster) | NA | PduT (40) |
See Data Set S1 in the supplemental material for the complete data set.
The closest homolog characterized by BLAST analysis. The representative shell proteins depicted in Fig. 3 were searched against PDU, EUT, carboxysome, and Planctomycetes and Verrucomicrobia microcompartment shell proteins in Salmonella enterica, Synechococcus elongatus PCC7942, Halothiobacillus neapolitanus, and Planctomyces limnophilus.
NA, not applicable.
The percent identities are to the GrpU orthologs in the Protein Data Bank.
DHAP, dihydroxyacetone phosphate.
Production of propanol is not necessarily required for NAD+ regeneration in GRM5, because of the proposed function of a lactaldehyde reductase.
In the model, the predicted positively charged pores are occluded (Fig. 3, model 11), but note that the best available structural template (CcmK1 from Thermosynechococcus elongatus) is only 35% identical.
Of the 78 GRM loci analyzed, only the GRM1 loci of D. psychrophila and Clostridium tetani appeared to have an incomplete set of shell proteins (see the supplemental material).
(i) BMC-Hs.
BMC-Hs are typically the most abundant component of a BMC shell (see Fig. S5 in the supplemental material), and it is presumed that the residues converging at the pores (pore motifs; Table 2), which have charges complementary to those of the metabolites (6, 62, 63), facilitate the majority of flux. Some general trends among BMC-Hs are apparent in the context of proposed substrates for each GRM locus type (Table 2). All GRMs require small negatively charged molecules (phosphate, acyl-phosphates) to cross the shell (Fig. 1). BMC-H proteins with positively charged pores, which would facilitate the diffusion of these metabolites, are observed in homology models for shell proteins from each GRM type (Fig. 3 and Table 2; see also Data Set S1 in the supplemental material for comparison).
FIG 3.
Electrostatic surface renderings of homology models of shell protein oligomers conserved among GRM loci. Both the concave and convex sides of the model of each shell protein assembly are shown. The GRM type (pore motifs) are indicated. The boxed GRM3 BMC-T serves as an example for shell protein trimers with three pores. Positive surface potentials (blue) and negative surface potentials (red) ranging from 10 to −10 kT/e are indicated, with the Boltzmann's constant (k), temperature (T), and the charge of an electron (e).
Only the GRM1 and GRM2 signature enzymes require a positively charged metabolite, choline, to cross the shell. Indeed, these loci encode one or two shell proteins that model as hexamers with negatively charged pores on their concave side and relatively neutral pores on their convex side (Fig. 3, models 1 and 4). Of all the organisms harboring GRM1 loci, only Desulfovibrio spp. lack this type of BMC-H. However, these loci encode a BMC-T protein that is predicted to form trimers with a pore motif similar to that of the hexamers that are apparently typical of GRM1 and GRM2 loci (see Data Set S1 in the supplemental material).
The only other BMC-H proteins that may form hexamers with negatively charged pores are orthologs of PduU (64) or EutS (65, 66), which are encoded by all GRM1 loci, three GRM3 loci, and one GRM5 locus. Crystal structures have shown that PduU and EutS have an additional N-terminal domain that folds into a β-barrel structure that caps the pore (64, 66). This β-barrel has been suggested to undergo a conformational change that removes the occlusion to permit metabolites to pass (64). Electrostatic surface analysis of the homology models of the GRM counterparts shows that the predicted pore of these hexamers is more or less uncharged and much larger (∼17 Å) than the others (Fig. 3, model 3). However, if the β-barrel is indeed able to undergo a conformational change (e.g., a twisting motion), a narrower (diameter, much less than 17 Å) and negatively charged channel could be exposed. Accordingly, when open, this type of BMC-H may allow the crossing of positively charged molecules.
Finally, GrpU, a BMC-H protein from a GRM3 locus, was recently shown to have within its pore an Fe-S cluster binding site formed by a G-X-C-P-Q motif; this could have implications for electron or Fe-S cluster transport across the shell (67). The majority of GRM1 loci (21 out of 34) and GRM3 loci (12 out of 16) encode a GrpU ortholog, while GRM2, GRM4, and GRM5 loci do not (Table 2; see also Data Set S1 in the supplemental material).
(ii) BMC-Ts.
BMC-T proteins, which form trimeric pseudohexamers, are considered relatively minor components of BMC shell facets. Structures of some BMC-Ts indicate that the residues surrounding the pores formed by the trimers can have alternate conformations to gate the pores (18, 68–71). A subset of BMC-Ts forms a distinct clade, and these pseudohexamers stack and are gated (68, 69). Notably, none of the GRM BMC-Ts are members of this stacking clade. Furthermore, one type of BMC-T protein, PduT of the PDU BMC, has been shown to coordinate a [4Fe-4S] cluster in its pore for electron channeling between the BMC lumen and the cytosol (72, 73). Of the 34 GRM1 loci analyzed, only 18 encode a PduT-like BMC-T with cysteine residues predicted to bind the [4Fe-4S] in the pore of the trimer, as do 4 out of the 16 GRM3 loci and all of the GRM5 loci (Table 2; see also Data Set S1 in the supplemental material).
Although the majority of GRM loci encode at least one BMC-T protein, all GRM2 loci are devoid of genes encoding BMC-T homologs. The same is true for most of the GRM1 loci found in Clostridium and Streptococcus spp. (Table 2; see also Data Set S1 in the supplemental material). Nevertheless, all GRM3 and GRM4 loci, including those that harbor a pduT gene ortholog, encode a BMC-T protein that is phylogenetically most closely related to PduB and EtuB of the PDU and ethanol-utilizing BMCs, respectively (see Fig. S5 in the supplemental material). The corresponding crystal structures were identified to be the best templates for homology modeling of GRM3 and GRM4 BMC-T homologs (Fig. 3, model 13). The models of the GRM3 pseudohexamers contain three weakly negatively charged pores (diameter, ∼3.7 Å) that may be involved in metabolite transport (18, 70). A small number of GRM1 loci (in Desulfovibrio and Desulfotalea spp.) encode yet another type of BMC-T protein (see Data Set S1 in the supplemental material), for which homology modeling also predicts three similar openings in the trimer.
(iii) BMC-Ps.
While they are required to seal icosahedral shells (37), BMC-P proteins are thought to play only a minor role in terms of shell permeability due to their low abundance (12 pentamers) per icosahedron. Accordingly, the amino acids surrounding the pores would presumably not be conserved. However, we found that the GRM BMC-P protein pore residues are generally conserved and are similar to those of carboxysome (63), EUT, and PDU BMC-P pores. The only exception to this is the BMC-P pore sequence of the GRM2 loci, E-Y-F/L-A-L, which is also exclusive to the GRM2 loci (see Data Set S1 in the supplemental material). Homology models show these pores to be occluded by the bulky aromatic residues. These models, like the topology of the BMC-P tree, are consistent with an evolutionary divergence of the GRM2 locus from the other GRM loci (see Fig. S5 in the supplemental material).
DISCUSSION
Prediction of GRM functions.
In contrast to other BMC in which the signature enzyme (e.g., propanediol dehydratase, ethanolamine ammonia-lyase) is sufficient to describe the function of the metabolosome, our analyses indicate that there are at least three different functional types of BMCs with a GRE as the signature enzyme (Fig. 1B). Moreover, these different types of GRE-associated microcompartments appear to differ from other BMCs and from one another in the details of their assembly, evident in the presence of atypical encapsulation peptides and some defunct catalytic domains, as well as distinctive shell protein complements.
All of the GRM1 loci, which encode tightly clustering signature and core enzymes (Fig. 2), likely represent choline-degrading BMCs (Fig. 1B); a GRM1 GRE has recently been experimentally verified to be a choline trimethylamine-lyase (33, 38). In our analysis, only the GRM1 GREs contain a predicted EP in a typical position, an N-terminal sequence extension. Furthermore, only the GRM1 loci encode two isoforms of AldDH, one of which seems to be catalytically defunct. What is the function of such dud enzymes? Perhaps these inactive dehydrogenases fulfill a structural role in the interior organization of the BMC analogous to the role of CcmM in β-carboxysome assembly (74). CcmM contains a carbonic anhydrase domain that is inactive in some strains (75), as well as multiple copies of domains resembling the small subunit of RuBisCO (76) that are involved in scaffolding of the carboxysome core (74). The GRM1 defunct AldDHs, by analogy, may serve a structural role, e.g., in coalescence of the enzymatic core.
While GRM2 GREs are situated phylogenetically near their GRM1 counterparts (Fig. 2A), they contain a unique N-terminal extension that resembles approximately half of the catalytic domain of the GRE. Recently, the crystal structure of a GRM2 GRE was solved; however, due to proteolysis the structure lacks the N-terminal extension domain (54). This extension may be involved in nucleating the core of the GRM2 metabolosome. Other characterized GREs (e.g., pyruvate formate-lyase and the vitamin B12-independent glycerol dehydratase) form homodimers (see Fig. S2A in the supplemental material) (27). On the basis of the homology model (see Fig. S2B in the supplemental material), the N-terminal extension of the GRM2 GREs could displace the second subunit of the homodimer. Multiple displacement events among neighboring GRE subunits could create a bolus of GREs (see Fig. S2C in the supplemental material) in a manner analogous to the coalescence of RuBisCO via RuBisCO small-subunit-domain mimics in β-carboxysome assembly (74). Alternatively, the N-terminal extension domain may fulfill the role of an EP (see Fig. S2D in the supplemental material).
The short branching length and position of the GRM2 GREs close to the GRM1 GREs indicate that these GREs also function as choline TMA-lyases; this was recently confirmed for the GRM2 GRE of Klebsiella pneumoniae (54). Moreover, it was demonstrated that strains harboring GRM2 loci were indeed able to form trimethylamine when choline was provided during growth (77). Amino acid sequence alignments and structural comparisons of both the GRM1 and GRM2 GREs show that their active-site residues are highly conserved. However, there are substantial differences between the complement of protein domains encoded by the GRM1 and GRM2 loci (Fig. 1B). The GRM2 loci do not encode the defunct AldDHs found in the GRM1 loci. The number of genes encoding shell proteins is relatively reduced in the GRM2 loci, whereas most GRM1 loci encode a BMC-T as well as an average of six (and up to eight) BMC-H proteins and all GRM2 loci harbor only 4 genes encoding BMC-H proteins and no genes for BMC-T proteins (Fig. 1B; see also Data Set S1 in the supplemental material). Moreover, the GRM1 and GRM2 loci differ in the type of BMC-Hs with positively charged pores. The GRM1 loci encode predominantly BMC-H proteins with L/F/Y-V-G-G pores, whereas the GRM2 loci encode BMC-Hs with K-V/I-G-S-type pores (Fig. 3 and Table 2; see also Data Set S1 in the supplemental material). All but one (C. ljungdahlii) of the GRM1 loci additionally encode a PduU-like BMC-H protein that forms hexamers with a negatively charged β-barrel that caps a relatively wide (∼17-Å) neutral pore (64). In summary, the differences between the GRM1 and GRM2 loci hint at different shell architectures and a different interior organization.
For the GRM3, GRM4, and GRM5 BMCs, our data indicate that their respective GREs function as 1,2-propanediol dehydratases. This was proposed earlier for a clostridial GRM locus (20), which in our classification is a GRM5. The clustering of a characterized vitamin B12-independent glycerol dehydratase (31) in the phylogenetic tree (Fig. 2A), forming a clade with the GREs of types GRM3, GRM4, and GRM5, corroborates this hypothesis. This type of glycerol dehydratase can also use 1,2-propanediol as the substrate (78); therefore, it is likely that the GRM3, GRM4, and GRM5 BMCs function similarly to the canonical PDU BMCs. Notably, only the vitamin B12-dependent signature enzyme has been replaced by the GRE; the three metabolosome core enzymes are similar to those of the canonical vitamin B12-dependent PDU BMC (Fig. 1B). The major differences among the GRM3, GRM4, and GRM5 loci are in the types of ancillary genes and the shell composition.
The GRM3 and GRM4 loci, but not the GRM5 loci, encode a BMC-T that, on the basis of homology modeling, contains three weakly negatively charged pores per trimer (Fig. 3 and Table 2; see also Data Set S1 in the supplemental material). In the template crystal structure (PduB from the PDU BMC), these pores contain glycerol molecules (70). Glycerol is physiochemically similar to 1,2-propanediol, the substrate of the PDU BMC and, according to our analysis, the GRM3 and GRM4 BMCs, indicating that this shell protein may form the conduit for 1,2-propanediol into these BMCs. In contrast, the GRM5 BMCs, which are also predicted to metabolize 1,2-propanediol, have only one shell protein predicted to form hexamers containing an open pore, a BMC-H with the K-I-G-S pore motif, which is also found in other GRM locus types (Fig. 3 and Table 2; see also Data Set S1 in the supplemental material). This implies that only negatively charged metabolites are able to cross the GRM5 shell. This is consistent with the hypothesis that fuculose phosphate and not propanediol is the substrate that must enter the GRM5 BMC. This is supported by the observation of an EP on the C terminus of the fuculose phosphate aldolase encoded in the GRM5 loci, indicating that it is encapsulated in the BMC. The phosphorylated cleavage product, dihydroxyacetone phosphate, could exit via the same pore while the other uncharged cleavage product, lactaldehyde, remains within the BMC and is further metabolized to propionyl phosphate (Fig. 1B). The latter metabolite could diffuse out of the shell through the K-I-G-S-type pores of the hexamers. If this proposed reaction sequence is correct, there would be no requirement for any uncharged metabolites (e.g., propanol) to cross the shell; furthermore, if lactaldehyde is converted to propanediol within the BMC, the NAD+ required for the AldDH is provided by that reaction (Fig. 1B). Therefore, the action of a propanol dehydrogenase would not be required. This could also account for why some GRM5 loci do not encode an ADH (see Table S2 in the supplemental material); the action of a propanol dehydrogenase could be detrimental to the redox balance within the GRM5 BMCs, if the resulting propanol diffused out of the compartment. The trapping of propanol within the BMC lumen would prevent the imbalance of the NADH/NAD+ recycling, as the buildup of propanol would result in ADH catalyzing the reverse reaction to propionaldehyde, regenerating the NADH that lactaldehyde reductase consumes (Fig. 1B). The lack of pores that allow diffusion of uncharged metabolites among the BMC-Hs of the GRM5 shell is consistent with this scenario. Furthermore, the encapsulation of the lactaldehyde reductase would be advantageous for organisms that are dependent on deriving energy (i.e., ATP) from fuculose phosphate degradation via the GRMs. All of the propionaldehyde produced can be channeled toward the ATP generated from propionyl phosphate (Fig. 1B). This may also enhance the metabolic flux through the BMC and thus render the utilization of fucose more (energy) efficient.
How larger cofactors cross the EUT and PDU BMC shells remains enigmatic, as does how SAM is supplied to the AEs within GRMs. Genes encoding SAM synthetases are found in only a few GRM3 loci (7). SAM could be synthesized in the cytosol and then diffuse into the BMC. Alternatively, if SAM is regenerated within the microcompartment lumen, as predicted for NAD+ (23, 24) and the vitamin B12 cofactor (61), its precursor, ATP, would be required to cross the shell, but ATP is a comparably large cofactor, so the question remains. It is possible that the BMC shell oligomers which we predict have occluded pores are gated and have conformational states that allow the passing of larger molecules. Notably, the PDU and EUT BMCs also require ATP to cross the shell in order to mature and regenerate the vitamin B12 cofactor (26). Furthermore, some carboxysomes are predicted to encapsulate a RuBisCO activase, which would require ATP as well (26, 79). To date, there are no experimental data demonstrating ATP entry into a BMC. This underscores the currently incomplete understanding of the properties of BMC shells.
Electron transport across the shell, which is required for the activation of the GREs by the AEs, is likewise not understood. The occurrence of shell proteins that are predicted to form assemblies with iron-sulfur clusters in the pores (PduT-like BMC-Ts or a GrpU-like BMC-H) offers one possibility (67, 72). The majority of GRM1, GRM3, and GRM5 loci encode at least one shell protein homolog potentially coordinating an Fe-S cluster in its predicted oligomeric state (Table 2; see also Data Set S1 in the supplemental material). However, these homologs are not found in GRM2 and GRM4 loci, indicating that these BMCs may not require electrons to cross the shell or that electron transfer proceeds via a different mechanism. If and how larger metabolites, oxygen, and electrons cross the shells of GRMs and other BMCs are fundamental questions to be answered about BMC function and for the engineering of BMCs for diverse applications in biotechnology.
Evolution of GRE-associated BMCs.
We have previously proposed that BMC loci are constructed of interchangeable modules, where components (e.g. signature enzymes) can be swapped among loci with minimal detriment to BMC function (26). The GRM loci provide naturally occurring examples of this kind of exchange. The composition of the GRM3 and GRM4 BMCs underscores that this occurs in nature, as they are the vitamin B12-independent counterparts of the PDU BMCs; only the signature, aldehyde-generating enzyme differs (Fig. 1B). The contents of the GRM5 loci also closely resemble those of the canonical PDU BCMs; in addition to the change in the signature enzyme, two upstream enzymes are included: fuculose phosphate aldolase and (a putative) lactaldehyde reductase (Fig. 1B). Likewise, the GRM1 (and, presumably, GRM2) BMCs resemble canonical EUT BMCs in function; only the acetaldehyde-generating signature enzyme and, concomitantly, its substrate (choline for ethanolamine) are replaced (33).
These observations raise an interesting evolutionary question: which signature enzyme, the GRE or the vitamin B12-dependent one, first came to be associated with a BMC? The GRE family of enzymes is presumably older than the vitamin B12-dependent enzymes, as the GREs utilize the comparatively much simpler amino acid radicals instead of the relatively elaborate vitamin B12 cofactor, which requires a rather complicated biosynthesis pathway (80, 81). However, we favor a model in which the vitamin B12-dependent BMCs arose first and later a GRE was substituted for the B12-dependent enzyme. Consistent with this hypothesis, many of the GRM loci encode the cobalamin reductase PduS (61, 82) but lack genes encoding any cobalamin-containing proteins/enzymes. Furthermore, PduS co-occurs only with its BMC-T interaction partner PduT (60). The latter was shown to coordinate an Fe-S cluster within its pore (72), which presumably conducts electron transfer from the cytosol to PduS in the lumen of the BMC. Because the enzymatic function of PduS is not required in GRMs, the electron transfer from PduT would presumably not be needed. However, these two genes are retained in a subset of GRM loci, where their products may serve structural roles or have been co-opted for a new function, such as transfer of electrons to the AEs.
Interestingly, all of the GRM-associated ADHs appear to be more closely related to the 1-propanol dehydrogenase PduQ of the PDU BMC than to the ethanol dehydrogenase EutG of the EUT BMC (see Fig. S4 in the supplemental material). Likewise, of the two distinct BMC-associated PTACs, the housekeeping PTA_PTB and PduL, only the latter is encoded by almost all of the GRM loci surveyed here. Collectively, these observations imply that the different GRMs are descended from PDU BMCs. However, some GRM loci also contain genes that appear to have originated from the EUT BMC. For example, an EutJ homolog is encoded by the majority of the GRM1 and GRM3 loci. There is no biochemical evidence for a specific role for EutJ, but its chaperonin-like nucleotide-binding domains hint at possible chaperone or adenosylcobalamin reactivation functions (83). Overall, the presence of genes homologous to components of both EUT and PDU BMC loci in GRM loci is also consistent with a mix-and-match evolution of the GRM loci by multiple gene duplication and gene transfer events. The latter is evident in a chimeric GRM1-GRM3 locus found in C. ljungdahlii and a PDU-GRM2 fusion locus in Escherichia fergusonii, which are described in the supplemental material.
Links between GRMs and environmental niche specialization.
The substrate of the GRM3 BMC, 1,2-propanediol, is a fermentation product of the common plant sugars fucose and rhamnose (84). Fucose is also found in glycans present in the intestinal tract (85), and these glycans are important for host-symbiont interactions (86). Fucosylated glycans can be found in other epithelial tissues as well (87), and the ability to use fucose as a carbon source is known to confer a competitive advantage to pathogenic bacteria (88). Accordingly, the GRM3 BMC may represent an energy-efficient strategy for the anaerobic utilization of propanediol derived from fucose and/or rhamnose fermentation, thus allowing the strain to be more successful during infection. Interestingly, some strains whose genomes encode the GRM3 locus are potentially able to aerobically degrade fucose and rhamnose by utilizing a fuculose phosphate aldolase that is ∼30% identical to the one encoded by GRM5 loci. The aerobic degradation does not proceed via propanediol (89), but it is conceivable that under anoxic conditions the same aldolase is used and propanediol is produced.
In anoxic marine and freshwater sediments, the decay of plant material produces rhamnose and fucose. Resident species, such as Rhodopseudomonas palustris and Rhodospirillum rubrum, likely utilize GRM3 for the catabolism of propanediol derived from these sugars. However, these species do not appear to encode a fuculose phosphate aldolase and, thus, could be dependent on other organisms providing either propanediol or lactaldehyde. The same is probably true for Shewanella species whose genomes encode the GRM4 loci; these are also facultative anaerobes isolated from marine and terrestrial environments (90).
On the other hand, the organisms whose genomes encode GRM5 loci are all obligate anaerobes. Ruminococcus spp. as well as Roseburia inulinivorans were isolated from human feces (91, 92), whereas Clostridium phytofermentans was found in forest soil (93). A simple explanation as to why only the GRM5 loci include the fuculose phosphate aldolase may be the strictly anaerobic nature of the GRM5-associated organisms. Most species whose genomes encode GRM3 and GRM4 loci are facultative anaerobes and would require the GRM only under anoxic conditions. Therefore, it would be ideal to regulate the expression of the fuculose phosphate aldolase independently from the expression of GRMs, as under oxic conditions fucose degradation proceeds through a different pathway that does not involve propanediol as an intermediate. Correspondingly, the GRM5 BMC in obligate anaerobes needs to be coexpressed with the aldolase; including its gene in a BMC locus/operon or polycistron would ensure cotranscription.
The phylogenetic distribution of the GRM1 and GRM2 loci hints at links to pathogenesis; GRM1 loci can be found in pathogenic Clostridium and Streptococcus species. Likewise, all species that encode GRM2 loci are known pathogens, mostly of the Enterobacteriaceae family. Furthermore, the GRM2 locus of the uropathogenic strain Escherichia coli 536 is located in a pathogenicity island (94). The ability to efficiently degrade choline would likely provide a nutritional advantage that could boost infectivity, as is known for other pathogens that metabolize propanediol or ethanolamine via the PDU and EUT BMCs (95–99). Interestingly, the genomes of many of these pathogenic organisms encode GRMs as well as other BMC types (Table 1). This indicates that these organisms may be able to switch between the use of compartments, depending on either the supply of the respective substrate (choline, ethanolamine, propanediol) or the availability of vitamin B12. Given that the vitamin B12 cofactor has to be repaired or regenerated frequently (31, 100) and the vitamin B12-dependent glycerol/diol dehydratases as well as the ethanolamine ammonia-lyases are prone to quick inactivation during the turnover of their respective substrates, the GRE signature enzyme and the GRM BMCs may provide a relatively parsimonious alternative to their EUT and PDU BMC counterparts. Moreover, the de novo biosynthesis of vitamin B12 is biochemically costly (80) and requires cobalt, which can be limiting. Both the biosynthesis and the repair of the vitamin B12 cofactor consume a substantial amount of ATP. In contrast, GREs circumvent these limitations; the synthesis of SAM as the substrate for the activation of the GREs requires much less energy because it is formed from methionine and only one ATP molecule. Moreover, GREs do not require constant repair or reactivation. Thus, a selective advantage for bacteria able to form GRMs would likely be in anoxic environments, where ATP is scarce, because the de novo synthesis of vitamin B12 would be too costly if environmental vitamin B12 sources are limiting.
The by-products of GRM-related metabolism may also have implications for human health. The catabolism of choline via GRMs results in the products trimethylamine (TMA) and acetaldehyde (Fig. 1B). In humans, bacteria in the gut are the sole source of TMA (101, 102), which is absorbed by the intestine and oxidized in the liver and kidneys to TMA oxide (TMAO) (103). TMAO has been linked to atherosclerosis and is one of the primary markers for this disease (102). The choline TMA-lyase of the GRM1 and GRM2 BMCs (33, 38, 54) is one of only two enzymes known to generate TMA, indicating that these BMCs may be a significant source of TMA in the human gut (77).
Conclusions.
Based on the in-depth bioinformatics analysis of this major, yet understudied, family of BMCs, we suggest that they play important roles in the environment and in pathogenesis. Because the GREs constitute a very diverse family of enzymes with various substrate and reaction specificities, they confer these distinct functions as the signature enzymes of microcompartments as either choline TMA-lyases or diol dehydratases. Our analysis also has implications for the biotechnological modification and design of BMCs, as not only the number and types of BMC constituents differ among the GRMs, but also new variants of putative encapsulation peptides as well as predicted novel domain interactions in assembly are apparent among the GRMs. Experimental studies are in order and under way to test our predictions about the biological function and the modes of assembly of the GRMs.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the U.S. Department of Energy, Basic Energy Sciences (DE-FG02-91ER20021), and the National Institutes of Health, NIAID (1R01AI114975-01).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02587-15.
REFERENCES
- 1.Lower BH, Bazylinski DA. 2013. The bacterial magnetosome: a unique prokaryotic organelle. J Mol Microbiol Biotechnol 23:63–80. doi: 10.1159/000346543. [DOI] [PubMed] [Google Scholar]
- 2.Gorby YA, Beveridge TJ, Blakemore RP. 1988. Characterization of the bacterial magnetosome membrane. J Bacteriol 170:834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerfeld CA, Heinhorst S, Cannon GC. 2010. Bacterial microcompartments. Annu Rev Microbiol 64:391–408. doi: 10.1146/annurev.micro.112408.134211. [DOI] [PubMed] [Google Scholar]
- 4.Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. 2014. Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 78:438–468. doi: 10.1128/MMBR.00009-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axen SD, Erbilgin O, Kerfeld CA. 2014. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 10:e1003898. doi: 10.1371/journal.pcbi.1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936–938. doi: 10.1126/science.1113397. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083–1086. doi: 10.1126/science.1151458. [DOI] [PubMed] [Google Scholar]
- 8.Sinha S, Cheng S, Sung YW, McNamara DE, Sawaya MR, Yeates TO, Bobik TA. 2014. Alanine scanning mutagenesis identifies an asparagine-arginine-lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol 426:2328–2345. doi: 10.1016/j.jmb.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA. 2015. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc Natl Acad Sci U S A 112:2990–2995. doi: 10.1073/pnas.1423672112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. 2015. Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol 4:444–453. doi: 10.1021/sb500226j. [DOI] [PubMed] [Google Scholar]
- 11.Shively JM, Ball F, Brown DH, Saunders RE. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584–586. doi: 10.1126/science.182.4112.584. [DOI] [PubMed] [Google Scholar]
- 12.Codd GA, Stewart WD. 1976. Polyhedral bodies and ribulose 1,5-diphosphate carboxylase of the blue-green alga Anabaena cylindrica. Planta 130:323–326. doi: 10.1007/BF00387840. [DOI] [PubMed] [Google Scholar]
- 13.Cannon GC, Heinhorst S, Kerfeld CA. 2010. Carboxysomal carbonic anhydrases: structure and role in microbial CO2 fixation. Biochim Biophys Acta 1804:382–392. doi: 10.1016/j.bbapap.2009.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Price GD, Coleman JR, Badger MR. 1992. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol 100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol 181:5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol 181:5317–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl Environ Microbiol 67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, Pickersgill RW. 2009. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem J 423:199–207. doi: 10.1042/BJ20090780. [DOI] [PubMed] [Google Scholar]
- 19.Seedorf H, Fricke WF, Veith B, Bruggemann H, Liesegang H, Strittmatter A, Miethke M, Buckel W, Hinderberger J, Li F, Hagemeier C, Thauer RK, Gottschalk G. 2008. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A 105:2128–2133. doi: 10.1073/pnas.0711093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, Schnell DJ, Leschine SB, Blanchard JL. 2013. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8:e54337. doi: 10.1371/journal.pone.0054337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erbilgin O, McDonald KL, Kerfeld CA. 2014. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 80:2193–2205. doi: 10.1128/AEM.03887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinsmade SR, Paldon T, Escalante-Semerena JC. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol 187:8039–8046. doi: 10.1128/JB.187.23.8039-8046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng S, Fan C, Sinha S, Bobik TA. 2012. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 7:e47144. doi: 10.1371/journal.pone.0047144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huseby DL, Roth JR. 2013. Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J Bacteriol 195:2864–2879. doi: 10.1128/JB.02179-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Jorda J, Yeates TO, Bobik TA. 2015. The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. J Bacteriol 197:2392–2399. doi: 10.1128/JB.00056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerfeld CA, Erbilgin O. 2015. Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol 23:22–34. doi: 10.1016/j.tim.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Selmer T, Pierik AJ, Heider J. 2005. New glycyl radical enzymes catalysing key metabolic steps in anaerobic bacteria. Biol Chem 386:981–988. [DOI] [PubMed] [Google Scholar]
- 28.Sawers G, Watson G. 1998. A glycyl radical solution: oxygen-dependent interconversion of pyruvate formate-lyase. Mol Microbiol 29:945–954. doi: 10.1046/j.1365-2958.1998.00941.x. [DOI] [PubMed] [Google Scholar]
- 29.Fontecave M. 1998. Ribonucleotide reductases and radical reactions. Cell Mol Life Sci 54:684–695. doi: 10.1007/s000180050195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leuthner B, Leutwein C, Schulz H, Horth P, Haehnel W, Schiltz E, Schagger H, Heider J. 1998. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol 28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 31.O'Brien JR, Raynaud C, Croux C, Girbal L, Soucaille P, Lanzilotta WN. 2004. Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochemistry 43:4635–4645. doi: 10.1021/bi035930k. [DOI] [PubMed] [Google Scholar]
- 32.Selmer T, Andrei PI. 2001. p-Hydroxyphenylacetate decarboxylase from Clostridium difficile. A novel glycyl radical enzyme catalysing the formation of p-cresol. Eur J Biochem 268:1363–1372. [DOI] [PubMed] [Google Scholar]
- 33.Craciun S, Balskus EP. 2012. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci U S A 109:21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frey M, Rothe M, Wagner AF, Knappe J. 1994. Adenosylmethionine-dependent synthesis of the glycyl radical in pyruvate formate-lyase by abstraction of the glycine C-2 pro-S hydrogen atom. Studies of [2H]glycine-substituted enzyme and peptides homologous to the glycine 734 site. J Biol Chem 269:12432–12437. [PubMed] [Google Scholar]
- 36.Wagner AF, Frey M, Neugebauer FA, Schafer W, Knappe J. 1992. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci U S A 89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S. 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4:e7521. doi: 10.1371/journal.pone.0007521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craciun S, Marks JA, Balskus EP. 2014. Characterization of choline trimethylamine-lyase expands the chemistry of glycyl radical enzymes. ACS Chem Biol 9:1408–1413. doi: 10.1021/cb500113p. [DOI] [PubMed] [Google Scholar]
- 39.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 44.Kallberg M, Wang HP, Wang S, Peng J, Wang ZY, Lu H, Xu JB. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Benkert P, Kunzli M, Schwede T. 2009. QMEAN server for protein model quality estimation. Nucleic Acids Res 37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, Rowe ML, Mulvihill DP, Prentice MB, Howard MJ, Warren MJ. 2014. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 3:454–465. doi: 10.1021/sb4001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinney JN, Salmeen A, Cai F, Kerfeld CA. 2012. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem 287:17729–17736. doi: 10.1074/jbc.M112.355305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan C, Cheng S, Sinha S, Bobik TA. 2012. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci U S A 109:14995–15000. doi: 10.1073/pnas.1207516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aussignargues C, Paasch BC, Gonzalez-Esquer R, Erbilgin O, Kerfeld CA. 2015. Bacterial microcompartment assembly: the key role of encapsulation peptides. Commun Integr Biol 8:e1039755. doi: 10.1080/19420889.2015.1039755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalnins G, Kuka J, Grinberga S, Makrecka-Kuka M, Liepinsh E, Dambrova M, Tars K. 2015. Structure and function of CutC choline lyase from human microbiota bacterium Klebsiella pneumoniae. J Biol Chem 290:21732–21740. doi: 10.1074/jbc.M115.670471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lanz ND, Booker SJ. 2012. Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes. Biochim Biophys Acta 1824:1196–1212. doi: 10.1016/j.bbapap.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 56.Selvaraj B, Pierik AJ, Bill E, Martins BM. 2014. The ferredoxin-like domain of the activating enzyme is required for generating a lasting glycyl radical in 4-hydroxyphenylacetate decarboxylase. J Biol Inorg Chem 19:1317–1326. doi: 10.1007/s00775-014-1189-3. [DOI] [PubMed] [Google Scholar]
- 57.Cobessi D, Tête-Favier F, Marchal S, Branlant G, Aubry A. 2000. Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J Mol Biol 300:141–152. doi: 10.1006/jmbi.2000.3824. [DOI] [PubMed] [Google Scholar]
- 58.Farres J, Wang TT, Cunningham SJ, Weiner H. 1995. Investigation of the active site cysteine residue of rat liver mitochondrial aldehyde dehydrogenase by site-directed mutagenesis. Biochemistry 34:2592–2598. doi: 10.1021/bi00008a025. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Leal NA, Sampson EM, Johnson CL, Havemann GD, Bobik TA. 2007. PduL is an evolutionarily distinct phosphotransacylase involved in B12-dependent 1,2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2. J Bacteriol 189:1589–1596. doi: 10.1128/JB.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parsons JB, Lawrence AD, McLean KJ, Munro AW, Rigby SE, Warren MJ. 2010. Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLoS One 5:e14009. doi: 10.1371/journal.pone.0014009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng S, Bobik TA. 2010. Characterization of the PduS cobalamin reductase of Salmonella enterica and its role in the Pdu microcompartment. J Bacteriol 192:5071–5080. doi: 10.1128/JB.00575-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeates TO, Crowley CS, Tanaka S. 2010. Bacterial microcompartment organelles: protein shell structure and evolution. Annu Rev Biophys 39:185–205. doi: 10.1146/annurev.biophys.093008.131418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kinney JN, Axen SD, Kerfeld CA. 2011. Comparative analysis of carboxysome shell proteins. Photosynth Res 109:21–32. doi: 10.1007/s11120-011-9624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crowley CS, Sawaya MR, Bobik TA, Yeates TO. 2008. Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure 16:1324–1332. doi: 10.1016/j.str.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka S, Sawaya MR, Yeates TO. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81–84. doi: 10.1126/science.1179513. [DOI] [PubMed] [Google Scholar]
- 66.Pitts AC, Tuck LR, Faulds-Pain A, Lewis RJ, Marles-Wright J. 2012. Structural insight into the Clostridium difficile ethanolamine utilisation microcompartment. PLoS One 7:e48360. doi: 10.1371/journal.pone.0048360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson MC, Wheatley NM, Jorda J, Sawaya MR, Gidaniyan SD, Ahmed H, Yang Z, McCarty KN, Whitelegge JP, Yeates TO. 2014. Identification of a unique Fe-S cluster binding site in a glycyl-radical type microcompartment shell protein. J Mol Biol 426:3287–3304. doi: 10.1016/j.jmb.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. 2013. The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem 288:16055–16063. doi: 10.1074/jbc.M113.456897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol 392:319–333. doi: 10.1016/j.jmb.2009.03.056. [DOI] [PubMed] [Google Scholar]
- 70.Pang A, Liang M, Prentice MB, Pickersgill RW. 2012. Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr 68:1642–1652. doi: 10.1107/S0907444912039315. [DOI] [PubMed] [Google Scholar]
- 71.Takenoya M, Nikolakakis K, Sagermann M. 2010. Crystallographic insights into the pore structures and mechanisms of the EutL and EutM shell proteins of the ethanolamine-utilizing microcompartment of Escherichia coli. J Bacteriol 192:6056–6063. doi: 10.1128/JB.00652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pang A, Warren MJ, Pickersgill RW. 2011. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallogr D Biol Crystallogr 67:91–96. doi: 10.1107/S0907444910050201. [DOI] [PubMed] [Google Scholar]
- 73.Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, Frank S, Smales CM, Lunsdorf H, Rambach A, Gass MH, Bleloch A, McClean KJ, Munro AW, Rigby SE, Warren MJ, Prentice MB. 2008. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem 283:14366–14375. doi: 10.1074/jbc.M709214200. [DOI] [PubMed] [Google Scholar]
- 74.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. 2013. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155:1131–1140. doi: 10.1016/j.cell.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 75.Cot SSW, So AKC, Espie GS. 2008. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol 190:936–945. doi: 10.1128/JB.01283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Price GD, Howitt SM, Harrison K, Badger MR. 1993. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J Bacteriol 175:2871–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinez-del Campo A, Bodea S, Hamer HA, Marks JA, Haiser HJ, Turnbaugh PJ, Balskus EP. 2015. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio 6:e00042-15. doi: 10.1128/mBio.00042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Raynaud C, Sarcabal P, Meynial-Salles I, Croux C, Soucaille P. 2003. Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc Natl Acad Sci U S A 100:5010–5015. doi: 10.1073/pnas.0734105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. 2013. Cyanobacterial-based approaches to improving photosynthesis in plants. J Exp Bot 64:787–798. doi: 10.1093/jxb/ers294. [DOI] [PubMed] [Google Scholar]
- 80.Warren MJ, Raux E, Schubert HL, Escalante-Semerena JC. 2002. The biosynthesis of adenosylcobalamin (vitamin B12). Nat Prod Rep 19:390–412. doi: 10.1039/b108967f. [DOI] [PubMed] [Google Scholar]
- 81.Martens JH, Barg H, Warren MJ, Jahn D. 2002. Microbial production of vitamin B12. Appl Microbiol Biotechnol 58:275–285. doi: 10.1007/s00253-001-0902-7. [DOI] [PubMed] [Google Scholar]
- 82.Sampson EM, Johnson CL, Bobik TA. 2005. Biochemical evidence that the pduS gene encodes a bifunctional cobalamin reductase. Microbiology 151:1169–1177. doi: 10.1099/mic.0.27755-0. [DOI] [PubMed] [Google Scholar]
- 83.Stojiljkovic I, Baumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177:1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Obradors N, Badia J, Baldoma L, Aguilar J. 1988. Anaerobic metabolism of the l-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J Bacteriol 170:2159–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robbe C, Capon C, Coddeville B, Michalski JC. 2004. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J 384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.El Aidy S, Van den Abbeele P, Van de Wiele T, Louis P, Kleerebezem M. 2013. Intestinal colonization: how key microbial players become established in this dynamic process: microbial metabolic activities and the interplay between the host and microbes. Bioessays 35:913–923. doi: 10.1002/bies.201300073. [DOI] [PubMed] [Google Scholar]
- 87.Becker DJ, Lowe JB. 2003. Fucose: biosynthesis and biological function in mammals. Glycobiology 13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 88.Stahl M, Friis LM, Nothaft H, Liu X, Li JJ, Szymanski CM, Stintzi A. 2011. l-Fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc Natl Acad Sci U S A 108:7194–7199. doi: 10.1073/pnas.1014125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baldomà L, Aguilar J. 1988. Metabolism of l-fucose and l-rhamnose in Escherichia coli: aerobic-anaerobic regulation of l-lactaldehyde dissimilation. J Bacteriol 170:416–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fredrickson JK, Zachara JM, Kennedy DW, Dong HL, Onstott TC, Hinman NW, Li SM. 1998. Biogenic iron mineralization accompanying the dissimilatory reduction of hydrous ferric oxide by a groundwater bacterium. Geochim Cosmochim Acta 62:3239–3257. doi: 10.1016/S0016-7037(98)00243-9. [DOI] [Google Scholar]
- 91.Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, Flint HJ. 2006. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol 56:2437–2441. doi: 10.1099/ijs.0.64098-0. [DOI] [PubMed] [Google Scholar]
- 92.Moore WEC, Johnson JL, Holdeman LV. 1976. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int J Syst Bacteriol 26:238–252. doi: 10.1099/00207713-26-2-238. [DOI] [Google Scholar]
- 93.Warnick TA, Methe BA, Leschine SB. 2002. Clostridium phytofermentans sp. nov., a cellulolytic mesophile from forest soil. Int J Syst Evol Microbiol 52:1155–1160. doi: 10.1099/00207713-52-4-1155. [DOI] [PubMed] [Google Scholar]
- 94.Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Oschlager T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hackert J, Dobrindt U. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A 103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Klumpp J, Fuchs TM. 2007. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 153:1207–1220. doi: 10.1099/mic.0.2006/004747-0. [DOI] [PubMed] [Google Scholar]
- 97.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kendall MM, Gruber CC, Parker CT, Sperandio V. 2012. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3:e00050-12. doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, Berchieri A Jr, Young J, Bumstead N, Barrow P. 2011. Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun 79:4105–4121. doi: 10.1128/IAI.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Toraya T. 2014. Cobalamin-dependent dehydratases and a deaminase: radical catalysis and reactivating chaperones. Arch Biochem Biophys 544:40–57. doi: 10.1016/j.abb.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 101.Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Treacy EP, Akerman BR, Chow LM, Youil R, Bibeau C, Lin J, Bruce AG, Knight M, Danks DM, Cashman JR, Forrest SM. 1998. Mutations of the flavin-containing monooxygenase gene (FMO3) cause trimethylaminuria, a defect in detoxication. Hum Mol Genet 7:839–845. doi: 10.1093/hmg/7.5.839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.