Abstract
Core-binding factor β (Cbfβ) is a subunit of the Cbf family of heterodimeric transcription factors which plays a critical role in skeletal development through its interaction with the Cbfα subunits, also known as Runt-related transcription factors (Runxs). However, the mechanism by which Cbfβ regulates cartilage and bone development remains unclear. Existing Cbfβ-deficient mouse models cannot specify the role of Cbfβ in skeletal cell lineage. Herein, we sought to specifically address the role of Cbfβ in cartilage and bone development by using a conditional knockout (CKO) approach. A mesenchymal-specific Cbfβ CKO mouse model was generated by using the Dermo1-Cre mouse line to specifically delete Cbfβ in mesenchymal stem cells, which give rise to osteoblasts and chondrocytes. Surprisingly, the mutant mice had under-developed larynx and tracheal cartilage causing alveolus defects which led to death shortly after birth from suffocation. Also, the mutant mice exhibited severe skeletal deformities from defective intramembranous and endochondral ossification, owing to delayed chondrocyte maturation and impaired osteoblast differentiation. Almost all bones of the mutant mice, including the calvariae, vertebrae, tibiae, femurs, ribs, limbs and sternums were defective. Importantly, we showed that Cbfβ was expressed throughout the skeleton during both embryonic and postnatal development, which explains the multiple-skeletal defects observed in the mutant mice. Consistently, Cbfβ deficiency impaired both chondrocyte proliferation and hypertrophy zone hypertrophy during growth-plate development in the long bones of mutant mice. Notably, Cbfβ, Runx1 and Runx2 displayed different expression patterns in the growth plates of the wildtype mice indicating that Cbfβ/Runx1 complex and Cbfβ/Runx2 complex may regulate chondrocyte proliferation and hypertrophy, respectively, in a spatial and temporal manner. Cbfβ deletion in the mesenchymal progenitors impacted bone development by dramatically down-regulating Collagen X (Col X) and Osterix (Osx), but had a dispensable effect on osteoclast development. Collectively, the results demonstrate that Cbfβ mediates cartilage and bone development by interacting with Runx1 and Runx2 to regulate the expressions of Col X and Osx for chondrocyte and osteoblast development. These findings not only reveal a critical role for Cbfβ in cartilage and bone development, but also facilitate the design of novel therapeutic approaches for skeletal diseases.
Keywords: Cbfβ, Osteoblast, Chondrocyte, Skeletal development, Ossification, Runx
1. Introduction
Core binding factors (Cbfs) are heterodimeric transcription factors which consist of the Cbf-alpha (Cbfα) and Cbf-beta (Cbfβ) subunits. The Cbfα subunits are encoded by the runt-related transcription factors (Runxs), which contain three members: Runx1, Runx2, and Runx3 [1]. Unlike the Cbfα subunits, the Cbfβ subunit is encoded by a single gene. The Cbfβ subunit is a non-DNA-binding factor that associates with the Runx proteins to mediate their DNA-binding affinities. Runx/Cbfβ heterodimeric transcription complexes play crucial roles in various developmental processes [2], including the development of the skeletal system partly by mediating gene expression. Runx1 is a pivotal transcription factor that mediates the development of the hematopoietic system and also regulates early chondrocyte formation during bone development [3]. Overexpression of Runx1 in mesenchymal stem cells has been shown to induce chondrocyte development [3]. As such, Runx1 deletion by the Prrx1 (paired-related homeobox transcription factor-1)-Cre mouse line [4] causes mineralization defect which affects the formation of the sternum. Runx2 is a master regulator of osteoblast differentiation and hence plays an important role in skeletal development [5-7]. Runx2 deficient (Runx2-/-) mice die after birth and exhibit severe skeletal defects from blocked intramembranous and endochondral ossification [5-7]. The role of Runx2 in osteoblasts is buttressed by the finding that calvarial cells derived from Runx2–/– mice fail to differentiate into osteoblasts, but these cells can still differentiate into adipocytes and chondrocytes [8]. Furthermore, by regulating the expressions of the receptor activator of nuclear factor қB ligand (Rankl) and osteoprotegrin (Opg), Runx2 can promote osteoclast formation and function which is also critical for skeletal development and bone homeostasis [9]. Finally, Runx3 plays a role during gastric epithelium growth and dorsal root ganglia proprioceptive neuron development [10] and can also cooperate with Runx2 to regulate chondrocyte development [11].
The Cbfβ subunit, the binding partner of the Runx proteins, also plays a central role in skeletal development. Mice deficient in Cbfβ die during embryonic development from a lack of definitive hematopoiesis and hemorrhage [12, 13]. The embryonic lethality of Cbfβ deficiency was circumvented by generating a knock-in mouse model expressing a Cbfβ-GFP fused protein (CbfβGFP/GFP knock-in mice) or by expressing Cbfβ under the control of hematopoietic specific promoters Tie2 or Gata1 in Cbfβ-defecient embryos [Cbfβ-/-Tg(Tek-GFP/Cbfβ) mice or Cbfβ-/-Tg(Gata1-Cbfβ) mice] [14-16]. These transgenic or knock-in mice exhibited numerous skeletal defects and died soon after birth. The skeletal defects observed in these transgenic mice were similar to those reported from the Runx2–/– mice, but were less severe because the bone defects observed in these transgenic mice resulted from delayed bone ossification, rather than a lack of bone ossification. Nonetheless, the role of Cbfβ in the development of chondrocytes and osteoblasts has not been specifically demonstrated. A greater understanding of the role of Cbfβ in the development of chondrocytes and osteoblasts should provide important insights into the role of Cbfβ during skeletal development.
We utilized the genetic approach of the Cre-loxP recombination system, which can delete genes flanked by loxP DNA through the expression of Cre-recombinase under the control of specific promoters, to specifically investigate the role of Cbfβ in the development of chondrocytes and osteoblasts. Toward this end, we used the Twist2 (Dermo1)-Cre mouse line [17] to generate a mesenchymal-specific Cbfβ conditional knockout (CKO) mouse model (Cbfβf/f Dermo1-Cre) to investigate the role of Cbfβ during skeletal development. Deletion of the Cbfβ gene in the mesenchymal progenitors, which gives rise to osteoblasts and chondrocytes, resulted in severe skeletal defects during embryonic development, but these mice died shortly after birth from respiratory distress.
2. Materials and methods
2.1 Generation of Cbfβ CKO mice
Cbfβf/f and Dermo1-Cre mice were purchased from Jackson Laboratory and were crossed to generate Cbfβf/+ Dermo1-Cre mice, which were intercrossed to obtain homozygous CKO (Cbfβf/f Dermo1-Cre) mice. The genotypes of the mice were determined by PCR. All mice were maintained under a 12-hour light–dark cycle with ad libitum access to regular food and water at the University of Alabama at Birmingham (UAB) Animal Facility. The study was approved by the UAB Institutional Animal Care and Use Committee and conformed to National Institutes of Health (NIH) guidelines.
2.2 Skeletal preparation
Tissue clarification was conducted with KOH as previously described [18]. For skeletal preparation, mice were skinned, eviscerated, and fixed in 95% ethanol before staining with Alcian blue and Alizarin red solutions. This was followed by tissue clarification with KOH as previously described. Finally, cartilage and bone mineralization were then characterized by different colors (blue and red, respectively).
2.3 Histology and tissue preparation
Histology and tissue preparation were performed as described previously [19]. Murine femurs and tibiae were harvested, skinned, and eviscerated before fixing in 4% paraformaldehyde (PFA) in PBS overnight. Samples were then dehydrated in ethanol and decalcified in 10% EDTA for 1 week. For paraffin sections, samples were dehydrated in ethanol, cleared in xylene, embedded in paraffin, sectioned at 6 μm with a Leica microtome, and then mounted on Superfrost Plus slides (Fisher). For frozen sections, samples were infiltrated in 30% sucrose, embedded in OCT, sectioned at 8 μm with a freezing microtome, and then mounted on Superfrost Plus slides (Fisher).
2.4 Hematoxylin & Eosin (H&E) staining
H&E staining was performed as described previously [20]. Mice were skinned and eviscerated, and then fixed in 4% PFA overnight. Specimens were dehydrated in ethanol and embedded in paraffin. Sections were cut at a thickness of 6 μm with a microtome and then mounted on Superfrost Plus slides (Fisher). Sections were deparaffinized and hydrated through a xylene and graded ethanol series, rinsed in hematoxylin, in 1% acid alcohol and ammonia-H2O, and then in eosin. Slides were dehydrated in graded ethanol and xylene.
2.5 Safranin O staining
Safranin O staining was performed as described previously [21]. Slides were deparaffinized and hydrated through a xylene and graded ethanol series, stained with Weigert's iron hematoxylin, rinsed in tap water, and counterstained with fast green solution. Slides were then stained in 0.1% Safranin O solution, dehydrated and mounted.
2.6 Alcian blue, Von Kossa and Trap staining
Alcian blue and Von Kossa staining were performed as described previously [22]. Mineralization was analyzed by Von Kossa staining. Slides were deparaffinized and hydrated and then incubated with 1% silver nitrate solution under ultraviolet light, unreacted silver was removed with 5% sodium thiosulfate, followed by staining in Alcian blue solution (pH 2.5), and counterstained with nuclear fast red.
For trap staining, paraffin sections were stained using Acid Phosphatase, Leukocyte (TRAP) kit (387A-1KT, Sigma) following manufacturer's instructions, counterstained with fast green, dehydrated, and mounted.
2.7 Immunofluorescence analysis
Osteoblast and chondrocyte genes were analyzed by immunofluorescence using the following primary antibodies: rabbit-anti-Col X (Abcam, ab58632), rabbit-anti-Opn (osteopontin) (Abcam, ab8448), anti-Cbfβ (Santa Cruz, sc-56751), rabbit-anti-Runx1 (Abcam, ab23980), rabbit-anti-Runx2 (Abcam, ab23981), rabbit-anti-Osx (Osterix) (Abcam, ab22552), and these secondary antibodies: FITC-goat-anti-mouse IgG(H+L) and TR-goat-anti-rabbit IgG (H+L). Imaging was taken by Leica Confocal Microscope and Zeiss fluorescent microscope.
2.8 Western blot analysis
Under a stereo microscope, white parts of the hindlimbs from newborn mice were identified as cartilage, dissected from bone tissue, washed with 1× PBS, immersed with 100μl 2×SDS sample buffer (100mM Tris-HCl, 4% SDS, 0.1% bromphenol blue, 20% glycerol, supplemented with DTT and protease inhibitors), lysed using an electronic homogenizer and denatured in 95°C for 10 minutes. Proteins were resolved on SDS-PAGE and electrotransferred to nitrocellulose membranes. Cbfβ, Col X and PCNA protein levels were analyzed using β-tubulin as a loading control with the following primary antibodies: rabbit-anti-Cbfβ (Santa Cruz, sc-56751), and mouse-anti-β-tubulin (DSHB, E7). Secondary blotting was performed using horseradish peroxidase-linked anti-rabbit IgG (7074) and horseradish peroxidase-linked anti-mouse IgG (7076) were from Cell Signaling.
2.9 Proliferation assay
Proliferating cell nuclear antigen (PCNA) immunostaining was performed using a commercial kit (93-1143; Zymed Laboratories, Inc.) with horseradish peroxidase reaction and subsequent detection by DAB (Vector Laboratories SK-4100).
2.10 In vivo osteoblastogenesis assays
Primary calvarial cells from newborn mice were isolated as previously described [23]. Next, 1.5×104 cells per well of 24-well plate were seeded. Cells were maintained in α-MEM supplemented with 10% FBS for 5 days until confluence and then in osteogenic medium [BGJb medium (Gibco, 12591) supplemented with 10% FBS, 50 μg/ml L-ascorbic acid (Sigma, A4544), and 5 mM β-glycerolphosphate (Sigma, G9891)] to induce osteoblast formation. Osteoblastogenesis was analyzed by alkaline phosphatase staining according to the manufacturer's manual (Sigma, A2356) on day 14 of osteoblastogenesis. Osteoblast mineralization was examined by Von Kossa staining on day 21 of osteoblastogenesis.
2.11 RNA extraction and Quantitative Real-Time PCR (qRT-PCR) analysis
mRNA was extracted from calvarial cells cultured in osteogenic medium for 14 days and 21 days using TRIzol (Invitrogen). 0.4ug total mRNA was reverse-transcribed into cDNA using SuperScript® VILO™ cDNA Synthesis Kit (Invitrogen) according to the manufacturer's manual. Expression of osteoblast marker genes was analyzed by quantitative PCR (qPCR) using StepOne™ Real-Time PCR System (Applied Biosystem) [19]. Primer sequences were as following, Cbfβ, 5′-GCCTTTGAAGAGGCTCGAAGAAG-3′, 5′-ACCGCCACCTAAGTTAGAACCAG-3′; Col1α1, 5′-CTTGGTGGTTTTGTATTCGATGAC-3′, 5′- GCGAAGGCAACAGTCGCT -3′; opn, 5′-CCCGGTGAAAGTGACTGATT-3′, 5′-TTCTTCAGAGGACACAGCATTC-3′; RANKL, 5′-TGAAAGGAGGGAGCACGAA-3′, 5′-ATCCAGCAGGGAAGGGTTG-3′; Opg, 5′-AGAGCAAACCTTCCAGCTGC-3′, 5′-CTGCTCTGTGGTGAGGTTCG-3′; Runx2-I, 5′-GGCGTCAAACAGCCTCTTCA-3′, 5′-GCTCACGTCGCTCATCTTGC-3′; Runx2-II, 5′-TCTCCCCCCGCCCCCACTTAC-3′, 5′-CCTCTCGCCCTCTCCTTCGCC-3′.
2.12 Statistical analysis
Data are presented as mean ± SD (n>3). Statistical significance was assessed using student's t-test, and p values less than 0.05 were considered significant. Histology data are representative of at least six mice per group. A total of 192 mice were euthanatized for analysis in this study.
3. Results
3.1 Newborn Cbfβf/f Dermo1-Cre mice had dwarfism with shortened limbs and under-developed skeletons from defective bone mineralization
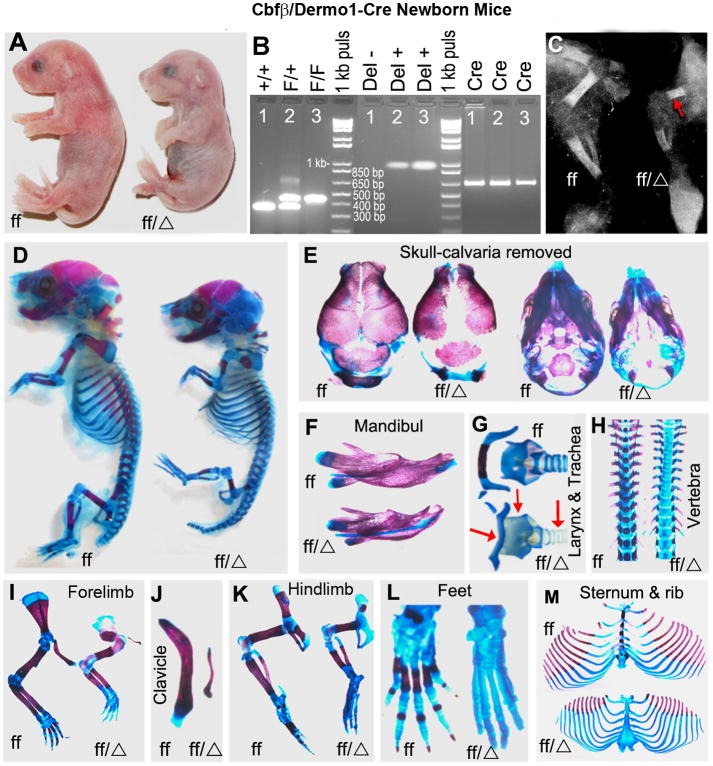
The skeletal system consists of many cell types including osteoblasts and chondrocytes, which derive from a common mesenchymal progenitors [24]. Previous studies have reported a critical role for Cbfβ in skeletal development by rescuing Cbfβ-/- embryos by overexpressing Cbfβ under the control of hematopoietic specific promoters (Tie 2 or Gata1) [Cbfβ-/-Tg(Tek-GFP/Cbfβ) mice or Cbfβ-/-Tg(Gata1-Cbfβ) mice] and by generating CbfβGFP/GFP knock-in mice [14-16]. To gain specific insights into the role of Cbfβ in chondrocytes and osteoblasts, we generated a mesenchymal-specific Cbfβ CKO (Cbfβf/f Dermo1-Cre) mouse model by breeding Cbfβf/f mice [25] with Twist2 (Dermo1)-Cre mice [17] (Fig. 1). The Dermo1 promoter is activated as early as embryonic (E) 9.5 day at the surface of mouse embryo and in mesodermal tissues such as branchial arches and somites [17]. Early during bone development, the Dermo1 promoter is first activated in condensed mesenchyme which gives rise to chondrocytes and osteoblasts. This is followed by its activation in chondrocytes and osteoblasts later during development. As such, the expression of Cre-recombinase via the Dermo1 promoter can excise the Cbfβ gene in the early mesenchymal lineage cells to assess its role in the development of the osteoblasts and chondrocytes. Similar to the Cbfβ-/-Tg(Tek-GFP/Cbfβ) mice, Cbfβ-/-Tg(Gata1-Cbfβ) mice and CbfβGFP/GFP knock-in mice [14-16], Cbfβf/f Dermo1-Cre mice circumvented the embryonic lethality of Cbfβ deficiency but died soon after birth. The newborn Cbfβf/f Dermo1-Cre mice had dwarfism with shortened limbs (Fig. 1A). The genotypes of mice were confirmed by PCR (Fig. 1B). X-ray analysis revealed delayed bone ossification and drastically reduced length of the limbs in the newborn Cbfβf/f Dermo1-Cre mice (Fig. 1C). Alizarin red and Alcian blue staining showed reduced calcification and multiple bone defects in the skeletons of the mutant mice (Fig. 1D). The mutant mice displayed widened fontanelles and undeveloped parietal, frontal and occipital bones (Fig. 1E). Also, the mutant mice had reduced ossification in several bones including the mandible (Fig. 1F), hyoid bone (Fig. 1G), vertebrae (Fig. 1H), forelimbs (Fig. 1I), clavicles (Fig. 1J), hindlimbs (Fig. 1K), and feet (Fig. 1L). Alcian blue staining in larynx and tracheal cartilage was lighter in the mutant mice compared to the wildtype (WT) mice, indicating the defects in airway tract cartilage formation (Fig. 1G). Notably, the mutant showed disproportionately shorter clavicles as compared to wild-type mice. Furthermore, whereas ossification of the sternums was drastically delayed in the mutant mice, the ribs appeared mostly normal but were of shorter length, which resulted from the reduced size of the mutant mice as compared to the WT mice (Fig. 1M). Overall, the skeletons of the mutant mice had severe developmental defects from reduced mineralization and delayed ossification, owing to defective intramembranous and endochondral ossification.
Fig.1.
Cbfβf/f Dermo1-Cre mice have underdeveloped skeletons with decreased bone mineralization. (A) Photographic analysis of newborn Cbfβf/f Dermo1-Cre (ff/Δ) and wildtype (WT, ff) mice. (B) PCR analysis for the presence, or deletion, of the Cbfβ gene in WT and Cbfβf/f Dermo1-Cre mice. F/+ or Cbfβf/+, indicates that the genome carries one copy of WT Cbfβ allele and one copy of floxed Cbfβ allele; F/F or Cbfβf/f, indicates that the genome carries two copies of floxed Cbfβ alleles; +/+ or Cbfβ+/+, indicates that the genome carries two copies of WT Cbfβ alleles; Cre, indicates that the genome carries the Cre gene; Del+, indicates that the genome carries the deleted Cbfβ allele; Del-, indicates that the genome doesn't carry the deleted Cbfβ allele. (C) X-ray analysis of the lower limbs of Cbfβf/f Dermo1-Cre and WT mice. The arrow shows decreased ossification in the femurs of Cbfβf/f Dermo1-Cre mice. (D-M) Whole-mount newborn skeletons from Cbfβf/f Dermo1-Cre and WT mice were stained with Alizarin red and Alcian blue to analyze bone and cartilage development. Overall, the skeletons of the Cbfβf/f Dermo1-Cre mice are less calcified than those of the WT mice (D). The parietal, frontal and occipital bones (E), mandible (F), hyoid bone (G), vertebrae (H), forelimbs (I), clavicles (J), hindlimbs (K), feet (L), sternum and ribs (M) are undercalcified in the mutant mice. Also, tracheal and larynx cartilage were underdeveloped in the mutant mice (G). The arrows in (G) from left to right indicates hyoid bone, larynx and trachea, respectively. The sutures and fontanelles are widened; the mandible is smaller; and the ossification of Meckel's cartilage is delayed in the Cbfβf/f Dermo1-Cre mice. The data are representative of eight mice per group.
3.2 Newborn Cbfβf/f Dermo1-Cre mice exhibited abnormal growth plate development
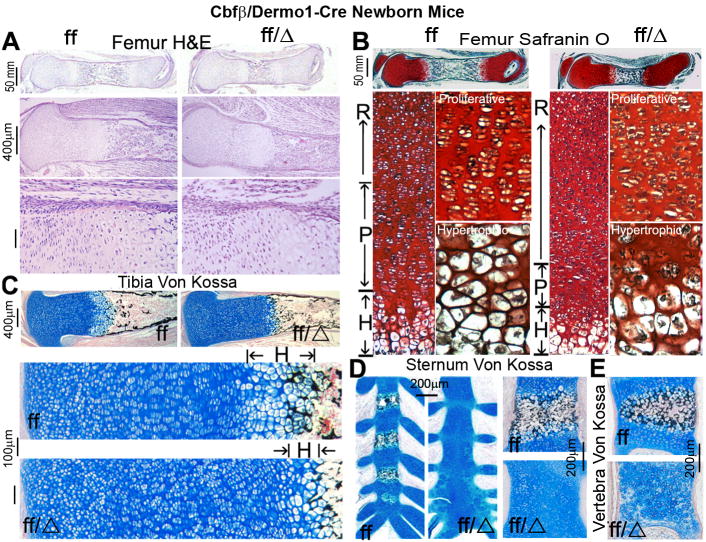
Endochondral bone formation originates from cartilage templates, which are then replaced by bone [26]. The delayed endochondral ossification observed in the Cbfβf/f Dermo1-Cre mice prompted us to examine the impact of the Dermo1-Cre-mediated Cbfβ deletion on the development of the growth plate. H&E and Safranin O staining of femurs revealed a greater relative area of growth plate over total length of the bone in the mutant mice (Fig. 2A,B). Also, the perichondral thickness in the femurs from the mutant mice was reduced in the region adjacent to the growth plate (Fig. 2A). The growth plates normally consist of many layers of chondrocytes at different stages of development, including the resting, proliferation and hypertrophic zones. Whereas the proliferative chondrocytes were well-organized in columns in the femurs from the WT mice, they were disturbed in the femurs from the mutant mice (Fig. 2A,B,C). The femurs from the mutant mice showed irregularly arranged and abnormally shaped chondrocytes. Notably, in comparison to the WT mice, the mutant mice showed a drastically elongated resting zone as well as shorter proliferative and hypertrophic zones in the growth plates, indicating a delay in the chondrocyte maturation (Fig. 2B,C). Furthermore, Von Kossa staining revealed a decreased mineralization in the tibiae (Fig. 2C), sternums (Fig. 2D), and vertebrae (Fig. 2E) of the mutant mice. The ossification centers of the vertebral bodies were barely detectable in the mutant mice. Moreover, the terminal hypertrophic chondrocytes were less calcified, and the ratio of calcified diaphysis was decreased in the mutant mice. Collectively, these results confirm that Cbfβ is critical for the development of the growth plate.
Fig. 2.
Cbfβf/f Dermo1-Cre mice have disorganized growth plates and delayed endochondral bone formation. (A) H&E staining of femoral sections from newborn Cbfβf/f Dermo1-Cre (ff/Δ) and wildtype (WT, ff) mice. (B) Safranin O staining of femoral sections from newborn Cbfβf/f Dermo1-Cre and WT mice. H&E and Safranin O staining show that newborn Cbfβf/f Dermo1-Cre mice have abnormal growth plate development and less calcified trabecular bone adjacent to the hypertrophic zone. (C-E) Von Kossa staining of the tibiae (C), sternums (D), and vertebrae (E) show decreased calcification in the Cbfβf/f Dermo1-Cre mice. R, resting zone; P, proliferating zone; and H, hypertrophic zone of the growth plate. The data are representative of six mice per group.
3.3 Cbfβf/f Dermo1-Cre mice display alveolar expansion defects which may derive from under-developed larynx and tracheal cartilage and display cardiac hypertrophy
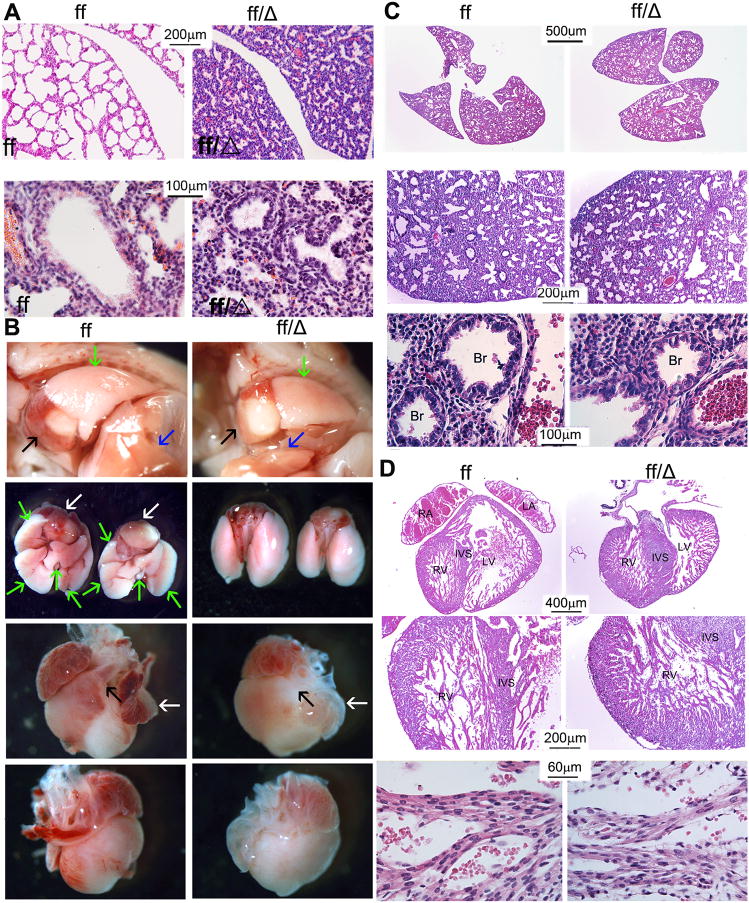
Further analysis demonstrated that the newborn Cbfβf/f Dermo1-Cre mice also had abnormal lung development (Fig. 3A). The lungs of newborn WT mice showed normally developed alveoli which were separated by connective tissue and capillaries, while the alveoli in the lungs of the newborn mutant mice were abnormally shaped. To determine whether this defects was derived from abnormal pulmonary development or defective airway tract cartilage (Fig. 1G), we examined the lungs during embryonic development (Fig. 3B,C). Although lungs of the mutant mice were slightly smaller, the gross morphology was similar to those of the WT mice (Fig. 3B upper 2 panels and C upper panels). WT and Cbfβf/f Dermo1-Cre embryos had similar pulmonary alveoli formation (Fig. 3C, the middle panels). They also shared the same bronchioles structure, lined with a layer of ciliated cuboidal epithelium and smooth muscle (Fig. 3C, the lower panels). These results indicate pulmonary development was normal in the mutant embryos. Cartilage rings in the airway tract hold the trachea and bronchi open and prevent their collapse during air exchange. Tracheomalacia, a human disease related to defected tracheal cartilage formation, can lead to respiratory distress. Similarly, Cbfβf/f Dermo1-Cre had under-developed larynx and tracheal cartilage (Fig. 1G), which caused respiratory distress and alveolar expansion failure. To examine whether Cbfβ deficiency affects other mesenchymal stem cell derived tissue, we examined the heart tissue (Fig. 3B,D). The mutant mice exhibited ventricle hypertrophy, indicating congenital heart disease in Cbfβf/f Dermo1-Cre mice (Fig.3B,D). However, the morphologies of the cardiomyocytes were similar among WT and mutant mice (Fig.3D the two lower panels). Collectively, these results indicate that Cbfβ is critical for the development and function of respiratory system and heart.
Fig. 3.
Cbfβf/f Dermo1-Cre mice have defective alveolar expansion and cardiac hypertrophy. (A) H&E staining of lung sections from newborn Cbfβf/f Dermo1-Cre (ff/Δ) and wildtype (WT, ff) mice reveal defective lung development in the mutant mice as compared to the WT mice. Higher magnification figures has been co-presented in the lower panels. (B) Photographic analysis of lung and heart from 18.5 days post coitum (18.5 dpc) Cbfβf/f Dermo1-Cre (ff/Δ) and WT (ff) mice. The 1st row shows the heart and lungs in the thoracic cavity (Black arrow, heart; blue arrow, liver; green arrow, lung). The 2nd row shows the underside view of heart and lungs (white arrow, heart; green arrow, lung). The 3rd row shows the left view of heart (right ventricle; black arrow, pulmonary artery; white arrow, left atrial). The 4th row shows the right view of heart (left ventricle). (C) H&E staining of lung sections from 18.5 days post coitum (18.5 dpc) Cbfβf/f Dermo1-Cre (ff/Δ) and WT (ff) mice. Higher magnification figures has been co-presnented in the lower panels. (D) H&E staining of cardiac sections from 18.5 days post coitum (18.5 dpc) Cbfβf/f Dermo1-Cre (ff/Δ) and WT (ff) mice. Higher magnification figures has been co-presnented in the lower panels. The data are representative of six mice in both WT and mutant group. Br, bronchiole; RV, right ventricle; LV, left ventricle; RA, right atrial; LA, left atrial; IVS, interventricle septal.
3.4 Cbfβ is highly expressed throughout the murine skeleton during skeletal development
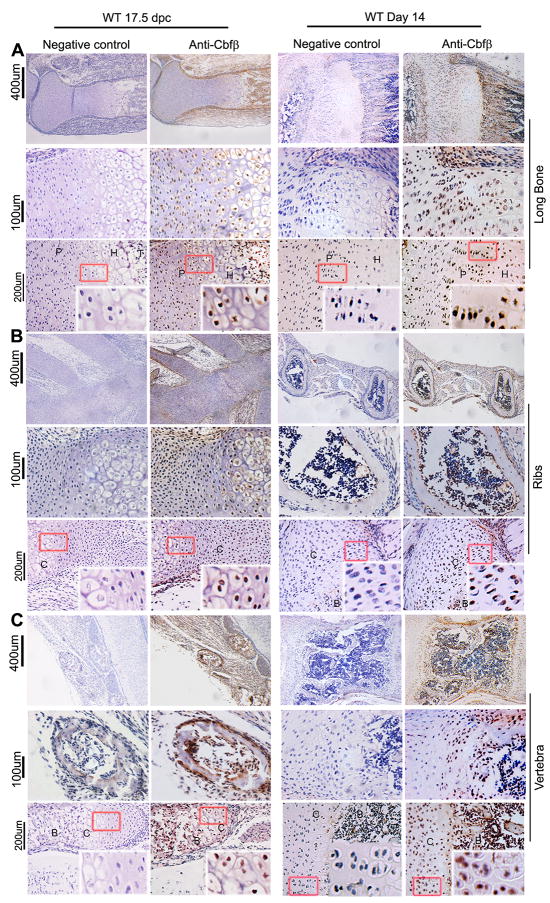
Given the drastic impact of Cbfβ deletion in early mesenchymal progenitors on skeletal development, we examined the expression pattern of Cbfβ in the murine skeleton by immunohistochemistry during embryonic and postnatal development to further evaluate its requirement for skeletal development (Fig. 4). Cbfβ was highly expressed in the long bones (Fig. 4A), ribs (Fig. 4B), and vertebrae (Fig. 4C) of the WT mice at 17.5 days post coitum (dpc) and day 14 post birth. During embryonic development, Cbfβ was highly detected in the perichondrium, periosteum, growth plates, and primary spongiosa of long bones as well as the periosteum, ossification centers, and the chondrocytes of the ribs. By two weeks of age, Cbfβ was mainly detected in the growth plates of long bones and the ossification centers of the ribs. However, while Cbfβ was mainly expressed in the bodies of the vertebrae during embryonic development, Cbfβ was mostly detected in the periosteum of the vertebrae by two weeks of age. Data indicate that Cbfβ is spatially and temporally expressed throughout the skeleton during both prenatal and postnatal development.
Fig. 4.
Cbfβ is highly expressed in the murine skeleton during embryonic and postnatal development. (A-C) Immunofluorescence staining with anti-Cbfβ antibody of paraffin sections from the long bones (A), ribs (B), and vertebrae (C) of wildtype (WT) mice at 17.5 days post coitum (17.5 dpc) and 14 days after birth (Day 14) show the expression of Cbfβ in the murine skeleton. The left panels serve as negative controls. Images with lower magnification are shown on top, and those with higher magnification are shown in the bottom. The data are representative of six WT mice.
3.5 Deletion of Cbfβ in early mesenchymal progenitors affects chondrocytes and osteoblasts development
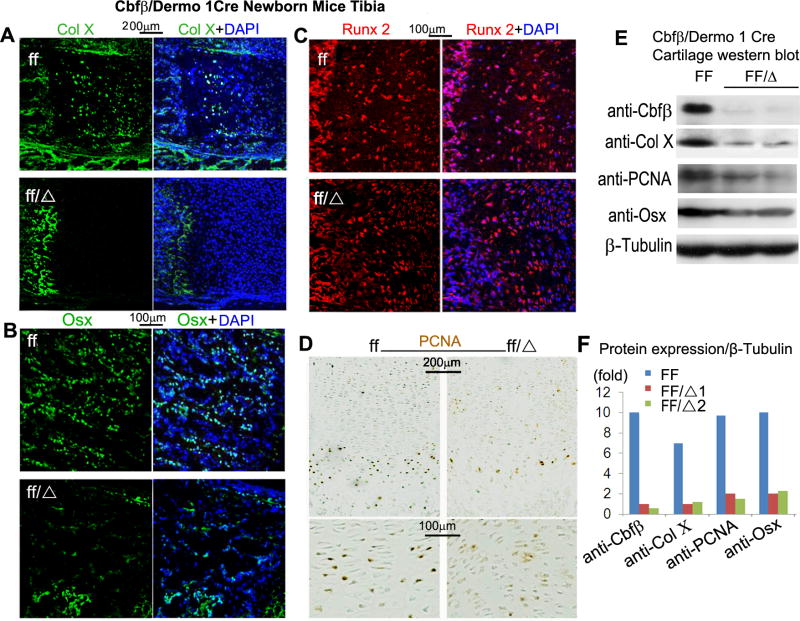
To further investigate the role of Cbfβ in skeletal development, we performed immunostaining analyses of Col X (Collagen X, a marker of chondrocyte hypertrophy), Osx (Osterix, a critical osteoblast gene) and Runx2. Col X expression was suppressed in the growth plates of the mutant mice (Fig. 5A). Whereas the expression of Osx, a target gene of Runx2, was repressed in the trabecular bone of the mutant mice (Fig. 5B), Runx2 expression was similar among the WT and mutant mice (Fig. 5C). Consistent with the decrease in the hypertrophic and proliferative chondrocytes observed in the mutant mice (Fig. 2), PCNA staining showed a decrease number of proliferating chondrocytes in the tibiae of the mutant mice (Fig. 5D). As expected, Western blot analysis confirmed that Cbfβ was effectively ablated in the cartilage of the mutant mice (Fig. 5E,F). Consistent with the immunostaining analyses, Western blot analysis showed that the levels of Col X, PCNA and Osx were reduced by at least five-fold in the mutant mice (Fig. 5E,F).
Fig. 5.
Cbfβf/f Dermo1-Cre mice exhibit delayed chondrocyte development and impaired osteoblast differentiation. (A-C) Immunofluorescence staining with anti-Col X (A), anti-Osx (B), or anti-Runx2 (C) antibodies of tibial paraffin sections from newborn Cbfβf/f Dermo1-Cre (ff/Δ) and wildtype (WT, ff) mice. (D) PCNA staining of tibial paraffin sections from newborn Cbfβf/f Dermo1-Cre and WT mice are shown. Blue staining from DAPI indicates cell nuclei in “A-D”. (E) Western blot analysis of the expression levels of Cbfβ, Col X, PCNA and Osxin the cartilage of Cbfβf/f Dermo1-Cre and WT mice. β-tubulin is used as loading control. (F) Quantification of “E” is shown. The data of A-D are representive of seven mice per group.
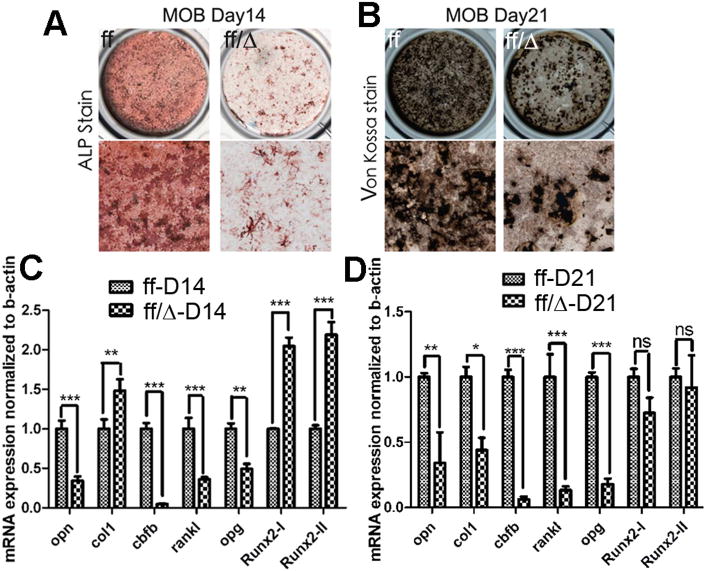
Next, we performed ALP activity and Von Kossa staining on calvarial cells from newborn mice cultured in hyperglycemic osteogenic medium (BGJB medium) to examine osteoblast differentiation and bone mineralization. ALP activity (Fig. 6A) and bone mineralization (Fig. 6B) were severely reduced in the mutant osteoblasts. qRT-PCR analysis revealed that expression of Opn (Osteopontin), an important osteoblast gene, as well as Rankl and Opg, were significantly repressed in the calvarial cells during osteoblast differentiation (Fig. 6C,D). However, Rankl/Opg ratio was not changed (Fig. S2A). Consistently, osteoclast development was not affected in the mutant mice (Fig. S2B). As expected, Cbfβ was barely detected in the calvarias of the mutant mice. The expression levels of Runx2-I and Runx2-II, two isoforms of Runx2, were significantly increased during the initial stage of the osteoblastogenesis in the mutant cells. However, their expression was similar among WT and mutant osteoblasts during the terminal stage of the osteoblastogenesis. The level of Col I, a marker of immature osteoblasts, was also increased during the initial stage of osteoblastogenesis but decreased during the terminal stage of the osteoblastogenesis.
Fig. 6.
Calvarial cells from Cbfβf/f Dermo1-Cre mice show impaired osteoblastogenesis and bone mineralization in vitro. (A-B) Calvarial cells from newborn Cbfβf/f Dermo1-Cre (ff/Δ) and wildtype (WT, ff) mice were submitted to osteoblastogenesis assays. Osteoblast differentiation was analyzed by ALP activity on day 14 (A) or by Von Kossa staining on day 21 (B). (C-D) mRNA expression levels of Opn, Col I, Cbfβ, Rankl, Opg, Runx2-I, and Runx2-II from calvarial cells of Cbfβf/f Dermo1-Cre and WT mice were analyzed on day 14 (C) and day 21 (D) of the osteoblast differentiation by qRT-PCR. Data were normalized to β-actin. Results are expressed as means ±SD, n>3 in each group. NS not significant, *p<0.05, **p<0.001, and ***<0.0001. MOB: murine osteoblast.
These results indicate Cbfβ deficiency affects the development of osteoblasts and chondrocytes by maintaining them in an immature stage and thus impact their terminal differentiation.
3.6 Cbfβ may interact with Runx1 and Runx2 to regulate chondrocyte proliferation and hypertrophy in a spatially and temporally specific manner
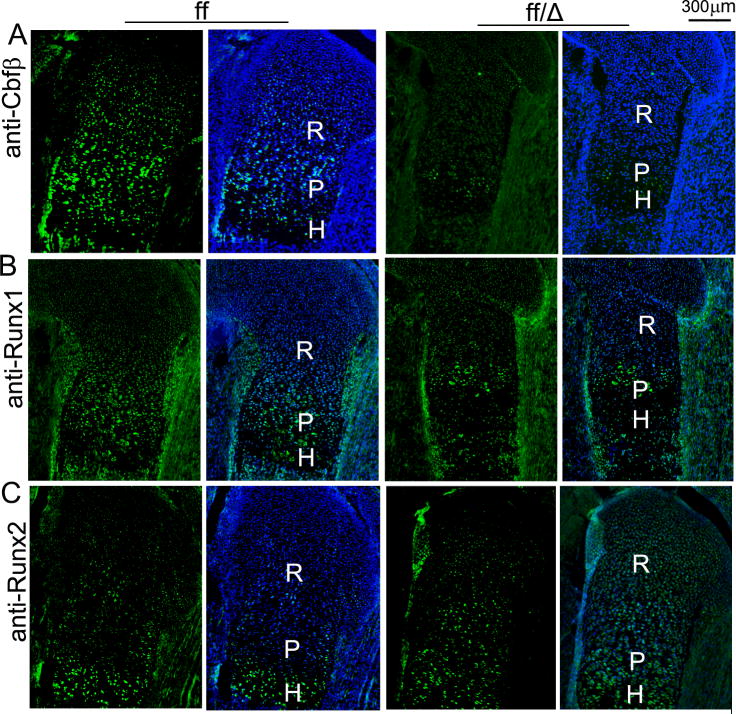
Runx proteins function by interacting with Cbfβ to induce gene expression during development, and different Runx proteins play different roles in growth plate development. Runx2 primarily regulate chondrocyte hypertrophy [27, 28] and Runx1 is involved in chondrocyte proliferation and lineage determination [29], but Cbfβ deficiency impaired both chondrocyte proliferation and maturation (Fig. 5). Thus, we examined the expression patterns of Cbfβ, Runx1 and Runx2 in the growth plates of WT mice (Fig. 7A-C, the left panels). Cbfβ was expressed throughout the growth plate, colocalizing with both Runx1 and Runx2 in WT mice. Whereas Runx1 was only detected in the proliferative and resting chondrocytes, Runx2 was expressed primarily by hypertrophic chondrocytes of the growth plates of WT mice. Predictably, the Cbfβ/Runx2 complex seems to play a major role in hypertrophic zone development while the Cbfβ/Runx1 complex mainly regulates the development of the proliferative zone of growth plates in WT mice. In addition, we also examined the expression of Cbfβ, Runx1 and Runx2 in the Cbfβf/f Dermo1-Cre mouse growth plate (Fig. 7, the right panels). Cbfβ was efficiently down-regulated in the Cbfβf/f Dermo1-Cre mice (Fig. 7A). Also, Runx1 expression was slightly down-regulated (Fig. 7B), but Runx2 expression was similar in the hypertrophic zone in the mutant mouse growth plates compared to the WT mouse growth plate (Fig. 7C). Although Runx2 expression was limited to the hypertrophic zone of the growth plates of WT mice, it was also weakly expressed in immature chondrocytes in the mutant mice (Fig. 7C). Furthermore, we examined whether Cbfβ may also function through both Runx1 and Runx2 during osteoblast differentiation by Western blot analysis (Fig. S1). The expressions of Cbfβ and Runx2 were specifically upregulated during osteoblast differentiation which led to the activation of Osteocalcin (Ocn) and activating transcription factor 4 (Atf4), two crucialosteoblast genes, during osteoblastogenesis. Suprisingly, Runx1 expression remained unchanged throughout osteoblastogenesis. Data indicate that while Cbfβ interacts with both Runx1 and Runx2 during chondrogenesis, it is likely to interact mainly with Runx2 during osteoblastogenesis.
Fig 7.
Analysis of expression of Runx1, Runx2 and Cbfβ in 18.5 dpc wildtype (WT) and Cbfβf/f Dermo1-Cre embryos. Femoral frozen sections of 18.5 dpc WT (ff) and Cbfβf/f Dermo1-Cre (ff/Δ) mice were submitted immunofluorescence analysis to examine the expression of Cbfβ (A), Runx1 (B) and Runx 2 (C) in the growth plates of these mice. Blue staining from DAPI indicates cell nuclei. R, resting zone. P, proliferation zone. H, hypertrophic zone. Data are representive of six mice per group.
4. Discussion
Previous studies through the global deletion of the Cbfβ gene in mice had led to embryonic lethality from defective hematopoiesis and hemorrhage [12, 13], which prevented the investigation of the role of the Cbfβ in skeletal development. The role of Cbfβ in skeletal development has recently been demonstrated through the generation of Cbfβ-/-Tg(Tek-GFP/Cbfβ) mice, Cbfβ-/-Tg(Gata1-Cbfβ) mice and CbfβGFP/GFP knock-in mice [14-16]. Whereas these transgenic mice died shortly after birth, they exhibited severe skeletal defects, revealing a crucial role for Cbfβ in skeletal development. Given that osteoblasts and chondrocytes originate from the mesenchymal but not hematopoietic progenitors, it can be argued that these studies did not unambiguously address the role of Cbfβ in chondrocytes and osteoblasts development. Hence, we utilized the Cre-loxP recombinase system to specifically restrict the Cbfβ gene in mesenchymal progenitors to investigate its role in osteoblasts and chondrocytes. The deletion of the Cbfβ in the undifferentiated mesenchymal stem cells causes numerous severe skeletal defects during embryonic development by affecting the development of both chondrocytes and osteoblasts. This Cbfβ CKO mouse model provides direct evidence for the role of Cbfβ in osteoblasts and chondrocytes during skeletal development.
4.1 Deletion of the Cbfβ gene in the early mesenchymal stem cells leads to death after birth but provides a direct evidence for the role of Cbfβ in chondrocytes and osteoblasts
Intramembranous bone formation results from osteogenesis of committed mesenchymal cells and gives rise to the flat bones of the skulls [26]. Endochondral ossification is responsible for the other bones of the body and results from the differentiation of committed mesenchymal cells into chondrocytes, which then undergo proliferation and maturation. We show that Cbfβ is expressed in a spatial and temporal manner in the murine skeleton during embryonic and postnatal skeletal development (Fig. 4). Cbfβ-deficiency is embryonically lethal [12, 13], so we were able to overcome this embryonic lethality [12, 13] by deleting the Cbfβ gene specifically in mesenchymal progenitors via the Dermo1-Cre mouse line (Fig. 1). Dermo1 is a transcription factor of the basic helix-loop-helix family that is highly expressed during embryonic development [30]. Dermo1-Cre recombinase activity is expressed by E9.5 in the mesoderm tissues, which gives rise to chondrocytes and osteoblasts [31]. The Dermo1-Cre mouse line provides a great tool for restricting genes in undifferentiated mesenchymal cells before the formation of limb bud. The mutant mice displayed severe bone defects during development, which are similar to Cbfβ-/-Tg(Tek-GFP/Cbfβ) mice, Cbfβ-/-Tg(Gata1-Cbfβ) mice and CbfβGFP/GFP knock-in mice [14-16], by impacting the development of osteoblasts and chondrocytes. These defects stem from defective intramembranous and endochondral bone formation. It was reported that deletion of Runx1 by Prx1-Cre mainly led to a delay in sternal development [8]. However, our deletion of the Cbfβ gene by Dermo1-Cre not only triggered a delay in sternal development but also affected the development of many bones including the bones of the skulls, tibiae, femurs, vertebrae and ribs. This can be explained by the fact that while the development of the sternums may primarily dependent on Runx1 through its interaction with Cbfβ, multiple Runx proteins may play roles in the development of the other bones of the body. Further, the deletion of Cbfβ in the mesenchymal progenitors also affects alveolus expansion through its role in larynx and tracheal cartilage development (Fig. 1G and 3). Nevertheless, the Cbfβf/f Dermo1-Cre mice died shortly after birth from respiratory distress, stemming from defective airway tract development.
4.2 Cbfβ may control osteoblastogenesis and chondrogenesis by regulating the expression of many genes that are critical for bone formation
Our mechanistic studies demonstrate that Cbfβ restriction in the mesenchymal progenitors affects the expression of many genes that are critical for skeletal development. The expression of Osx, a master gene for osteoblastogenesis, was drastically repressed in the mutant mice, which provide an explanation for the impact of this deletion on bone formation. Consistently, while the expression of Col X, a marker of chondrocyte maturation, was drastically repressed. It was reported that Runx2 gene expression is auto-regulated in part by negative feedback inhibition of Runx2/Cbfβ complex binding to its own promoter[32].Consistently with posture, there was a significant up-regulation of Runx2-I and Runx2-II expression at D14 mutant osteoblasts (Fig. 6C). However, in growth plates, trabecular bones, and D21 osteoblasts, Runx2 have similar expression between WT and mutant group (Fig. 6D, 5C), indicating that this negative feedback inhibition machinery may work in specific cell types and/or at specific differentiation stages. Consistent with that posture, our data reveal that while Cbfβ is expressed in the entire growth plate of WT mice, Runx1 is only expressed in the resting zone and the proliferative zone of the growth plate and Runx2 is detected only in the hypertrophic zone (Fig. 7). We believe that the Runx1/Cbfβ complex may play a stronger role in chondrocyte proliferation than the Runx2/Cbfβ complex, since both Cbfβ and Runx1 were expressed in the proliferation zone. The notion is further supported by fact that Runx2 is usually considered a positive regulator of chondrocyte maturation rather than a stimulator of chondrocyte proliferation [27, 28]. This finding indicates that Cbfβ promotes skeletal development by interacting with Runx1 and Runx2 in a spatially and temporally specified manner, which is critical for inducing gene expression during different stages of bone formation. This finding also supports the recent report that the Runx1/Cbfβ complex is important for cartilage growth and repair in osteoarthritis and fracture healing [29, 33].
Considering the high glucose concentration (10g/L) in the BGJb medium, our in-vitro osteoblastogenesis system (Fig. 6) only reflects the pathophysiological context of hyperglycemia, rather than the normal physiological cell responses. Wang et al. recently reported that hyperglycemia diverts dividing osteoblastic precursor cells to an adipogenic pathway and induces synthesis of a hyaluronan matrix, which is adhesive for monocytes [34]. However, using BGJb medium as the osteogenic medium because this is a well-established [35] and widely used. We noted that the Wang et al. [34] were using bone marrow stromal cells, while our data was obtained using calvarial cells, which may have resulted in the cells being at different differentiation stages. Our data showed high mineralization on Day 21 and did not show any obvious adipogenesis in our calvarial osteoblast primary culture system, indicating that the findings presented in Figure 6 is still significant.
5. Conclusions
Our study reveals that Cbfβ is critical for embryonic skeletal development as its deletion in the early stage of skeletal development results in death shortly after birth and show many developmental deformities. These mutant mice exhibite numerous bone defects from skulls to sternums, vertebrae, tibiae, and femurs. Our results also indicate that Runx1/Cbfβ and Runx2/Cbfβ may regulate chondrocyte proliferation and hypertrophy, respectively. Taken together, this work provides important insights into the role of Cbfβ in the development of the chondrocytes and osteoblasts during skeletal development. Further understanding of the role of Cbfβ in postnatal bone development may provide a greater understanding into the role of Cbfβ in skeletal development and many skeletal disorders stemming from defective bone development.
Supplementary Material
Highlights.
Cbfβ is expressed highly through skeletons in both embryonic and postnatal mice.
Cbfβ mutant impairs airway tract cartilage development resulted in newborn mice death.
Cbfβ deletion impaired almost all of the cartilages and bones in the mutant mice.
Cbfβ deletion de-regulates expression of the skeleton genes including ColX and Osx.
Cbfβ interacting with Runx1 and 2 regulates chondrocyte proliferation and hypertrophy
Acknowledgments
We thank Ms. Suzy Newton and Ms. Christie Paulson for their excellent assistance with the manuscript. We appreciate the assistance of the Center for Metabolic Bone Disease (P30 AR046031), Small Animal Phenotyping Core, Metabolism Core, and Neuroscience Molecular Detection Core Laboratory (P30 NS0474666) at UAB. This work was supported by the NIH grants AR-44741 (Y.P.L.) and AR-055307 (Y.P.L.).
Abbreviations
- Cbfβ
core-binding factor β
- Cbfα
core-binding factor α
- Runxs
runt-related transcription factors
- Col
collagen
- Osx
osterix
- CKO
conditional knockout
- Prrx1
paired-related homeobox transcription factor-1
- Rankl
receptor activator of nuclear factor қB ligand
- Opg
osteoprotegerin
- UAB
University of Alabama at Birmingham
- NIH
National Institutes of Health
- H&E
Hematoxylin & Eosin
- Opn
osteopontin
- Sc
Santa Cruz
- ab
Abcam
- E
embryonic day
- WT
wildtype
- R
resting zone
- P
proliferating zone
- H
hypertrophic zone
- dpc
days post coitum
- Br
bronchiole
- RV
right ventricle
- LV
left ventricle
- RA
right atrial
- LA
left atrial
- IVS
interventricular septum
Footnotes
Conflict of interest: The authors declare no competing financial interests.
References
- 1.Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994;23:425–32. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- 2.Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int. 2003;27:315–24. doi: 10.1016/s1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Belflower RM, Dong YF, Schwarz EM, O'Keefe RJ, Drissi H. Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J Bone Miner Res. 2005;20:1624–36. doi: 10.1359/JBMR.050516. [DOI] [PubMed] [Google Scholar]
- 4.Kimura A, Inose H, Yano F, Fujita K, Ikeda T, Sato S, Iwasaki M, Jinno T, Ae K, Fukumoto S, Takeuchi Y, Itoh H, Imamura T, Kawaguchi H, Chung UI, Martin JF, Iseki S, Shinomiya K, Takeda S. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010;137:1159–67. doi: 10.1242/dev.045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 6.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 7.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi H, Gao Y, Ueta C, Yamaguchi A, Komori T. Multilineage differentiation of Cbfa1-deficient calvarial cells in vitro. Biochem Biophys Res Commun. 2000;273:630–6. doi: 10.1006/bbrc.2000.2981. [DOI] [PubMed] [Google Scholar]
- 9.Enomoto H, Shiojiri S, Hoshi K, Furuichi T, Fukuyama R, Yoshida CA, Kanatani N, Nakamura R, Mizuno A, Zanma A, Yano K, Yasuda H, Higashio K, Takada K, Komori T. Induction of osteoclast differentiation by Runx2 through receptor activator of nuclear factor-kappa B ligand (RANKL) and osteoprotegerin regulation and partial rescue of osteoclastogenesis in Runx2-/- mice by RANKL transgene. J Biol Chem. 2003;278:23971–7. doi: 10.1074/jbc.M302457200. [DOI] [PubMed] [Google Scholar]
- 10.Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21:3454–63. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, Komori T. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–63. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci U S A. 1996;93:12359–63. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe AH, Speck NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, Komori T. Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat Genet. 2002;32:633–8. doi: 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- 15.Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, Nuckolls GH, Speck NA. The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat Genet. 2002;32:645–9. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- 16.Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, Muenke M, Lian JB, Yang Y, Nuckolls GH, Stein GS, Liu PP. Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat Genet. 2002;32:639–44. doi: 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- 17.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–74. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 18.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 19.Yang S, Hao L, McConnell M, Xuedong Z, Wang M, Zhang Y, Mountz J, Reddy M, Eleazer P, Li YP, Chen W. Inhibition of Rgs10 Expression Prevents Immune Cell Infiltration in Bacteria-induced Inflammatory Lesions and Osteoclast-mediated Bone Destruction. Bone Research. 2013;1:267–81. doi: 10.4248/BR201303005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Wang Y, Abe Y, Cheney L, Udd B, Li YP. Haploinsuffciency for Znf9 in Znf9+/- mice is associated with multiorgan abnormalities resembling myotonic dystrophy. J Mol Biol. 2007;368:8–17. doi: 10.1016/j.jmb.2007.01.088. [DOI] [PubMed] [Google Scholar]
- 21.Zhen G, Wen C, Jia X, Li Y, Crane JL, Mears SC, Askin FB, Frassica FJ, Chang W, Yao J, Carrino JA, Cosgarea A, Artemov D, Chen Q, Zhao Z, Zhou X, Riley L, Sponseller P, Wan M, Lu WW, Cao X. Inhibition of TGF-beta signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med. 2013;19:704–12. doi: 10.1038/nm.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S, Li YP. RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation. Genes Dev. 2007;21:1803–16. doi: 10.1101/gad.1544107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aubin JE, Turksen K, Heersche JNM. Osteoblastic cell lineage. In: Nod M, editor. Cellular and Molecular Biology of Bone. New York: Academic Press; 1993. p. 1. London: Academic Press. [Google Scholar]
- 25.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204:1749–55. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provot S, Schipani E. Molecular mechanisms of endochondral bone development. Biochem Biophys Res Commun. 2005;328:658–65. doi: 10.1016/j.bbrc.2004.11.068. [DOI] [PubMed] [Google Scholar]
- 27.Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, Kishimoto T, Komori T. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem. 2000;275:8695–702. doi: 10.1074/jbc.275.12.8695. [DOI] [PubMed] [Google Scholar]
- 28.Drissi MH, Li X, Sheu TJ, Zuscik MJ, Schwarz EM, Puzas JE, Rosier RN, O'Keefe RJ. Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J Cell Biochem. 2003;90:1287–98. doi: 10.1002/jcb.10677. [DOI] [PubMed] [Google Scholar]
- 29.Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, Schultz PG. A stem cell-based approach to cartilage repair. Science. 2012;336:717–21. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Cserjesi P, Olson EN. Dermo-1: a novel twist-related bHLH protein expressed in the developing dermis. Dev Biol. 1995;172:280–92. doi: 10.1006/dbio.1995.0023. [DOI] [PubMed] [Google Scholar]
- 31.Franco HL, Casasnovas J, Rodriguez-Medina JR, Cadilla CL. Redundant or separate entities?--roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011;39:1177–86. doi: 10.1093/nar/gkq890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drissi H, Luc Q, Shakoori R, Chuva De Sousa Lopes S, Choi JY, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, Van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–50. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 33.Soung do Y, Talebian L, Matheny CJ, Guzzo R, Speck ME, Lieberman JR, Speck NA, Drissi H. Runx1 dose-dependently regulates endochondral ossification during skeletal development and fracture healing. J Bone Miner Res. 2012;27:1585–97. doi: 10.1002/jbmr.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang A, Midura RJ, Vasanji A, Wang AJ, Hascall VC. Hyperglycemia diverts dividing osteoblastic precursor cells to an adipogenic pathway and induces synthesis of a hyaluronan matrix that is adhesive for monocytes. J Biol Chem. 2014;289:11410–20. doi: 10.1074/jbc.M113.541458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aronow MA, Gerstenfeld LC, Owen TA, Tassinari MS, Stein GS, Lian JB. Factors that promote progressive development of the osteoblast phenotype in cultured fetal rat calvaria cells. J Cell Physiol. 1990;143:213–21. doi: 10.1002/jcp.1041430203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.