Abstract
Across the span of the last 75+ years, technological and conceptual advances in genetics have found rapid implementation at the beginning of human life. From karyotype testing, to molecular cytogenetics, to gene panel testing and now whole exome and whole genome sequencing, each iterative expansion of our capability to acquire genetic data on the next generation has been implemented quickly in the clinical setting. In tandem, our continuously expanding ability to acquire large volumes of genetic data has generated its own challenges in terms of interpretation, clinical utility of the information, and concerns over privacy and discrimination; for the first time, we are faced with the possibility of having complete access to our genetic data from birth, if not shortly after conception. Here we discuss the evolution of the field towards this new reality and we consider the potentially far-reaching consequences and, at present, an unclear path towards developing best practices for implementation.
Congenital genetic disorders place a disproportionally high burden on families and the health care system and, in many instances, have long-term health ramifications. Structural birth defects alone, a significant fraction of which are underscored by genetic and genomic lesions, have consistently been the leading cause of mortality in the first postnatal year1. Importantly, although mortality in the first year of life has decreased by 50% over the past 30 years, there is little evidence of continued improvement; according to a 2007 census, birth defects alone accounted for 21% of all U.S. infant deaths1,2. At the same time, many of the infants with genetic and genomic lesions that now survive into childhood are in acute need of better prognostic and prophylactic paradigms that can anticipate health challenges that manifest later, even though these might appear to be unrelated from the clinical problems identified in early life1,2.
The significant contribution of genetic disease in young children is not restricted to structural defects present at birth. According to one census, 71% of the 4,224 children admitted to Cleveland’s Rainbow Babies and Children’s Hospital in 1996 had a disorder of genetic causality or susceptibility, while 96% of admissions due to chronic disorders were of genetic origin. $50 million of the $62 million charges were driven by genetic disorders3. It is obvious that these data are not representative of the contribution of genetics in the general population, since these measurements were derived from a specialty hospital. However, even in general hospitals not enriched for unusual or particularly challenging cases, 5–10% of admissions are driven by genetic defects4.
In other regions of the world, pediatric genetic disorders are an even more acute logistical challenge, for which current solutions have not been able to scale: in India, for example, it is estimated that birth defects prevalence varies from 6–7% of live births, and 70% of these defects are preventable5. Taken together, these epidemiological studies, daunting as they might appear at first, suggest that improvements in diagnosis and management of young children with pediatric genetic disorders would have a major impact across the globe.
Diagnosis and ultimately management of known genetic disorders can be considered broadly in two tiers: (1) population-level screening (that includes asymptomatic individuals) and (2) targeted analysis of individuals with suspected genetic disease. For both paradigms, progressive advances in technology have changed (improved, for the most part) analytic ability, while striving to balance the acute need to diagnose early with the need to protect privacy, control costs, and devise appropriate plans for interpreting and reporting ambiguous results. Here, we will summarize the state of the art for both population level and individual targeted approaches and we will discuss how the implementation of newer, and cheaper, sequencing paradigms have both accelerated our diagnostic ability and also presented us with new dilemmas about how to implement genetic screening at the population level during the early developmental stages encompassing the fetal, neonate, infant and childhood stages of life.
Population-based screening
Many countries and cultures have implemented genetic screening for a variety of purposes, typically targeting specific subsets of genetic disorders of concern to the local population. For example, a number of nations practice mandatory genetic testing for β-thalassemia carrier status6, while Rh factor blood typing (a phenotype-based genetic test) is likewise common in pregnant females7 because of the impact of these data to the clinical management of subsequent pregnancies. In some Orthodox Jewish communities, more extensive testing for common founder mutations has been implemented in an effort to reduce the burden of catastrophic, typically pediatric-period diagnosed genetic disorders such as Tay Sachs8.
The implementation of these tests tends to be decided on the relative benefit to the individual and to the population versus the risk of loss of privacy, emotional distress, and stigmatization9. In the developed world, restricting genetic tests to disorders for which diagnosis can lead to therapeutic intervention has been a key driver for shaping the targets of the most common type of population-based genetic screening: newborn screening for metabolic disorders.
The implementation of New Born Screening (NBS) 50 years ago changed the life of newborns that would otherwise perish10. NBS is a public health program targeting the early identification of infants at risk for developing conditions for which treatments are available. Beginning with a pioneer test to prevent newborns at risk from developing Phenylketonuria (PKU), a rare recessively inherited disorder that causes severe mental retardation, NBS currently includes a screening panel for the detection of 29 core and 25 secondary targeted diseases11. NBS, in addition to its tangible impact in improving outcomes, is also an exemplar of how harnessing appropriate technology can both expand the scope of tests and reduce time and cost. Early NBS on Guthrie blood spots required one test (blood spot) for one disorder and had a turnover of multiple days. In later years, the advent of mass spectrometry enabled the accurate measurement of metabolites relevant to a subset of metabolic disorders. More recently, miniaturization technologies, such as microfluidics, have raised the promise of decreasing cost and time further, while increasing sensitivity to measuring analytes12.
While there is consensus that NBS is beneficial and is saving lives with current applications, there is robust debate about how best to expand NBS. In addition to cost considerations, analytic sensitivity and specificity are an aspect that requires constant careful attention. For example, in 2006 the state of New York sought to identify infants at risk for developing Krabbe disease and began screening newborns for this condition13. Krabbe disease is a rare, devastating neurological disorder that affects 1:100,000 newborns for whom diagnosis is not straightforward, in part because enzyme activity of GALC and/or mutational analysis of the encoded gene do not reliably predict phenotype14. Furthermore, the only available treatment, hematopoietic stem cell transplantation15, has reported some effectiveness only when performed before clinical symptoms develop; the procedure alone carries a lethality as high as 10%13. Dilemmas similar to Krabbe disease will continue to arise as we expand newborn screening to disorders for which treatments are not readily available.
Targeted genetic testing
Differing from population-based screening, targeted genetic testing typically represents a multi-tiered approach offered either pre-symptomatically or post-symptomatically. Pre-symptomatic testing is offered to individuals with a known genetic burden and/or acute risk factors, such as familial disease burden. Although such testing is often thought of in the context of cancer susceptibility in adults (e.g. BRCA1/2 testing in women with a family history), prenatal testing for aneuploidies in cases of advanced maternal age or specific genetic/genomic lesions known to be present in other family members also fall under this category, with the list of single-gene tests offered expanding with the accelerating discovery of the molecular causes of Mendelian disorders16. By contrast, post-symptomatic genetic testing is now offered with increasing regularity to individuals suspected clinically to be affected by genetic disease, ranging from gene-specific to panel-specific to whole exome/genome testing, depending on the complexity of the clinical presentation17.
In either category, genetic testing has evolved and expanded as new technologies have become available. A classic exemplar is molecular cytogenetic testing, which traces its roots to chromosome studies in patients with Down syndrome. Soon after whole chromosome ploidies could be detected, G-banding staining of chromosomal spreads in the 1950s allowed the detection of large rearrangements in the genome, including multi Mb-long deletions and duplications, as well as inversions and balanced translocations. With the advent of molecular cytogenetics, including FISH (fluorescent in situ hybridization), array-based comparative genomic hybridization (aCGH)18 and, more recently, genotyping-based platforms that can also look at dosage changes the increase in speed and resolution, chromosome studies are becoming almost routine testing in cases of suspected genetic disorders in young children. These advances have been beneficial: pathogenic genomic lesions have been discovered at an increasing rate19, informing disease gene discovery and accelerating diagnosis.
Of note, the expanded capability of testing multiple genomic sites (from whole chromosomes to sub-MB deletions and duplications) is accompanied by the inherent increase in the generation of results of uncertain clinical significance. The interpretation of such data and their delivery to healthcare providers, patients and their families, remains a contentious issue that has become amplified further by the clinical implementation of whole exome and whole genome sequencing (WES and WGS respectively)20,21.
Whole exome/whole genome sequencing in the clinical and research setting
Since the completion of the first draft of the human genome in 2001, technological advances in massively parallel sequencing have been transformational in the delivery of genomic data to clinical care providers, transitioning the community from single-gene Sanger sequencing based tests to next-gen platforms. This transformation has been driven, in part, by extraordinary reduction in test costs and turnaround time. WES was introduced as a viable technology in 200522,23, with proof of principle application in genomic medicine reported four years later to identify the molecular cause of the rare recessive disorder, Miller syndrome24, paving the road to a new approach towards gene discovery and molecular diagnosis25. Since then, WES has been adopted into the clinic to identify genes responsible for rare genetics disorders in children and adults with suspected genetic disorders. The clinical application of WES remains non-uniform, primarily because of variable cost recovery models in various healthcare and insurance systems (public and private)26. However, as cost consideration becomes minimized, the expectation is that WES or whole genome sequencing (WGS) will become the standard of choice.
The a priori diagnostic success rates of WES/WGS and their potential impact on healthcare choices of patients and families will continue to drive testing choices. At the moment, both are modest but by no means negligible; WES studies of approximately 2,500 patients referred by physicians who suspected a genetic condition had a diagnostic rate of ~25%, which is markedly higher than previously reported by other traditional genetics methods such as karyotype analysis27,28. For specific subsets of disorders in which the genetic architecture is understood better, yields can reach 50%, as exemplified by studies of intellectual disability29,30,31. Both these numbers are sure to improve as our understanding of the genetic basis of disease continues to evolve.
In the neonatal setting, in addition to diagnostic yield, speed is also a significant consideration. In that context, WES/WGS implementation finds similarities with the evolution from Guthrie cards to mass spectrometry, the fundamental premise being that application of NGS as early as possible can have has high clinical impact. In one example, focusing on individuals in a Neonatal Intensive Care Unit (NICU), WGS and interpretation of genomic data was performed within a 50 hours period, providing a definitive or likely molecular diagnosis for four of five neonates enrolled in this study32. Another recent study included 30 neonates diagnosed by ultrasound with a diverse range of fetal abnormalities and it revealed a 10% detection rate of de novo mutations in causative genes, and 17% detection rate of likely causative genes that required further confirmatory analysis33. These detection rates are lower than those reported for older children and are probably hampered by analytic limitations including the fact that a) the clinical phenotype has not been fully manifested yet; and b) that a subset of patients might be affected by non-genetic causes. Nonetheless, it is reasonable to argue that even a 10–20% success rate is invaluable if it allows for an intervention window that would otherwise be nonexistent.
Analytical tools and in vivo model organism provide further mutational burden of proof
Given the amount of data produced by NGS, variant interpretation remains a major challenge. To facilitate the analysis and narrow down the list of candidate pathogenic sites, databases that contain frequent single nucleotide polymorphisms (SNPs) or recurrent mutations associated with human genetic disorders have been compiled. For example, HGMD34 and ClinVar35 are populated by curated mutational data from the literature and from Clinical genetic testing laboratories. In parallel, the Exome Variant (EVS)36 (6503 samples) and the 1000 Genomes37 database, provide aggregate data on the frequency of alleles found in various human populations. More recently, the Exome Aggregation Consortium (ExAC), populated by uniformly-called exome data from ~61,000 individuals, was created by investigators willing to share genetic data from different sequencing projects to the scientific community38. Although almost all of these samples are thought to be bereft of individuals with severe genetic disorders, they do contain data from individuals with psychiatric traits. As such, the presence, at low frequency, of candidate pathogenic variants remains difficult to interpret.
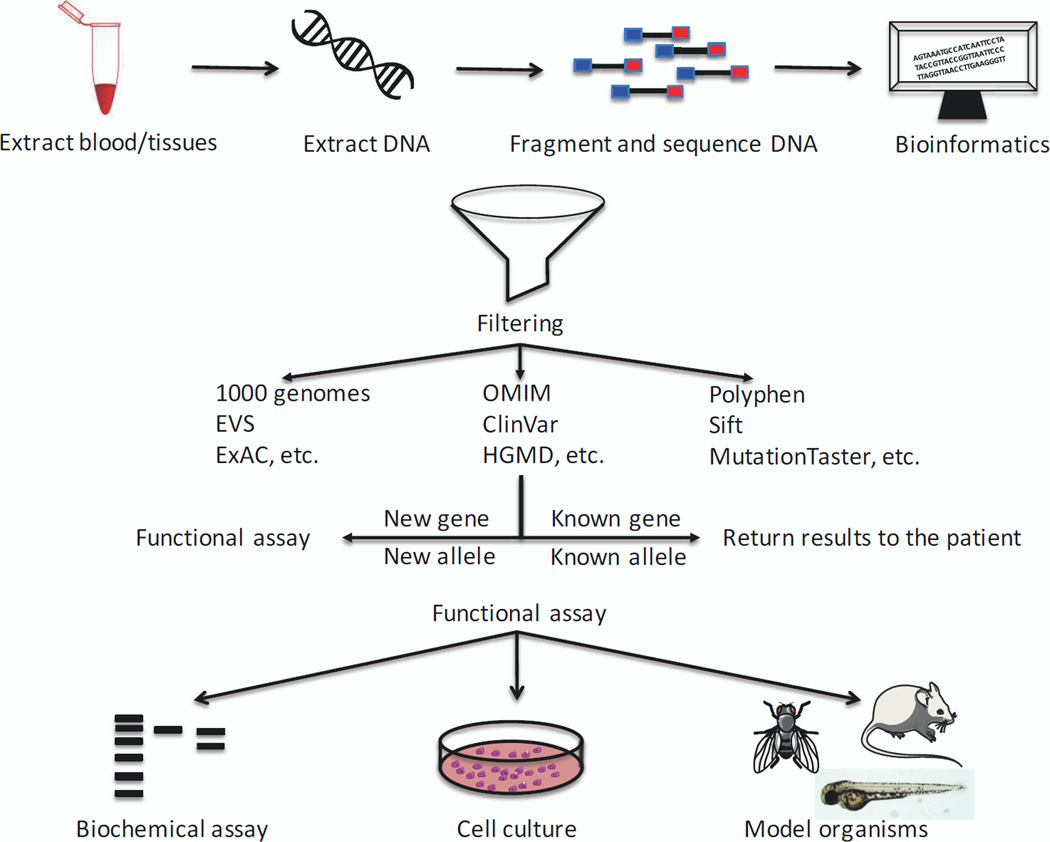
More broadly, although population frequency of candidate pathogenic alleles can offer potential clues to pathogenicity, as can evolutionary constraint, both make assumptions about a) the extent of phenotypic data available in database exomes; b) the genetic architecture of disease with regard to penetrance and expressivity; and c) the relevance of evolutionary constraint. All three can give rise to false negatives and, consequently, overfiltering. Ultimately, despite the continuous improvement of computational prediction tools such as Polyphen, SIFT, MutationTaster39–42, to name but a few commonly-used ones, direct biological evidence of pathogenicity remain irreplaceable. In that context, model organism such as flies, worm, zebrafish and mice have become invaluable tools for testing human mutations consequently contributing substantially to the mutational data burden of proof43 (Fig. 1).
Figure 1. Next-generation sequence workflow complemented with functional studies.
Upon consent of family members, collection of either blood or tissue is performed, followed by extraction of DNA. DNA is then fragmented and a sequence library is subjected to massively parallel sequencing. Resulting reads are processed and curated human genetic database to identify candidate variants that are/are not associated with human genetics diseases. The identification of genes or alleles not previously implicated in human genetic disease requires functional assays to test variants pathogenicity.
This process is exemplified by the case of a 15-month-old boy who presented with signs consistent with a Crohn disease-like illness who was not responding to traditional therapy, exome sequencing identified over 10,000 variants. Further filtration to identify rare, non-synonymous variants predicted to cause alterations in immune phenotypes narrowed the list of candidates to one novel, hemizygous missense mutation in the X-linked inhibitor of apoptosis gene with a known central role in the proinflammatory response and bacterial sensing through NOD signaling. The likely causal relationship between the child’s phenotype and this particular variant was borne out with functional testing, which demonstrated presence of the mutation led to an increased susceptibility to activation-induced cell death and defective responsiveness to NOD2 ligands, consistent with loss of normal protein function. Based on this finding, the child had an allogeneic hematopoietic progenitor cell transplant which is the recommended treatment for X-linked inhibitor of apoptosis deficiency44. Functional testing of novel variants in genes previously not suspected to be associated with a child’s abnormal phenotype will possibly lead to enhanced understanding of pathophysiology and developmental biology of the phenotype, and ideally, to strategies to treat the current phenotype and the predict and prevent disease manifestations that are predicted to occur later in life.
Who should be sequenced at birth?
One of the key questions facing the medical genomics, pediatric and neonatal intensive care communities is whether NGS should become a component of NBS. Indeed, given the evolution of the technology, NGS is now beginning to be implemented prenatally through the use of maternal-fetal testing for aneuploidies25. The purpose of NBS is to predict severe health conditions for which there are available treatments, and by doing so to improve the child’s (and hopefully the family’s) quality of life. NGS embraces the same concept as NBS: to be predictable, preventive, and personalized. Still, despite recent technological advances and decline of cost to perform WES/WGS, this cost does not reflect the “real cost” of clinical interpretation of genomic data, clinical follow-up, and periodical revision of the data45.
The question remains: who should be sequenced at birth? A recent survey showed that 74% of parents have an overall interest in having newborns undergo WES/WGS as long as it provides accurate information to prevent a child from developing illnesses46. However, before it becomes universal, it will most likely be primarily used for infants born with detectable anomalies (structural defects or overt neurological conditions are likely to be most common) and will therefore be a test triggered by evidence of pathology, not as a screen for asymptomatic infants. As discussed earlier, many clinics have already adopted this technique to determine the cause of pediatric disorders. While potentially providing a diagnosis for these children, WES/WGS can also uncover clinically important secondary findings, i.e. genetic findings unrelated to the primary indication of sequencing. In fact, for every patient that undergoes WES/WGS, the American College of Medical Genetics and Genomics (ACMG) recommends to report findings on an additional list of 56 clinically “actionable” genes47, a list that is sure to evolve as our understanding of causality and patho-mechanism continues to improve. Because the quality of clinical exome data may be suboptimal for certain regions of the exome depending on the platform and software program that perform alignment and base calling used, returning these 56 “actionable genes,” potentially false positive or false negatives, can have irreversible implications on someone’s life, especially infants48,49.
What history has taught us with regard to the implementation of genetic/genomic technologies in the newborn setting is that NGS will, inevitably, become applied in NBS. Such an event will represent a step-increase in our approach to pre-symptomatic genomic screening; it will lead to fundamentally different opportunities to promote early detection, and will offer the opportunity of better management and effective treatment throughout someone’s life, including late-onset disorders. At the same time, such approaches will also expose individuals to risks of information they might not wish to own, to abuse of genetic data by third parties and, if employed as a standard screen, with de facto threaten free choice in terms of accessing this type of information. Importantly, NGS-based NBS would mean that, by definition, we would start screening sites in the genome that are not amenable to treatment; even if such sights were not interpreted actively by the NBS program, such data will unavoidably become available to families. Before changes in policy are implemented, objective data are needed to inform what will surely become a polarized debate in the medical professional and advocacy communities.
An ongoing pilot study funded by the National Institute of Child Health and Development (NICHD) intends to study the effects of implementing NGS into NBS. Perhaps once such studies have been completed we will have a better understanding on the implications of this technology in the life of every newborn and we can then apply to others safely.
Acknowledgments
We thank Erica Davis for her critical review of the manuscript. This work was supported by P50DK096415 (NK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mathews TJ, MacDorman MF. Infant mortality statistics from the 2006 period linked birth/infant death data set. National vital statistics reports : from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2010 Apr 30;58(17):1–31. [PubMed] [Google Scholar]
- 2.Singh GKvD P. Infant Mortality in the United States, 1935–2007: Over Seven Decades of Progress and Disparities. US Department of Health and Human Services, H.R.a.S.A. 2010 [Google Scholar]
- 3.McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children's hospital. American journal of human genetics. 2004 Jan;74(1):121–127. doi: 10.1086/381053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Science translational medicine. 2011 Jan 12;3(65):65ra64. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma R. Birth defects in India: Hidden truth, need for urgent attention. Indian journal of human genetics. 2013 Apr;19(2):125–129. doi: 10.4103/0971-6866.116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cousens NE, Gaff CL, Metcalfe SA, Delatycki MB. Carrier screening for beta-thalassaemia: a review of international practice. European journal of human genetics : EJHG. 2010 Oct;18(10):1077–1083. doi: 10.1038/ejhg.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avent ND. RHD genotyping from maternal plasma: guidelines and technical challenges. Methods in molecular biology. 2008;444:185–201. doi: 10.1007/978-1-59745-066-9_14. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan F. Tay-Sachs disease carrier screening: a model for prevention of genetic disease. Genetic testing. 1998;2(4):271–292. doi: 10.1089/gte.1998.2.271. [DOI] [PubMed] [Google Scholar]
- 9.de Jong A, Dondorp WJ, Frints SG, de Die-Smulders CE, de Wert GM. Advances in prenatal screening: the ethical dimension. Nature reviews. Genetics. 2011 Sep;12(9):657–663. doi: 10.1038/nrg3036. [DOI] [PubMed] [Google Scholar]
- 10.Therrell BL, Adams J. Newborn screening in North America. Journal of inherited metabolic disease. 2007 Aug;30(4):447–465. doi: 10.1007/s10545-007-0690-z. [DOI] [PubMed] [Google Scholar]
- 11.McCabe LL, McCabe ER. Expanded newborn screening: implications for genomic medicine. Annual review of medicine. 2008;59:163–175. doi: 10.1146/annurev.med.59.110106.132016. [DOI] [PubMed] [Google Scholar]
- 12.Millington DS, Sista R, Eckhardt A, et al. Digital microfluidics: a future technology in the newborn screening laboratory? Seminars in perinatology. 2010 Apr;34(2):163–169. doi: 10.1053/j.semperi.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffner PK, Caggana M, Orsini JJ, et al. Newborn screening for Krabbe disease: the New York State model. Pediatric neurology. 2009 Apr;40(4):245–252. doi: 10.1016/j.pediatrneurol.2008.11.010. discussion 253–245. [DOI] [PubMed] [Google Scholar]
- 14.Wenger DA, Rafi MA, Luzi P. Molecular genetics of Krabbe disease (globoid cell leukodystrophy): diagnostic and clinical implications. Human mutation. 1997;10(4):268–279. doi: 10.1002/(SICI)1098-1004(1997)10:4<268::AID-HUMU2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 15.Escolar ML, Poe MD, Provenzale JM, et al. Transplantation of umbilical-cord blood in babies with infantile Krabbe's disease. The New England journal of medicine. 2005 May 19;352(20):2069–2081. doi: 10.1056/NEJMoa042604. [DOI] [PubMed] [Google Scholar]
- 16.Bodurtha J, Strauss JF., 3rd Genomics and perinatal care. The New England journal of medicine. 2012 Jan 5;366(1):64–73. doi: 10.1056/NEJMra1105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saudi Mendeliome G. Comprehensive gene panels provide advantages over clinical exome sequencing for Mendelian diseases. Genome biology. 2015;16:134. doi: 10.1186/s13059-015-0693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crotwell PL, Hoyme HE. Advances in whole-genome genetic testing: from chromosomes to microarrays. Current problems in pediatric and adolescent health care. 2012 Mar;42(3):47–73. doi: 10.1016/j.cppeds.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Iyer J, Girirajan S. Gene discovery and functional assessment of rare copy-number variants in neurodevelopmental disorders. Briefings in functional genomics. 2015 May 13; doi: 10.1093/bfgp/elv018. [DOI] [PubMed] [Google Scholar]
- 20.Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. Jama. 2014 Mar 12;311(10):1035–1045. doi: 10.1001/jama.2014.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klitzman R, Appelbaum PS, Chung W. Return of secondary genomic findings vs patient autonomy: implications for medical care. Jama. 2013 Jul 24;310(4):369–370. doi: 10.1001/jama.2013.41709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shendure J, Ji H. Next-generation DNA sequencing. Nature biotechnology. 2008 Oct;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 23.Metzker ML. Sequencing technologies - the next generation. Nature reviews. Genetics. 2010 Jan;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 24.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nature genetics. 2010 Jan;42(1):30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009 Sep 10;461(7261):272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Ruivenkamp CA, Hoffer MJ, et al. Next-generation diagnostics: gene panel, exome, or whole genome? Human mutation. 2015 Jun;36(6):648–655. doi: 10.1002/humu.22783. [DOI] [PubMed] [Google Scholar]
- 27.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. The New England journal of medicine. 2013 Oct 17;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. Jama. 2014 Nov 12;312(18):1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014 Jul 17;511(7509):344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 30.Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Science translational medicine. 2014 Dec 3;6(265):265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor JC, Martin HC, Lise S, et al. Factors influencing success of clinical genome sequencing across a broad spectrum of disorders. Nature genetics. 2015 Jul;47(7):717–726. doi: 10.1038/ng.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Science translational medicine. 2012 Oct 3;4(154):154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carss KJ, Hillman SC, Parthiban V, et al. Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Human molecular genetics. 2014 Jun 15;23(12):3269–3277. doi: 10.1093/hmg/ddu038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenson PD, Ball EV, Mort M, et al. Human Gene Mutation Database (HGMD): 2003 update. Human mutation. 2003 Jun;21(6):577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 35.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic acids research. 2014 Jan;42:D980–D985. doi: 10.1093/nar/gkt1113. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: [Accessed September, 2015]. http://evs.gs.washington.edu/EVS/ [Google Scholar]
- 37.Genomes Project C. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Exome Aggregation Consortium (ExAC) Cambridge, MA: [Accessed September, 2015]. http://exac.broadinstitute.org. [Google Scholar]
- 39.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nature methods. 2010 Aug;7(8):575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 40.Flanagan SE, Patch AM, Ellard S. Using SIFT and PolyPhen to predict loss-of-function and gain-of-function mutations. Genetic testing and molecular biomarkers. 2010 Aug;14(4):533–537. doi: 10.1089/gtmb.2010.0036. [DOI] [PubMed] [Google Scholar]
- 41.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nature methods. 2010 Apr;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 43.Davis EE, Frangakis S, Katsanis N. Interpreting human genetic variation with in vivo zebrafish assays. Biochimica et biophysica acta. 2014 Oct;1842(10):1960–1970. doi: 10.1016/j.bbadis.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genetics in medicine: official journal of the American College of Medical Genetics. 2011 Mar;13(3):255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 45.Beckmann JS. Can We Afford to Sequence Every Newborn Baby's Genome? Human mutation. 2014 Dec 25; doi: 10.1002/humu.22748. [DOI] [PubMed] [Google Scholar]
- 46.Goldenberg AJ, Dodson DS, Davis MM, Tarini BA. Parents' interest in whole-genome sequencing of newborns. Genetics in medicine: official journal of the American College of Medical Genetics. 2014 Jan;16(1):78–84. doi: 10.1038/gim.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Directors ABo. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genetics in medicine: official journal of the American College of Medical Genetics. 2015 Jan;17(1):68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 48.Park JY, Clark P, Londin E, Sponziello M, Kricka LJ, Fortina P. Clinical exome performance for reporting secondary genetic findings. Clinical chemistry. 2015 Jan;61(1):213–220. doi: 10.1373/clinchem.2014.231456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark MJ, Chen R, Lam HY, et al. Performance comparison of exome DNA sequencing technologies. Nature biotechnology. 2011 Oct;29(10):908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]