Abstract
Core binding factor beta (Cbfβ) is essential for embryonic bone morphogenesis. Yet, the mechanisms by which Cbfβ regulates chondrocyte proliferation and differentiation as well as postnatal cartilage and bone formation remain unclear. Hence, using the paired-related homeobox transcription factor 1-Cre (Prx1-Cre) mice, mesenchymal stem cell-specific Cbfβ-deficient (Cbfβf/f Prx1-Cre) mice were generated to study the role of Cbfβ in postnatal cartilage and bone development. These mutant mice survived to adulthood but exhibited severe sternum and limb malformations. Sternum ossification was largely delayed in the Cbfβf/fPrx1-Cre mice and the xiphoid process was non-calcified and enlarged. In newborn and 7-day-old Cbfβf/fPrx1-Cre mice, the resting zone was dramatically elongated, the proliferation zone and hypertrophic zone of the growth plates were drastically shortened and disorganized, and trabecular bone formation was reduced. Moreover, in one-month-old Cbfβf/fPrx1-Cre mice, the growth plates were severely deformed and trabecular bone was almost absent. In addition, Cbfβ deficiency impaired intramembranous bone formation both in vivo and in vitro. Interestingly, while the expression of Indian hedgehog (Ihh) was largely reduced, the expression of Parathyroid hormone-related protein (PTHrP) receptor (PPR) was dramatically increased in the Cbfβf/fPrx1-Cre growth plate, indicating that that Cbfβ deficiency disrupted the Ihh-PTHrP negative regulatory loop. Chromatin immunoprecipitation (ChIP) analysis and promoter luciferase assay demonstrated that the Runx/Cbfβ complex binds putative Runx-binding sites of the Ihh promoter regions, and also Runx/Cbfβ complex directly up-regulates Ihh expression at the transcriptional level. Consistently, the expressions of Ihh target genes, including CyclinD1, Ptc and Pthlh, were down-regulated in Cbfβ-deficient chondrocytes. Taken together, our study reveals not only that Cbfβ is essential for chondrocyte proliferation and differentiation for the growth and maintenance of the skeleton in postnatal mice, but also that it functions in up-regulating Ihh expression to promoter chondrocyte proliferation and osteoblast differentiation and inhibiting PPR expression to enhance chondrocyte differentiation.
Keywords: genetic animal models, signaling pathways, development, osteoblasts, growth plate, Indian hedgehog
Introduction
Core binding factors (CBFs) are heterodimeric transcription factors composed of two subunits: the core binding factor alpha (CBFα) and core binding factor beta (CBFβ) (1). CBFα subunits are encoded by three gene, namely, Runt-related transcription factor 1 (Runx1) (Cbfα2), Runx2 (Cbfα1) and Runx3 (Cbfα3) (2), each of which plays important roles in skeletal development. Runx2 is a key transcription factor associated with osteoblast differentiation and chondrocyte hypertrophy (3), Runx3 is indispensable for endochondral ossification if the Runx2 gene dosage is reduced (4), and Runx1 cooperates with Runx2 to regulate sternal morphogenesis (5,6). The non-DNA-binding subunit, Cbfβ, cooperates with Cbfα to form DNA-protein complexes and protects the Cbfα subunits from degradation (7). Cbfβ−/− embryos died from an absence of fetal liver hematopoiesis at mid-gestation (8,9). This barrier had impeded further research in understanding the role of Cbfβ in skeletal development until the generation of the three mice models (CbfβGFP/GFP knock-in mice, and Tek-GFP/Cbfβ and Gata1-Cbfβ transgenic mice) in 2002 (10-12). CbfβGFP/GFP knock-in mice died soon after birth. Cbfβ−/− embryos that were rescued by Tek-GFP/Cbfβ [Cbfβ−/−Tg(Tek-GFP/Cbfβ)] and Gata1-Cbfβ [Cbfβ−/−Tg(Gata1-Cbfβ)] transgene died around birth. These mouse models enabled the study of Cbfβ’s role in embryonic skeletal development (10-12). The role of Cbfβ in postnatal bone formation was unexplored until the generation of Cbfβ/Runx2 double transgenic mice, which exhibited severe osteopenia (13). However, the physiological defects caused by Cbfβ deficiency in postnatal mice have not yet been clarified. To further explore the role of Cbfβ in skeletal development, we generated mesenchymal stem cell (MSC)-specific Cbfβ conditional knockout mice by crossing Cbfβf/f mice (14) with (paired-related homeobox transcription factor 1) Prx1-Cre mice (15). Cre expression driven by the Prx1-promoter was first detected in the forelimb mesenchyme at embryonic day (E) 9.5 and then in all MSCs at E10.5 (15). Cbfβf/fPrx1-Cre mice survived into adulthood, Cbfβf/fPrx1-Cre mice displayed short limbs, short statures, inhibited osteoblastogenesis, inhibited chondrocyte differentiation, and impaired trabeculae formation. In addition, Cyclin D1, Indian hedgehog (Ihh) and parathyroid hormone-related protein receptor (PPR) expression were dysregulated in the growth plates of the Cbfβf/f Prx1-Cre mice.
Materials and Methods
Generation of Cbfβ conditional knockout mice
Cbfβf/f mice (B6.129P2-Cbfβtm1Itan/J) mice (14) and Prx1-Cre (B6.Cg-Tg(Prx1-Cre)1Cjt/J) (15) (Jackson Laboratory) were crossed to generate Cbfβf/+Prx1-cre mice, and their progeny were intercrossed to obtain Cbfβf/fPrx1-cre mice. Mice were housed in the animal room of University of Alabama at Birmingham (UAB) (Birmingham, AL, USA). All research procedures using mice were approved by the UAB Animal Care and Use Committee and conformed to NIH guidelines.
Statistical analysis and data quantification analysis
All data were presented as the mean ± standard deviation (SD). Statistical significance was assessed using Student’s t-test performed with the SPSS 16.0 software (SPSS Incorporation, Chicago, IL, USA). P-values <0.05 were considered significant, and labeled *p<0.05, **p<0.01, ***p<0.001 in the graphs. The results are representative of at least six individual experiments (n≧6).
Results
Spatial and temporal expression of Cbfβ in wild-type (WT) mouse skeleton
Expression of Cbfβ in skeletons at postnatal one-day-old (P1) mice was detected by immunohistochemistry (IHC) using paraffin sections. The results showed that Cbfβ is highly expressed in proliferative chondrocytes, hypertrophic chondrocytes, and osteoblasts in long bones (Supplementary Fig. S1A), vertebrae (Supplementary Fig. S1B), and ribs (Supplementary Fig. S1C).
MSC-specific Cbfβ-deficient mice exhibited dwarfism with skeletal malformation and delayed ossification
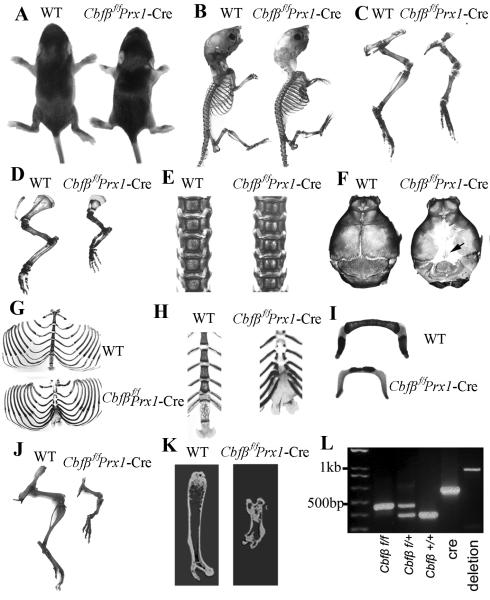
MSC-specific Cbfβ-deficient mice were generated by crossing Cbfβf/f mice with Prx1-cre mice, and genotypes were confirmed by PCR analysis (Fig. 1L). Efficient ablation of Cbfβ expression in chondrocytes and osteoblasts in ribs, vertebrae, and long bones in Cbfβf/fPrx1-Cre mice was confirmed by IHC (Supplementary Fig. S2A-C). Although a small portion (~10%) of homozygotes died of asphyxiation problems soon after birth, most survived into adulthood. Male homozygotes were fertile. Female homozygotes were able to conceive, although dystocia would occur, probably due to an abnormal pelvis (Fig. 2A, upper panel and lower panel). Postnatal seven-day-old (P7) Cbfβf/fPrx1-Cre mice exhibited dwarfism (Fig. 1A, B) with shortened limbs (Fig. 1A-D) compared to WT cohorts. Bone morphology of P7 mice was analysed by Alizarin red S/Alcian blue staining (Fig. 1B-I). Although limbs were shortened, the epiphyseal cartilage (blue color) was elongated (Fig. 1C,D) in the Cbfβf/fPrx1-Cre mice. In addition, mutant mice had widened sutures and enlarged fontanelles, with delayed ossification of the parietal and frontal bones (Fig. 1F). Ossification of the sternum (Fig. 1H) and hyoid bone (Fig. 1I) were also delayed in mutant mice compared to WT mice. The xiphoid process was non-calcified and abnormally enlarged (Fig. 1H). Spines and ribs didn’t show notable morphological changes in P7 mutant mice (Fig. 1E, G), but they did show delayed ossification in P1 mutant mice (Supplementary Fig. S2D, E). Thus, defects may be compensated for as the mice aged. Finally, the short-limb deformity observed in the mutant mice (Fig. 1A-D) persists with age, which was confirmed again by Alizarin red S/Alcian blue (Fig. 1J) and micro-CT analysis (Fig. 1K) of one-month-old (P30) mice. Notably, X-ray analysis showed that six-week-old Cbfβf/fPrx1-Cre mice displayed several characteristics of cleidocranial dysplasia (e.g. short stature, short limb, and absent clavicles) (Fig. 2A). Taken together, these results indicate that loss of Cbfβ results in dwarfism, limb and sternum malformation, and delayed skeletal ossification during postnatal skeletal development.
Fig. 1. Cbfβf/fPrx1-Cre mice had dwarfism with shortened limbs.
(A) Gross morphology of postnatal 7-day-old (P7) Cbfβf/fPrx1-Cre and wild-type (WT) mice. (B-I) Skeletal analysis by Alizarin red S/Alcian blue staining of P7 Cbfβf/fPrx1-Cre and WT mice. Long bones were shorter (C,D), sutures and fontanelles were widened (black arrow) (F), and ossification of parietal bones (F), frontal bone (F), sternum (H) and hyoid bone (I) was delayed in Cbfβf/fPrx1-Cre mice. Development of ribs (G) and spine (E) was not affected in Cbfβf/fPrx1-Cre mice. (J) Skeletal analysis by Alizarin red S/Alcian blue staining of P30 Cbfβf/fPrx1-Cre and WT mouse limb. (K) Micro-CT analysis of P30 Cbfβf/fPrx1-Cre and WT mouse femur. (L) PCR was used to determine Cbfβ alleles (f/f, f/+, +/+, or deletion) and the presence of Cre.
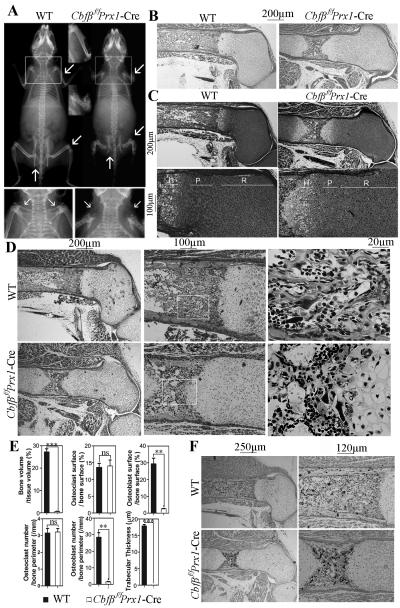
Fig. 2. Cbfβ deficiency resulted in cleidocranial dysplasia-like phenotype in adult mice and skeletal defects in newborn mice.
(A) Whole body X-ray of 6-week-old mice showed cleidocranial dysplasia-like phenotype of Cbfβf/fPrx1-Cre mice, including shortened stature, shortened long bones (upper lane, marked by arrowhead), and absent clavicles (lower lane marked by arrowhead and upper lane marked by arrowhead and square). (B) H&E staining of paraffin sections of femurs from newborn Cbfβf/fPrx1-Cre mice and WT mice. (C) Safranin O staining of paraffin sections of femurs from newborn Cbfβf/fPrx1-Cre mice and WT mice. (D) Goldner’s trichrome staining of paraffin sections of femurs from newborn Cbfβf/fPrx1-Cre mice and WT mice. (E) Quantification data of Goldner’s trichrome staining were presented as mean ± SD, n≧6, ns (non-significant), **p<0.01, ***p<0.001 versus WT. (F) TRAP staining of paraffin sections of femurs from newborn Cbfβf/fPrx1-Cre mice and WT mice. P: proliferation zone. R: resting zone. H: hypertrophic zone.
Loss of Cbfβ impaired skeletal development in newborn mice
Next, to examine the role of Cbfβ in the differentiation of chondrocytes, osteoblasts, and osteoclasts in newborn mice, hemotoxylin and eosin (H&E) staining, Safranin O staining, Goldner’s trichrome staining, and TRAP staining were performed on paraffin sections of femurs (Fig. 2B-F). Compared with WT mice, newborn Cbfβf/fPrx1-Cre mice had elongated growth plates and shortened diaphysis (Fig. 2B, C). Although the resting zone was elongated, the proliferation zone was shortened and the proliferative columns, which were presented in the WT growth plates, were disrupted in newborn mutant mice (Fig. 2C). The hypertrophic zone was also slightly deformed in newborn mutant mice (Fig. 2C). Furthermore, the number and thickness of trabecular bones and the number of osteoblasts were significantly reduced in newborn Cbfβf/fPrx1-Cre mice (Fig. 2D, E), the osteoclast number did not change (Fig. 2D-F). These results indicate that Cbfβ is important for chondrocyte proliferation and maturation in newborn mice and that it may play a lesser role in osteoclasts.
Loss of Cbfβ impaired growth plate development in P7 mice
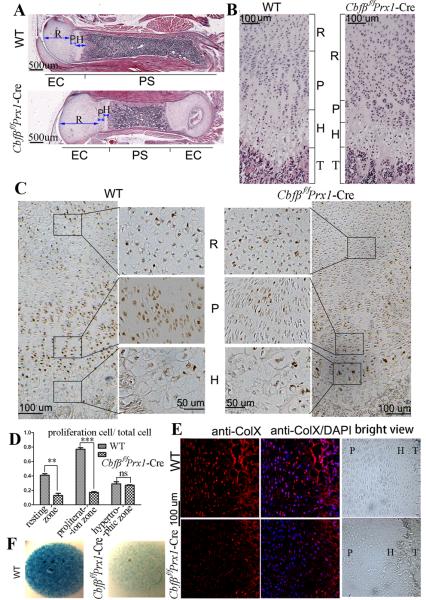
Continuous postnatal skeletal development is required for normal development toward adulthood. H&E staining on paraffin sections of P7 mouse femur showed that growth plate and trabecular bone development were delayed in mutant mice (Fig. 3A, B). Comparable with the newborn mutant mice (Fig. 2B, C), P7 mutant mice also had shortened femurs, elongated growth plates, shortened diaphysis (Fig. 3A), elongated resting zones, and shortened and disorganized proliferation zones (Fig. 3B). While the hypertrophic zone was slightly deformed in newborn mutant mice (Fig. 2C), it was notably shortened in the P7 Cbfβf/fPrx1-Cre mice (Fig. 3B). Moreover, proliferating cell nuclear antigen (PCNA) staining showed that the number of proliferating chondrocytes was decreased (Fig. 3C, D), and immunofluorescent (IF) staining showed that collagen type X (ColX) expression was reduced in mutant mice compared to WT (Fig. 3E). These results confirmed the retardation of chondrocyte proliferation and hypertrophy in Cbfβf/fPrx1-Cre mice. Micromass culture of growth plate chondrocytes from newborn mice and Alcian blue staining confirmed that chondrocyte differentiation in vitro was also drastically delayed in the absence of Cbfβ (Fig. 3F). Taken together, these results demonstrate that Cbfβ plays important roles in growth plate formation in the early stages of postnatal development.
Fig. 3. Cbfβ deficiency retards the development of primary spongiosa and delays chondrocyte proliferation and maturation.
(A, B) H&E staining of paraffin sections of femurs from P7 Cbfβf/fPrx1-Cre mice and WT mice. Femur and primary spongiosa were shortened, while the epiphyseal growth plate was elongated in Cbfβf/fPrx1-Cre mice. Columnar structure of proliferative chondrocyte zone was lost in Cbfβf/fPrx1-Cre mice. (C) PCNA staining of paraffin sections of femurs from P7 Cbfβf/fPrx1-Cre mice and WT mice. Second and third columns show magnified images of areas in black boxes. (D) The ratio of proliferating cells in total cells in the three chondrocyte zones of WT and Cbfβf/fPrx1-Cre mice from (C). Data were presented as mean ± SD, n≧6, ns (non-significant), **p<0.01, ***p<0.001 versus the same zone in WT mice. (E) IF staining using frozen sections of P7 mouse femurs showed that ColX expression was decreased in Cbfβf/fPrx1-Cre mice. Bright field views were co-presented on the right panel. (F) Micromass culture of growth plate chondrocytes of newborn mice. PS: primary spongiosa. EC: epiphyseal cartilage. R: resting zone. P: proliferation zone. H: hypertrophic zone. T: trabecular bone.
Loss of Cbfβ blocked Ihh-cyclin D1 signalling and the Ihh-PTHrP negative feedback loop
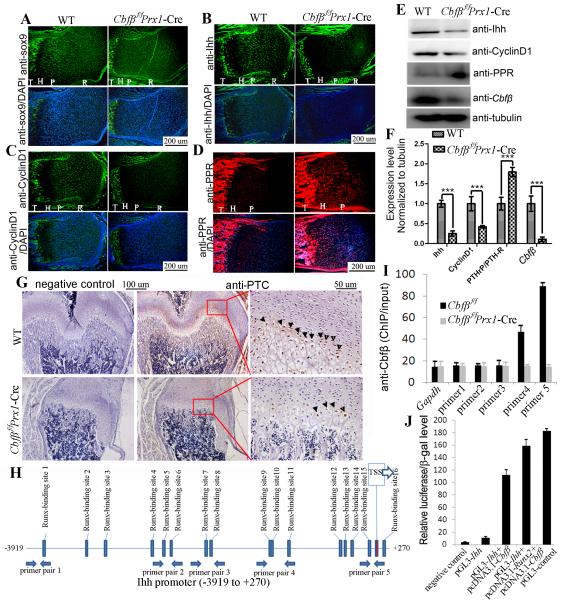
Protein expression in the growth plates was detected by IF staining using P7 mouse femur sections (Fig. 4A-D, bright field views are co-presented in Supplementary Fig. S3A-D) and western blot using proteins from newborn mouse femoral cartilage (Fig. 4E, F). Expression of SRY-related high mobility group-Box gene 9 (Sox9), a key transcription factor in the chondrocyte lineage, was similar in WT and Cbfβf/fPrx1-Cre mice (Figs. 4A; Supplementary Fig. S3A). However, expression of Ihh was dramatically reduced in Cbfβf/fPrx1-Cre mice (Fig. 4B, E, F; Supplementary Fig. S3B). Cyclin D1, a cell-cycle-regulating protein downstream of Ihh (16), was also decreased in the proliferation zone of Cbfβf/fPrx1-Cre femurs (Fig. 4C, E, F; Supplementary Fig. S3C), which may inhibit chondrocyte proliferation. Consistently, expression of other Ihh targeted genes besides Cyclin D1 [eg. Pthlh and Patched (PTC)] were also reduced in the growth plates of Cbfβf/fPrx1-Cre mice. qRT-PCR using mRNA from newborn mouse femur showed that Pthlh expression was reduced by 30% in Cbfβf/fPrx1-Cre mice (Supplementary Fig. S5B). IHC staining using P7 mouse femur sections showed that PTC expression was detected in the pre-hypertrophic zone in the growth plates in WT mice, but that it was greatly reduced in mutant mice (Fig. 4G). Ihh induces PTHrP expression in periarticular cells around pre-hypertrophic chondrocytes, which in turn suppresses chondrocyte differentiation through a feedback regulatory process denoted as the “Ihh-PTHrP negative feedback loop” (17). Interestingly, while Ihh and Pthlh expression were reduced, PPR expression was increased in the Cbfβf/fPrx1-Cre growth plate. Up-regulation of PPR in Cbfβf/fPrx1-Cre mice may increase the sensitivity of chondrocytes to PTHrP and retard chondrocyte hypertrophy even in the presence of a permissive dose of PTHrP resulting from decreased Ihh expression. In conclusion, Cbfβ deficiency may affect chondrocyte proliferation by inhibiting Ihh-cyclin D1 signalling and interfere with normal chondrocyte hypertrophy by disturbing the Ihh-PTHrP negative feedback loop.
Fig. 4. Expression of Sox9, Ihh, CyclinD1, PTHrP-R, and Cbfβ in chondrocytes of Cbfβf/fPrx1-Cre mice and WT mice.
Similar Sox9 expression (A), decreased Ihh expression (B), decreased Cyclin D1 expression (C), and increased PPR expression (D) was observed in femur growth plates of P7 Cbfβf/fPrx1-Cre mice compared to that of WT mice, as detected by IF staining using frozen sections. (E) The expression of Ihh, CyclinD1, PPR and Cbfβ was confirmed by Western blot using protein lysates of femoral cartilage from Cbfβf/fPrx1-Cre and WT newborn mice. (F) Protein levels in (E) were quantified and normalized to tubulin. (G) The expression of Patched (PTC) in the pre-hypertrophic zone of the growth plates in P7 Cbfβf/fPrx1-Cre mice was reduced compared with that in WT mice, as detected by IHC staining using paraffin sections. Third column shows the magnified images of areas in red boxes. Arrowheads indicate positive stains. R: resting zone. P: proliferation zone. H: hypertrophic zone. T: trabecular bone. (H) Schematic display of the Ihh promoter region (−3919/+270). TSS (transcriptional start site), predicted Runx-binding sites and ChIP primer positions were indicated in the figure. (I) ChIP was performed using WT chondrocyte lysates, anti-Cbfβ antibody, and primers as indicated in (H). (J) The Ihh promoter region (−1287/+162) was inserted into the pGL3-basic vector. Primary Cbfβf/fPrx1-Cre chondrocytes were transfected with pGL3-control, pGL3-Ihh+pcDNA3.1a-Cbfβ, or pGL3-Ihh+pcDNA3.1a-Cbfβ+pcNDA3.1a-Runx2. The β-GAL-expression plasmid was also co-transfected. Luciferase activity was detected 48 hours post-transfection, and normalized to β-GAL activity. Data were presented as mean ± SD, n≧6, ns (non-significant), ***p<0.001.
Runx/Cbfβ complex regulated Ihh expression by binding to the Ihh promoter directly
In order to determine if the Runx/Cbfβ complex binds the promoter region of Ihh, chromatin immunoprecipitation (ChIP) assay was performed using anti-Cbfβ antibody and the primers shown in Fig. 5A. The ChIP input value using each primer pair represents the binding efficiency of its adjacent region. There were 16 predicted Runx-binding sites localizing in the Ihh promoter region (−3919/+27) (Fig. 4H). Primer pairs 4 and 5 resulted in the highest values (Fig. 4I), indicating that Runx2/Cbfβ complex may bind sites 9-16 most efficiently. A 1.4-kb Ihh promoter region (−1287/+162), which contains Runx-binding sites 9-16, was cloned into the pGL3-basic vector. Luciferase activity driven by the Ihh promoter (−1287/+162) was low in the Cbfβf/fPrx1-Cre chondrocytes and robustly increased (10-fold) after the expression of Cbfβ (Fig. 4J). Ectopic expression of Runx2 further promoted the luciferase signal. In conclusion, we believe that the Runx/Cbfβ heterodimer directly binds to the Runx-binding sites of the Ihh promoter regions and up-regulates Ihh expression at the transcriptional level.
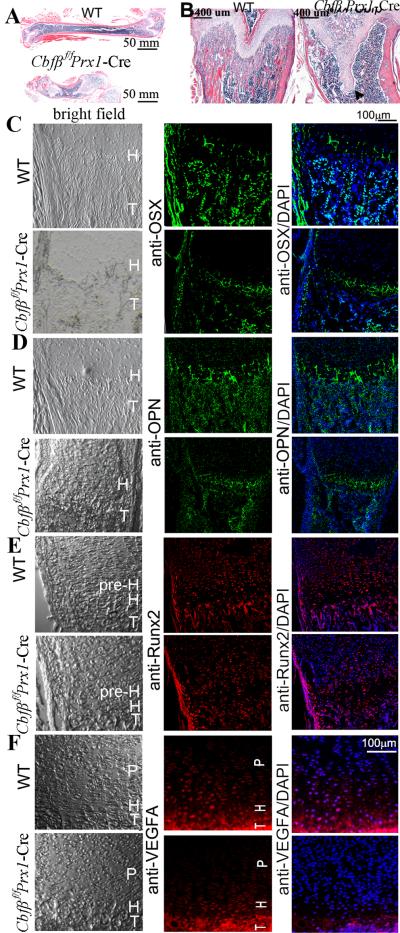
Fig. 5. Mice lacking Cbfβ had delayed ossification.
(A,B) H&E staining of the femurs of P30 (postnatal one-month-old) WT and Cbfβf/fPrx1-Cre mice. The femur was shortened and trabecular bone was lost in Cbfβf/fPrx1-Cre mice. The epiphyseal growth plate protruded deep into the diaphysis abnormally (marked by arrowhead). (C-F) IF staining using frozen sections of femurs of P7 (C-E) and newborn (F) WT and Cbfβf/fPrx1-Cre mice. In the Cbfβf/fPrx1-Cre mice, expression of OSX (C), OPN (D) and VEGFA (F) was decreased, but expression of Runx2 (E) was not changed. Bright field views of C-F are co-presented on the left panel. T: trabecular bone. H: hypertrophic zone. P: proliferation zone. pre-H: pre-hypertrophic zone.
Trabecular bone formation is impaired in MSC-specific Cbfβ-deficient mice
Delayed ossification was observed in P1 (Supplementary Fig. S2D, E), P7 (Fig. 1), and 6-week-old (Fig. 2A) Cbfβf/fPrx1-Cre mice. This was further confirmed by the H&E staining of P30 mice paraffin sections, which revealed a dramatic loss of trabecular bone in mutant mice (Fig. 5A, B). Notably, the growth plates were also severely deformed, protruding deep into the diaphysis (Fig. 5B, arrow). To investigate the potential role of Cbfβ in osteoblasts in vivo, IF staining using mouse femur sections was performed (Fig. 5C-F; Supplementary Fig. S4). Expression of Runx2 (a master transcription factor in the osteoblast lineage) was similar in P7 WT and Cbfβf/fPrx1-Cre growth plates and trabecular bone, and increased in mutant perichondrium (Fig. 5E). Expression of Runx3, detected mainly in hypertrophic and pre-hypertrophic chondrocytes, was also similar between E18.5-day and E16.5-day WT and mutant femur (Supplementary Fig. S4). However, Osx (Osterix) (Fig. 5C) and Opn (Osteopontin) (Fig. 5D), which are osteoblast-related genes, showed reduced expression in P7 Cbfβf/fPrx1-Cre trabecular bone compared with WT, indicating that osteoblastogenesis may be influenced. In addition, vascular endothelial growth factor (VEGF) expression was down-regulated in the growth plates of newborn mutant mice (Fig. 5F). Decreased VEGF expression results in reduced angiogenesis which may affect the population of MSC for normal trabecular bone formation. Taken together, these data demonstrate that Cbfβ not only regulates chondrocyte proliferation and maturation, but that it also influences trabecular bone formation for normal skeleton morphogenesis.
Loss of Cbfβ impaired osteoblastogenesis of calvarial cells in vitro
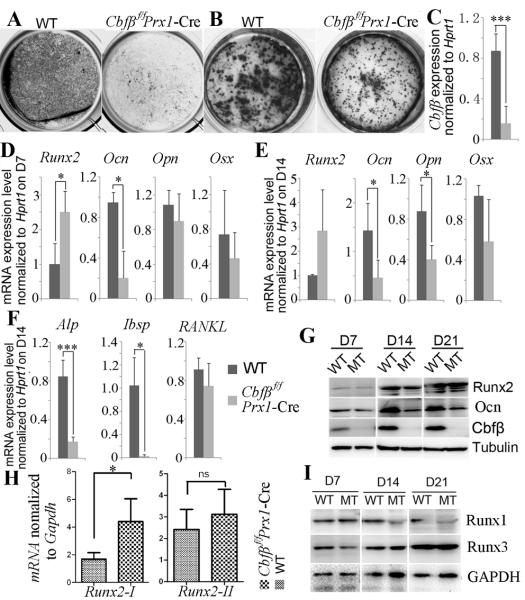
To further confirm the role of Cbfβ in bone ossification, we investigated the role of Cbfβ in osteoblastogenesis in vitro using osteoblast derived from calvarial cell primary culture (Fig. 6). Deletion of Cbfβ expression in Cbfβf/fPrx1-Cre osteoblasts was confirmed by quantitative RT-PCR (qRT-PCR) (Fig. 6C) and Western blot (Fig. 6G). Cbfβf/fPrx1-Cre osteoblasts showed reduced alkaline phosphatase (ALP) activity (Fig. 6A) and mineralization (Fig. 6B) compared to WT cells, indicating that osteoblastogenesis was impeded in mutant cells. qRT-PCR revealed that the osteoclastogenesis related cytokines receptor activator of nuclear factor-kB ligand (RANKL) was similarly expressed in Cbfβf/fPrx1-Cre and WT osteoblasts (Fig. 6F). Expression of the osteoblast-related gene Ocn (Osteocalcin) was decreased in Cbfβf/fPrx1-Cre cells on day 7 of osteoblastogenesis (Fig. 6D), and the expression of Ocn, Opn, Alp and integrin binding sialoprotein (Ibsp) was also decreased in Cbfβf/fPrx1-Cre cells on day 14 of osteoblastogenesis (Fig. 6E, F). However, Runx2 expression was increased (Fig. 6D, E), especially isoform 1 of Runx2 (Runx2-I) (Fig. 6H). The expression of Osx, another master transcription factor of osteoblast differentiation, was also similar in WT and Cbfβ-deficient cells (Fig. 6D, E). Considering that Osx expression was decreased in Cbfβf/fPrx1-Cre trabecular bone (Fig. 5C), the mechanism by which Runx2 regulates Osx expression may be different in calvaria and long bones. Western blot revealed that protein levels of Runx2 and Runx3 were not affected, but that expression of Ocn and Runx1 was reduced in Cbfβf/fPrx1-Cre osteoblasts on day 14 and 21 of osteoblastogenesis. When Runx1 expression was selectively rescued in the endothelial and hematopoietic systems of Runx1−/− embryos (Runx1Re/Re mice), these mice survived until birth and displayed disrupted mineralization in some skull elements (6), indicating Runx1 play a role in calvarial osteoblastogenesis. Thus, Runx1 hypo-sufficiency in Cbfβf/fPrx1-Cre cells may partially contribute to the impeded osteoblast differentiation. Taken together, these results indicate that Cbfβ deficiency affects osteoblast differentiation in vitro.
Fig. 6. Cbfβ is required for osteoblastogenesis of calvarial cells.
Calvarial cells from Cbfβf/fPrx1-Cre and WT newborn mice were used for primary culture. Osteoblastogenesis was detected by (A) ALP staining on day 14 of osteoblastogenesis and mineralization was detected by (B) Von Kossa staining on day 21 of osteoblastogenesis. (C) Cbfβ expression levels in calvarial cells were detected by qRT-PCR and normalized to Hprt1. (D, E) Expression of Runx2, Opn, Ocn, and Osx in calvarial cells on day 7 (D7) (D) and day 14 (D14) (E) of osteoblastogenesis was detected by qRT-PCR and normalized to Hprt1. (F) Expression of Alp, Ibsp and RANKL in calvarial cells on day 14 of osteoblastogenesis was detected qRT-PCR and normalized to Hprt1. (G) Western blot was applied to detect the protein levels of Runx2, Ocn, and Cbfβ in WT and mutant (MT) calvarial cells on day 7, 14, and 21 (D7, D14 and D21) of osteoblastogenesis. (H) Expression levels of Runx2-I and Runx2-II in calvaria were determined by qRT-PCR and normalized to Gapdh. (I) Western blot was applied to detect the protein levels of Runx1 and Runx3 in WT and mutant calvarial cells on day 7, 14, and 21 of osteoblastogenesis. Results were presented as mean ± SD, n≧6, ns (non-significant), *p<0.05, ***p<0.005.
Discussion
The Ihh-PTHrP negative feedback loop maintains chondrocyte proliferation and counters chondrocyte hypertrophy (17). We report that Cbfβ plays a dual function in this loop by regulating the expression of Ihh and PPR (Fig. 4B, D), thereby affecting the balance of chondrocyte proliferation and maturation. The Runx/Cbfβ complex binds the promoter region of the Ihh gene so as to regulate its expression (Fig. 5H-J). Consistently, diminished Ihh expression was also observed in Runx2−/− and Runx2−/−Runx3−/− mice (4). It is therefore possible that Cbfβ also regulates the Ihh-PTHrP negative feedback loop by interacting with Runx2 and Runx3, thereby regulating chondrocyte proliferation and maturation. Thus, Cbfβ deficiency results in impaired growth plates development (Figs 2, 3) and severe skeletal malformation (Fig. 1).
Runx2 is a master regulator of the commitment and differentiation of pluripotent MSCs to osteoblasts (3). As a subunit of the CBF complex, Cbfβ interacts with Runx2 to stabilize its interaction with DNA. pGL3-3XOSE2 was constructed by inserting three OSE2 (18), a Runx2 binding sequence, into the pGL3-promoter. If only Runx2 and Cbfβ were co-expressed, luciferase driven by the promoter with the OSE2 sequence increased 1.7 folds (Supplementary Fig. S5A), indicating that Runx activities were deregulated in the Cbfβ-deficient cells. Thus, Cbfβf/fPrx1-Cre mice at birth displayed a similar, but less severe, phenotype to Runx2−/− mice (3) (eg, reduced ossification and inhibited chondrocyte hypertrophy). Consistently, expression of Runx2 targeted genes, ColX (Fig. 3E) and Ocn (Fig. 6D, E), were also down-regulated in Cbfβ-deficient mice. Although Cbfβ has been reported to protect the Cbfα subunits from degradation (7), only Runx1 expression was reduced in Cbfβf/fPrx1-Cre calvarial cells compared to WT (Fig. 6I), and Runx2 and Runx3 were similarly expressed in WT and Cbfβf/fPrx1-Cre calvarial cells and long bones (Figs 6G, I, 5E; Supplementary Fig. S4). In fact, Runx2 expression was higher in Cbfβf/fPrx1-Cre perichondrium compared to WT (Fig. 5E). Expression of Runx2-I, a specific Runx2 isoform expressed in the perichondrium and proliferating chondrocytes (19), was also increased in Cbfβf/fPrx1-Cre calvaria (Fig. 6H). Thus, the protection mechanism of Runx proteins may vary depending on the cell type and the differentiation stage. In addition, the high expression of Runx2 in the mutant mice where it would not normally have such high expression may indicate the immaturity of the cells.
While the Runx2−/− skeletal system showed a complete lack of ossification (3), Cbfβ-deficient mice only have delayed ossification. The delayed mineralization observed in the ribs and spines in some mice at birth became less severe as the mice aged. This indicates that Runx2 remains partially active without Cbfβ. The in vitro promoter assay showed that the two isoforms of Runx2 (i.e. Runx2-I and Runx2-II) retained some transcriptional activity in the absence of Cbfβ (13). qRT-PCR showed that Runx2-I expression was increased in Cbfβ-deficient calvaria (Fig. 6H). The partially rescued phenotype of Cbfβ-deficient mice may be a combined action of the remaining activity of Runx2-I and Runx2-II.
Cbfβ may form a complex with Runx2 or Runx3 to regulate Ihh expression, and thereby regulate chondrocyte proliferation and maturation. Recent studies have shown that the Cbfβ/Runx1 complex plays an important role in chondrogenesis and chondrocyte proliferation (20). Soung et al. reported that Runx1 is required for endochondral ossification during skeletal development (21). Moreover, Runx1 and Cbfβ both have much higher expression levels in MSCs and chondrocytes than Runx2 and Runx3 do (21). In addition, Runx2 is usually considered as a positive regulator in chondrocyte maturation rather than in chondrocyte proliferation (22,23). All these studies indicate that the Runx1/Cbfβ complex may exert a more important role than Runx2/Cbfβ and Runx3/Cbfβ in chondrocyte proliferation. To our knowledge, our study is the first report of Cbfβ regulating chondrocyte proliferation. Additional studies are needed to characterize the mechanism underlying the roles of Runx/Cbfβ complexes in regulating the chondrocyte proliferation in vivo.
In summary, we investigated the role of Cbfβ in postnatal cartilage and bone development. We found that Cbfβ is a key factor for chondrocyte proliferation, chondrocyte differentiation, and the maintenance of growth plates and trabecular bone in postnatal mice. Cbfβ up-regulated Ihh and down-regulated PPR in postnatal growth plates and thereby controlled the proliferation of chondrocytes to pre-hypertrophic chondrocytes. Our results also indicate that Cbfβ may interact with Runx1 to regulate chondrocyte proliferation.
Supplementary Material
Acknowledgements
We would like to thank Mr. Matthew McConnell and Dr. Joel Jules for their extensive reading, discussion, and excellent assistance with this manuscript. We thank Ms. Christie Paulson for her excellent assistance with the manuscript. We thank Mr. Matthew McConnell for his excellent assistance with bioinformatics work and ChIP primer design. We appreciate the assistance of the Center for Metabolic Bone Disease at the University of Alabama at Birmingham (UAB) (P30 AR046031). We are also grateful for the assistance of the Small Animal Phenotyping Core, Metabolism Core, and Neuroscience Molecular Detection Core Laboratory at UAB (P30 NS0474666). This work was supported by NIH grants AR-055307 (Y.P.L.) and R01-AR-44741(Y.P.L.).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
All authors state that they have no conflicts of interest.
Authors' roles: Study design: YPL and WC. Study conduct: FT, MW, WC, JM, GZ, BG. Data collection and analysis: MW, FT, WC, LD, JM, GZ, BG, LW and YPL. Drafting manuscript: MW, WC, FT and YPL. Revising manuscript: MW, WC and YPL. All authors approved the final version of the manuscript for submission. WC and YPL take responsibility for the integrity of data analysis.
References
- 1.Speck NA, Terryl S. A new transcription factor family associated with human leukemias. Crit Rev Eukaryot Gene Expr. 1995;5(3-4):337–64. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 2.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23(2):425–32. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 3.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18(8):952–63. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura A, Inose H, Yano F, Fujita K, Ikeda T, Sato S, Iwasaki M, Jinno T, Ae K, Fukumoto S, Takeuchi Y, Itoh H, Imamura T, Kawaguchi H, Chung UI, Martin JF, Iseki S, Shinomiya K, Takeda S. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010;137(7):1159–67. doi: 10.1242/dev.045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liakhovitskaia A, Lana-Elola E, Stamateris E, Rice DP, van 't Hof RJ, Medvinsky A. The essential requirement for Runx1 in the development of the sternum. Dev Biol. 2010;340(2):539–46. doi: 10.1016/j.ydbio.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Huang G, Shigesada K, Ito K, Wee HJ, Yokomizo T, Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20(4):723–33. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93(22):12359–63. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87(4):697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 10.Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, Muenke M, Lian JB, Yang Y, Nuckolls GH, Stein GS, Liu PP. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32(4):639–44. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 11.Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, Nuckolls GH, Speck NA. The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat Genet. 2002;32(4):645–9. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32(4):633–8. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- 13.Kanatani N, Fujita T, Fukuyama R, Liu W, Yoshida CA, Moriishi T, Yamana K, Miyazaki T, Toyosawa S, Komori T. Cbf beta regulates Runx2 function isoform-dependently in postnatal bone development. Dev Biol. 2006;296(1):48–61. doi: 10.1016/j.ydbio.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 14.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204(8):1749–55. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 16.Long F, Zhang XM, Karp S, Yang Y, McMahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development. 2001;128(24):5099–108. doi: 10.1242/dev.128.24.5099. [DOI] [PubMed] [Google Scholar]
- 17.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–6. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 18.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15(4):1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao ZS, Hjelmeland AB, Quarles LD. Selective deficiency of the “bone-related” Runx2-II unexpectedly preserves osteoblast-mediated skeletogenesis. J Biol Chem. 2004;279(19):20307–13. doi: 10.1074/jbc.M401109200. [DOI] [PubMed] [Google Scholar]
- 20.Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, Schultz PG. A stem cell-based approach to cartilage repair. Science. 2012;336(6082):717–21. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- 21.Soung do Y, Talebian L, Matheny CJ, Guzzo R, Speck ME, Lieberman JR, Speck NA, Drissi H. Runx1 dose-dependently regulates endochondral ossification during skeletal development and fracture healing. J Bone Miner Res. 2012;27(7):1585–97. doi: 10.1002/jbmr.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, Kishimoto T, Komori T. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem. 2000;275(12):8695–702. doi: 10.1074/jbc.275.12.8695. [DOI] [PubMed] [Google Scholar]
- 23.Drissi MH, Li X, Sheu TJ, Zuscik MJ, Schwarz EM, Puzas JE, Rosier RN, O'Keefe RJ. Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J Cell Biochem. 2003;90(6):1287–98. doi: 10.1002/jcb.10677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.