Abstract
Polymorphisms in chemokine receptors, serving as HIV co-receptors, and their ligands are among the well-known host genetic factors associated with susceptibility to HIV infection and/or disease progression. Papua New Guinea (PNG) has one of the highest adult HIV prevalences in the Asia-Pacific region. However, information regarding the distribution of polymorphisms in chemokine receptor (CCR5, CCR2) and chemokine (CXCL12) genes in PNG is very limited. In this study, we genotyped a total of nine CCR2-CCR5 polymorphisms, including CCR2 190G>A, CCR5 −2459G>A and Δ32, and CXCL12 801G>A in PNG (n = 258), North America (n = 184), and five countries in West Africa (n = 178). Using this data, we determined previously characterized CCR5 haplotypes. In addition, based on the previously reported associations of CCR2 190, CCR5 −2459, CCR5 open reading frame, and CXCL12 801 genotypes with HIV acquisition and/or disease progression, we calculated composite full risk scores, considering both protective as well as susceptibility effects of the CXCL12 801 AA genotype. We observed a very high frequency of the CCR5 −2459A allele (0.98) in the PNG population, which together with the absence of Δ32 resulted in a very high frequency of the HHE haplotype (0.92). These frequencies were significantly higher than in any other population (all P-values < 0.001). Regardless of whether we considered the CXCL12 801 AA genotype protective or susceptible, the risk scores were significantly higher in the PNG population compared with any other population (all P-values < 0.001). The results of this study provide new insights regarding CCR5 variation in the PNG population, and suggest that the collective variation in CCR2, CCR5, and CXCL12 may increase the risk of HIV/AIDS in a large majority of Papua New Guineans.
Keywords: CCR2, CCR5, CXCL12, HIV, Papua New Guinea
1. Introduction
Papua New Guinea (PNG), an Oceania country located ~160 km north of Australia, has one of the highest prevalences of HIV and sexually transmitted infections in the Asia-Pacific region (Coghlan et al., 2011; Vallely et al., 2010; Vallely et al., 2014). In a meta-analysis of community-based studies (n = 9), pooled HIV prevalence was 1.8% in men; 2.6% in women; and 11.8% among female sex workers (Vallely et al., 2010). In a recent study conducted at two sexual health clinics, HIV prevalence in men and women combined was 3.3% (Vallely et al., 2014). Based on the most recent HIV estimates (Global AIDS Report 2012, Papua New Guinea), the national prevalence of HIV among the adult population 15–49 years is 0.8% (0.55–0.94%) in 2010-2011, and is expected to increase to 1.0% by 2020, based on current model. HIV prevalence among pregnant women 15–24 years attending antenatal care clinics was 0.6% in 2011, indicating that there is HIV transmission occurring among sexually active women in this age group and their heterosexual partners. Genetic characterization of HIV-1 in PNG shows that the predominant subtype is C (>90%), and the remaining is subtype B (Anyiwo et al., 2010; Ryan et al., 2007).
Limited data exists from PNG regarding the distribution of host genetic markers known to be associated with clinical outcomes of HIV/AIDS (Clark and Dean, 2004; Martinson et al., 2000; Su et al., 1999; Su et al., 2000). Despite small sample sizes (n = 21–96), these studies provide important information regarding the distribution of a 32-base pair (bp) deletion (Δ32, rs333) in the open reading frame (ORF) of chemokine (C-C motif) receptor 5 (CCR5), a single nucleotide polymorphism (SNP, 190G>A [Val64Ile], rs1799864) in the ORF of chemokine receptor CCR2, and a SNP (801G>A, rs1801157) in the 3’-untranslated region (3’-UTR) of chemokine (C-X-C motif) 12 (CXCL12 [stromal cell-derived factor 1, SDF-1]) in PNG. CXCL12 is the only natural ligand for chemokine receptor CXCR4. A variety of studies conducted outside PNG have examined the associations between these polymorphisms and HIV infection/disease progression, and have revealed that the CCR5 Δ32 (Dean et al., 1996; Martin et al., 1998; Zimmerman et al., 1997) and the CCR2 190A (Hendel et al., 1998; Ioannidis et al., 2001; Kostrikis et al., 1998; Mulherin et al., 2003; Mummidi et al., 1998; Passam et al., 2005; Smith et al., 1997) alleles were associated with protection against HIV infection and/or delayed disease progression, compared with the CCR5 ORF wild type (wt) and the CCR2 190G alleles respectively. However, the association between CXCL12 801G>A and HIV infection/disease progression is complex; the mutant allele, 801A, may be protective (Hendel et al., 1998; Passam et al., 2005; Tiensiwakul, 2004; Winkler et al., 1998) as well as susceptible (Daar et al., 2005; Mummidi et al., 1998; Petersen et al., 2005).
Among the studies that provided information regarding the distribution of CCR5 Δ32, CCR2 190G>A, and CXCL12 801G>A in PNG (Clark and Dean, 2004; Martinson et al., 2000; Su et al., 1999; Su et al., 2000), only one study included the CCR5 promoter SNP −2459G>A (59029G>A/303G>A, rs1799987) (Clark and Dean, 2004). In studies conducted elsewhere, the −2459G allele was associated with delayed HIV disease progression (Martin et al., 1998; McDermott et al., 1998). Furthermore, two of the PNG studies computed the relative hazard (RH) values to evaluate the risk of AIDS onset on the basis of two- and three-locus genotypes (Su et al., 1999; Su et al., 2000). However, those RH values were computed considering only the protective effects of the CXCL12 801 AA genotype (Winkler et al., 1998), and not the susceptibility effects of the genotype. Based on this limited assessment, PNG populations had the lowest predicted RH values compared with worldwide populations, indicating potentially the highest protection from AIDS onset, or even HIV infection (Su et al., 1999; Su et al., 2000).
The main aim of the present study was to further understand how CCR2, CCR5, and CXCL12 variation may influence the risk of HIV infection/disease progression in PNG. For this aim, we genotyped a total of nine CCR2-CCR5 polymorphisms, including CCR5 −2459G>A, in two populations from PNG, two major racial groups in North America, and in populations from five countries in West Africa. Using this data, we determined previously characterized CCR5 haplotypes (Gonzalez et al., 1999; Mummidi et al., 2000). We also genotyped CXCL12 801G>A in all study populations. Based on the previously reported associations of CCR2 190, CCR5 −2459, CCR5 ORF, and CXCL12 801 genotypes with HIV acquisition and/or disease progression, we calculated composite full risk scores, considering both protective as well as susceptibility effects of the CXCL12 801 AA genotype. Together, these results provide new insights regarding CCR5 variation in the PNG population, and suggest that the collective variation in CCR2, CCR5, and CXCL12 may elevate the risk of HIV/AIDS as opposed to conferring protection indicated by previous studies.
2. Materials and methods
2.1. Study populations and genomic DNA extraction
The study populations and sample collection procedures have been previously described (Kasehagen et al., 2007; Mehlotra et al., 2002; Mehlotra et al., 2000; Zimmerman et al., 1992; Zimmerman et al., 1999). These samples were collected as parts of malaria epidemiological studies in PNG (Kasehagen et al., 2007; Mehlotra et al., 2002; Mehlotra et al., 2000) and onchocerciasis epidemiological study in West Africa (Zimmerman et al., 1992). Blood samples were collected from Papua New Guineans (n = 258) from Dreikikir (Mehlotra et al., 2000) and the Wosera (Kasehagen et al., 2007; Mehlotra et al., 2002), both areas in the East Sepik Province, and from healthy, adult North American random blood donors (n = 184) belonging to two major racial groups: Caucasian (CA) and African American (AFA) (Zimmerman et al., 1999). DNA samples were collected from West Africans (n = 178) from five countries: Senegal, Guinea, Sierra Leone, Ivory Coast, and Ghana (Zimmerman et al., 1992). The number of samples in each subgroup is provided in Table 1. The PNG samples were collected from individuals speaking Urat (Dreikikir) or Abelam (the Wosera), which belong to Torricelli language family and Sepik language family respectively. The race of the North American individuals was self-identified. All West African samples were collected from the West Atlantic subgroup of the Niger-Congo language family. No information is available regarding HIV exposure and infection status of our study populations. All samples were collected under protocols including the procedures for informed consent approved by the corresponding institutional review boards (UHCMC IRB 08-03-33; PNGIMR IRB 0801; PNG MRAC 07.28).
Table 1.
Number of individuals in populations/subgroups.
| Population | Subgroup | n |
|---|---|---|
| Papua New Guinean | 258 | |
| Dreikikir | 173 | |
| The Wosera | 85 | |
| North American | 184 | |
| Caucasian | 89 | |
| African American | 95 | |
| West African | 178 | |
| Senegal | 9 | |
| Guinea | 35 | |
| Sierra Leone | 53 | |
| Ivory Coast | 45 | |
| Ghana | 36 |
Genomic DNA was extracted from the PNG and North American blood samples using the QIAamp® 96 DNA blood kit (QIAGEN, Valencia, CA).
2.2. Polymerase chain reaction (PCR)
PCR primers and conditions to selectively amplify CCR2 ORF (327 bp), CCR5 promoter (1118 bp), and CCR5 ORF (312 [wt] or 280 [Δ32] bp) regions have been described elsewhere (Mehlotra et al., 2011). Previously described primers were used to amplify the CXCL12 region (301 bp) harboring 801G>A (Passam et al., 2005) under the following amplification conditions: 95°C 2 min, 95°C 30 sec 64°C 30 sec 72°C 30 sec (39×), 72°C 3 min. The PCR buffer and method used to perform agarose gel electrophoresis have been described elsewhere (Mehlotra et al., 2011).
2.3. SNP genotyping and CCR5 haplotype determination
After PCR amplification, genotyping of CCR2 190G>A and CCR5 −2459G>A was performed using a high-throughput, oligonucleotide ligation detection reaction-fluorescent microsphere assay (LDR-FMA) on the Bio-Plex multiplex suspension array system (Bio-Rad Laboratories, Hercules, CA), as described elsewhere (Mehlotra et al., 2011). Using the same assay, we performed genotyping of six other CCR5 promoter (−2773A>G [rs2856758], −2554G>T [rs2734648], −2135T>C [rs1799988], −2132C>T [rs41469351], −2086A>G [rs1800023], and −1835C>T [rs1800024]) and CXCL12 801G>A SNPs. All seven sets of LDR primer sequences and LDR conditions for CCR5 and CXCL12 SNP genotyping are provided in the supplementary Table S1. The mean and 95% confidence interval (CI) of log-transformed fluorescent values corresponding to each genotype are presented in the supplementary Table S2.
The CCR5 haplotype nomenclature system has been previously described (Gonzalez et al., 1999; Mummidi et al., 2000) (Table S3). Using a total of nine CCR2-CCR5 polymorphisms, haplotypes could be organized into seven evolutionarily distinct human haplogroups (HH) that are designated HHA, -B, -C, -D, -E, -F (F*1 and F*2), and -G (G*1 and G*2). We followed this nomenclature system to determine CCR5 haplotypes in study populations.
2.4. CCR5 promoter sequencing
Sanger sequencing of CCR5 promoter amplicons from the PNG (n = 87; Dreikikir [n = 63] and the Wosera [n = 24]), CA (n = 12), AFA (n = 14), and West African (n = 15) samples was performed using a modified Applied Biosystems BigDye® Terminator v3.1 Cycle sequencing kit protocol. Sequencing reactions were performed on ABI 3730 DNA Analyzer system (Applied Biosystems, Foster City, CA). Sequences were analyzed using Sequencher v4.6 (Gene Codes Corporation, Ann Arbor, MI).
2.5. Genotype and composite risk scores
We assigned a risk score of 2, 1, or 0 to each of the CCR2 190, CCR5 −2459, CCR5 ORF, and CXCL12 801 genotypes (Table 2). These arbitrary risk scores are based on the previously reported associations of these genotypes with HIV acquisition and/or disease progression. We first assigned these risk scores to the CCR5 −2459 and CCR5 ORF genotypes, where a score of 2 denotes susceptibility, 1 denotes partial protection, and 0 denotes protection. It is important to note that the same risk scores assigned to genotypes of different polymorphisms should not be interpreted as being associated with the same effects. For example, the effect of a risk score of 2, assigned to CCR5 −2459 AA, is not the same as the effect of a risk score of 2 assigned to CCR5 ORF wtwt. However, compared with the GG and Δ32Δ32 genotypes, respectively, both were considered susceptible genotypes (Dean et al., 1996; Martin et al., 1998; McDermott et al., 1998; Zimmerman et al., 1997) and were therefore assigned a risk score of 2.
Table 2.
Risk scores associated with various genotypes.
| Risk score | CCR2 190 |
CCR5 −2459 |
CCR5 ORF |
CXCL12 801 (I) |
CXCL12 801 (II) |
CXCL12 801 (III) |
CXCL12 801 (IV) |
|---|---|---|---|---|---|---|---|
| 2 | - | AAa | wtwta | - | - | - | - |
| 1 | GGb | GAc | wtΔ32c | GGb, GAb | GGb | AAa | GAa, AAa |
| 0 | GAd, AAd | GGd | Δ32Δ32d | AAd | GAd, AAd | GGb, GAb | GGb |
Roman numerals I, II, III, and IV represent four different scenarios, described in Materials and methods.
Susceptibility/risk
Reference genotype
Partial protection
Protection
Most previous studies have shown a protective effect of the CCR2 190A allele (GA+AA compared with GG) on HIV disease progression (Hendel et al., 1998; Ioannidis et al., 2001; Kostrikis et al., 1998; Mulherin et al., 2003; Mummidi et al., 1998; Passam et al., 2005; Smith et al., 1997). We therefore assigned a risk score of 0 to the GA and AA genotypes, and 1 to the GG genotype, which was used as reference. The findings regarding the association of CXCL12 801 genotypes with HIV infection/disease progression are complex, and present four different scenarios: AA is protective compared with GG+GA (scenario I) (Hendel et al., 1998; Winkler et al., 1998); GA+AA is protective compared with GG (scenario II) (Passam et al., 2005); AA is susceptible compared with GG+GA (scenario III) (Mummidi et al., 1998); and GA+AA is susceptible compared with GG (scenario IV) (Daar et al., 2005; Petersen et al., 2005). Accordingly, we assigned a risk score of 1 (susceptibility) or 0 (protection) to these genotypes (Table 2).
After assigning a risk score to each genotype, we first calculated composite partial risk scores, where risk scores of CCR2 190, CCR5 ORF, and CXCL12 801 (only scenario I or II) genotypes were included. This analysis is similar to the one performed previously (Su et al., 2000), as it does not include CCR5 −2459 genotypes and considers only the protective effects of the CXCL12 801 AA or GA+AA genotype. Composite partial risk scores ranged from 2 to 4, and some examples are provided in the supplementary Table S4. We then calculated composite full risk scores, where CCR5 −2459 genotypes were included together with CCR2 190, CCR5 ORF, and CXCL12 801 genotypes. All four scenarios of CXCL12 801 genotypes (I, II, III, or IV) were considered. Composite full risk scores ranged from 3 to 6. Finally, given the complexities of CXCL12 801 genotype-phenotype associations, we calculated a composite simple risk score of each individual, which does not include CXCL12 801 genotype. Thus, this is the sum of risk scores assigned to CCR2 190, CCR5 −2459, and CCR5 ORF genotypes of that individual. Composite simple risk scores ranged from 3 to 5. For simplification, we defined composite simple risk scores 3, 4, and 5 as low, medium, and high risk respectively.
2.6. Statistical analysis
From the genotype data, CCR2, CCR5, and CXCL12 allele frequencies were calculated and the Hardy-Weinberg exact test was performed for each population using GenePop (http://genepop.curtin.edu.au/). An online 2 × 2 contingency table for Fisher’s exact test (http://www.langsrud.com/fisher.htm) was used to calculate differences in allele and haplotype frequencies between populations, and a 2-tailed P < 0.05 was considered significant.
Arlequin v3.11 (www.cmpg.unibe.ch/software/arlequin3/) was used to compute CCR5 promoter haplotype diversity (H; mean value ± standard error [S.E.]) and pairwise population differentiation (PA-Fst) based on the SNP genotype as well as nucleotide sequence data. Both H and Fst values range from 0 (no diversity, no differentiation) to 1 (every individual has a different haplotype, fixed difference between populations).
Distributions of composite risk scores were compared to find differences among as well as between study populations (PNG, CA, AFA, and West African). First, the composite risk score of each population was analyzed using Bartlett’s test of the homogeneity of variances. If this test showed heterogeneity of the variances, then non-parametric tests, robust to both non-homogeneity of variances as well as unequal sample sizes, were used. To find differences among populations using a non-parametric method, the Kruskal-Wallis test was used, and if a significant difference was detected, then the Games-Howell pairwise comparison test was used. Composite partial risk scores (both scenarios I and II), composite full risk scores (scenarios II and IV), and composite simple risk scores were compared using this approach. On the other hand, if Bartlett’s test showed homogeneity of variances, then a simple one-way ANOVA and Tukey’s Honestly Significant Difference (HSD) test were used. Composite full risk scores (scenarios I and III) were compared using this approach. All these analyses were conducted in R (http://www.r-project.org/), and a P < 0.05 was considered significant.
3. Results
3.1. CCR2, CCR5, and CXCL12 allele frequencies
Distribution of CCR2, CCR5, and CXCL12 allele frequencies in the PNG, North American, and West African populations is presented in Table 3. In the PNG populations, CCR2 190G>A had a low to moderate prevalence, many of the CCR5 promoter SNPs were rare or absent, CCR5 Δ32 was absent, whereas CCR5 −2459G>A and CXCL12 801G>A were highly prevalent. The CCR5 −2459A (0.98, cumulative) and CXCL12 801A (0.64, cumulative) allele frequencies in the PNG populations were significantly higher than in any other population (P-values < 0.001).
Table 3.
Distribution of CCR2, CCR5, and CXCL12 allele frequencies in various populations.
| Population |
CCR2 190G>A |
CCR5 −2733A>G |
CCR5 −2554G>T |
CCR5 −2459G>A |
CCR5 −2135T>C |
CCR5 −2132C>T |
CCR5 −2086A>G |
CCR5 −1835C>T |
CCR5 Δ32 |
CXCL12 801G>A |
|---|---|---|---|---|---|---|---|---|---|---|
| f(G)a | f(A)a | f(G)a | f(G)a | f(T)a | f(C)a | f(A)a | f(C)a | f(Δ32) | f(G) | |
| Papua New Guinean | ||||||||||
| Dreikikir | 0.95 | 1 | 0.98 | 0.02 | 0.02 | 1 | 0.98 | 0.95 | 0 | 0.35 |
| The Wosera | 0.89 | 1 | 0.99 | 0.01 | 0.01 | 1 | 0.99 | 0.89 | 0 | 0.38 |
| Cumulative | 0.93 | 1 | 0.98 | 0.02 | 0.02 | 1 | 0.98 | 0.93 | 0 | 0.36 |
| Caucasian | 0.95 | 0.83 | 0.66 | 0.41 | 0.41 | 1 | 0.66 | 0.94 | 0.10 | 0.78 |
| African American | 0.85 | 0.94 | 0.65 | 0.55 | 0.55 | 0.81 | 0.84 | 0.83 | 0.02 | 0.94 |
| West African | ||||||||||
| Senegal | 0.83 | 0.89 | 0.56 | 0.67 | 0.67 | 0.72 | 0.83 | 0.83 | 0 | 0.95 |
| Guinea | 0.88 | 0.99 | 0.67 | 0.67 | 0.67 | 0.72 | 0.95 | 0.86 | 0 | 0.99 |
| Sierra Leone | 0.96 | 0.94 | 0.66 | 0.71 | 0.71 | 0.73 | 0.94 | 0.88 | 0 | 0.94 |
| Ivory Coast | 0.89 | 0.96 | 0.61 | 0.60 | 0.60 | 0.70 | 0.91 | 0.80 | 0 | 0.98 |
| Ghana | 0.81 | 0.93 | 0.71 | 0.50 | 0.50 | 0.79 | 0.93 | 0.78 | 0 | 0.96 |
| Cumulative | 0.89 | 0.95 | 0.65 | 0.62 | 0.63 | 0.73 | 0.93 | 0.84 | 0 | 0.96 |
frequency (allele, present in HHA haplotype [Table S3])
In addition, inter-population differences in allele frequencies were observed. For example, CCR5 −2132C>T was highly prevalent in the populations of African ancestry, whereas Δ32 was prevalent in the CA population. The CA and AFA/West African populations were significantly different at CCR5 −2733A>G, −2086A>G, −1835C>T, and Δ32, and CXCL12 801G>A (P-values < 0.05).
We analyzed all polymorphisms among West African samples for each represented country. The cumulative allele frequencies in West Africans were largely comparable to those in AFA. The relatively small number of samples (varying from 9 to 53) from each country limited allele frequency comparisons. In subsequent analyses, all West African samples were analyzed as one population.
We calculated expected genotype numbers for each polymorphism in the PNG (two populations), North American (two racial groups), and West African (one population) populations. For all polymorphisms analyzed, the expected genotype numbers did not differ significantly from the observed genotype numbers in any of the populations (data not shown). Overall, these allele frequency results are in agreement with those previously reported for comparable populations (Clark and Dean, 2004; Gonzalez et al., 1999; Gonzalez et al., 2001; Martinson et al., 2000; Su et al., 1999; Su et al., 2000).
3.2. CCR5 haplotype frequencies and diplotype profiles
Based on a total of nine CCR2-CCR5 polymorphisms, we determined CCR5 haplotypes in study populations as previously described (Gonzalez et al., 1999; Mummidi et al., 2000). For this and subsequent analyses, we combined the two PNG populations. The haplotype frequencies are presented in Table 4. As expected from the allele frequency data (Table 3), the PNG population had the highest frequency of HHE (0.92), which was significantly higher than in any other population (P-values < 0.001). HHB was rare, and inter-population differences in other haplotype frequencies were observed. For example, HHD was highly prevalent in the populations of African ancestry, whereas Δ32-carrying HHG*2 was prevalent in the CA population. The frequencies of HHA and HHC were significantly different between the CA and AFA/West African populations (P-values < 0.001).
Table 4.
CCR5 haplotype frequencies in various populations.
| Population | HHA | HHB | HHC | HHD | HHE | HHF*1 | HHF*2 | HHG*1 | HHG*2 |
|---|---|---|---|---|---|---|---|---|---|
| Papua New Guinean | 0 | 0 | 0.02 | 0 | 0.92 | 0 | 0.06 | 0 | 0 |
| Caucasian | 0.06 | 0 | 0.34 | 0 | 0.37 | 0.01 | 0.05 | 0.07 | 0.10 |
| African American | 0.20 | 0 | 0.16 | 0.19 | 0.22 | 0.03 | 0.15 | 0.04 | 0.02 |
| West African | 0.29 | 0.01 | 0.07 | 0.25 | 0.15 | 0.06 | 0.11 | 0.05 | 0 |
HH stands for human haplogroups, as described by Gonzalez et al. (1999) and Mummidi et al. (2000).
CCR5 diplotype profiles are presented in the supplementary Table S5. Given the frequency of HHE in the PNG population (0.92), it was not unexpected to find that 84% individuals were homozygous for this haplotype, and 12% were HHE/HHF*2 heterozygous. HHD/any and HHG*2/any diplotypes were common in the populations of African ancestry and in the CA population respectively.
3.3. CCR5 promoter sequencing
The PNG population had a very high frequency of −2459G>A, but many of the other promoter SNPs were rare or absent. Therefore, we performed sequencing of the promoter region in the PNG samples (n = 87), and compared those sequences with the reference sequence (GenBank accession number NT_022517, nucleotide coordinates 46,360,142-46,366,206) and with the sequences from the North American (n = 26) and West African (n = 15) samples. After alignment and trimming, sequences of 1009 nucleotides were analyzed from all samples. These nucleotides correspond to positions 46,360,151 to 46,361,159 (NT_022517) and −2752 to −1744, the latter based on the previously described nucleotide numbering system (Mummidi et al., 2000). This analysis revealed that (1) the genotypes of all seven SNPs in all PNG, North American, and West African samples were concordant with the sequence data, (2) no other polymorphism was found in any of the North American and West African samples, and (3) in three PNG samples from Dreikikir, a new SNP was identified. Following the previously described nucleotide numbering system (Mummidi et al., 2000), three PNG individuals were found to be heterozygous CG at position −1918. Thus, the frequency of −1918C>G in the samples from Dreikikir (n = 63) and in the total PNG population sequenced (n = 87) was 0.024 and 0.017 respectively. All three individuals were HHE homozygous and were not related. The CCR5 promoter sequence of a PNG individual, carrying −1918CG within the HHE haplotype, has been deposited at GenBank under the accession number KP313871.
3.4. CCR5 promoter haplotype diversity and population differentiation
The PNG population had a very different CCR5 promoter allele-frequency profile compared with all the other populations (Table 3). Therefore, we determined CCR5 promoter haplotype diversity and differentiation in all populations. For this analysis, we used the SNP genotype as well as nucleotide sequence data. The CCR5 promoter H and PA-Fst values are presented in Table 5 and Table 6 respectively. H values obtained by using the nucleotide sequence data were somewhat lower than those obtained by using the SNP genotype data for all populations (Table 5). No such pattern was noticed regarding the PA-Fst values (Table 6). The PNG population showed much lower haplotype diversity compared with all the other populations (Table 5), but much higher differentiation in all pairwise comparisons (Table 6).
Table 5.
CCR5 promoter haplotype diversity in various populations.
| Population | H (mean ± S.E.)a | H (mean ± S.E.)b |
|---|---|---|
| Papua New Guinean | 0.155 ± 0.021 | 0.142 ± 0.036 |
| Caucasian | 0.874 ± 0.011 | 0.790 ± 0.045 |
| African American | 0.912 ± 0.01 | 0.862 ± 0.028 |
| West African | 0.888 ± 0.009 | 0.825 ± 0.044 |
Haplotype diversity based on the seven SNPs shown in Table 3.
Haplotype diversity based on 1009 nucleotide sequences.
Table 6.
CCR5 promoter haplotype-based pairwise population differentiation.
| West African | AFA | CA | |
|---|---|---|---|
| Papua New Guinean | 0.509a | 0.566 | 0.588 |
| 0.640b | 0.603 | 0.513 | |
| West African | 0.011 | 0.084 | |
| 0.007 | 0.082 | ||
| AFA | 0.051 | ||
| 0.057 |
First row, population differentiation based on the seven SNPs shown in Table 3.
Second row, population differentiation based on 1009 nucleotide sequences.
CA = Caucasian
AFA = African American
3.5. Composite risk scores
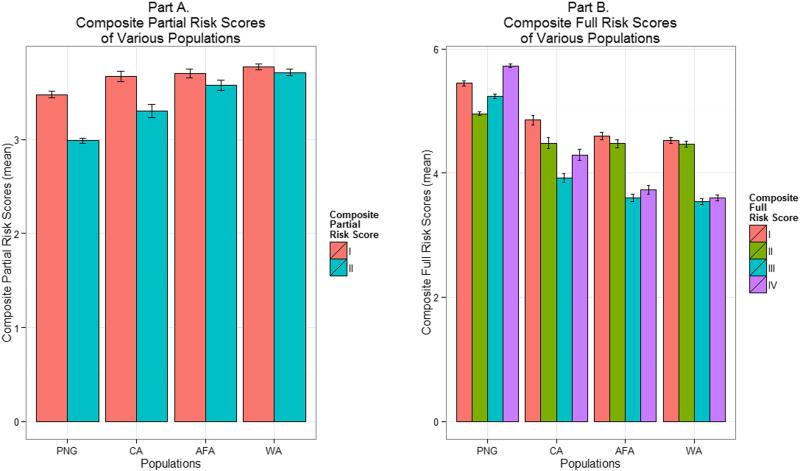
Composite partial risk scores of all study populations are presented in Figure 1A. These risk scores are without CCR5 −2459 genotypes and include CXCL12 801 genotype risk scores only from scenario I or II (AA or GA+AA protective). The composite partial risk scores were significantly lower in the PNG population compared with any other population (P-values ≤ 0.035, Games-Howell test). This is consistent with the findings of the previous study (Su et al., 2000).
Figure 1.
A: Representation of composite partial risk scores (as mean ± S.E.) of various study populations. PNG, Papua New Guinea; CA, Caucasian; AFA, African American; WA, West African. In scenarios I and II, the CXCL12 801 genotypes AA and GA+AA, respectively, were considered protective.
B: Representation of composite full risk scores (as mean ± S.E.) of various study populations. PNG, Papua New Guinea; CA, Caucasian; AFA, African American; WA, West African. In scenarios I and II, the CXCL12 801 genotypes AA and GA+AA, respectively, were considered protective, whereas in scenarios III and IV, these genotypes, respectively, were considered susceptible.
Full spectrum of composite full risk scores of all study populations is presented in Figure 1B. These risk scores include CCR5 −2459 genotypes as well as CXCL12 801 genotype risk scores from all scenarios (I, II, III, or IV). The overall profile of the PNG population was noticeably different from all the other populations. In contrast to the composite partial risk scores, the composite full risk scores were significantly higher in the PNG population compared with any other population when the 801A allele was considered protective (scenario I, P-values < 0.001, Tukey’s HSD test; scenario II, P-values < 0.001, Games-Howell test). Similarly, these risk scores were also significantly higher in the PNG population compared with any other population when the 801A allele was considered susceptible (scenario III, P-values < 0.001, Tukey’s HSD test; scenario IV, P-values < 0.001, Games-Howell test).
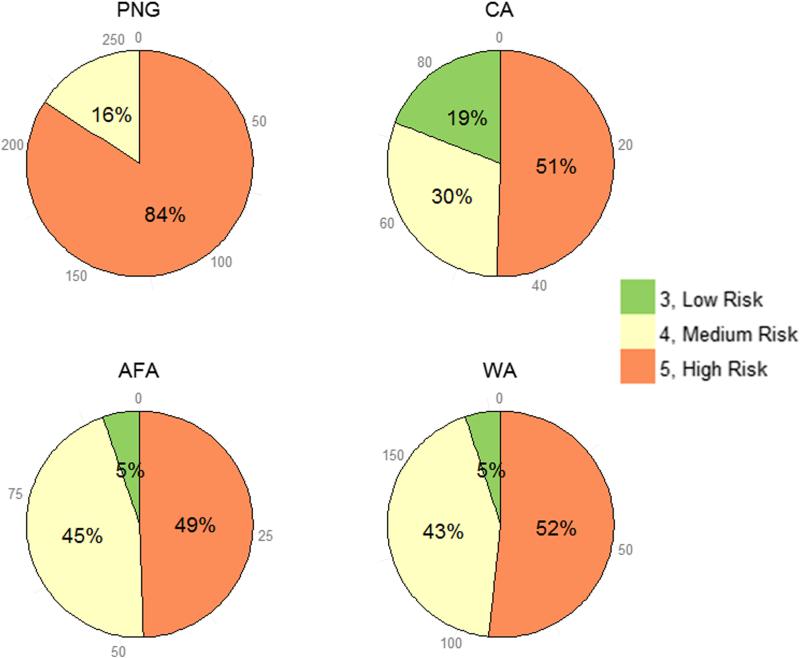
Proportions of composite simple risk scores, which do not include CXCL12 801 genotypes and are the sum of risk scores assigned to CCR2 190, CCR5 −2459, and CCR5 ORF genotypes, in all study populations is presented in Figure 2. Unlike all the other populations, most individuals in the PNG population had high risk (84% with a risk score of 5) and no individual had low risk (a risk score of 3). In the other populations, individuals with high risk varied from 5–19%. Proportions of individuals with low (a risk score of 3) or high (a risk score of 5) risk were also noticeably different between the CA and AFA/West African populations.
Figure 2.
Proportions of composite simple risk scores in various study populations. PNG, Papua New Guinea; CA, Caucasian; AFA, African American; WA, West African. For simplification, composite simple risk scores 3, 4, and 5 were defined as low, medium, and high risk respectively.
Distributions, means, and medians of composite simple risk scores of all study populations are presented in Figure 3. These distributions were significantly different (P-values < 0.001, Kruskal-Wallis test). Pairwise comparisons showed that the risk score was significantly higher in the PNG population compared with any other population (P-values < 0.001, Games-Howell test). Consistent with the proportions of individuals with a risk score of 3 or 5 in the CA vs. AFA/West African population (Figure 2), the risk score was significantly higher in the CA population compared with the AFA (P = 0.035, Games-Howell test) or West African (P = 0.0009, Games-Howell test) population.
Figure 3.
Violin plots showing distributions, means, and medians of composite simple risk scores of various study populations. PNG, Papua New Guinea; CA, Caucasian; AFA, African American; WA, West African. The violin plot is a cross between a box plot and a kernel density plot. These violin plots show not only the mean and median risk scores of each population, but also make it simple to see the differences in risk score value distributions of populations.
4. Discussion
Utilizing samples from PNG, North America, and West Africa, this study provides evidence that the collective variation in CCR2, CCR5, and CXCL12 may affect the occurrence of HIV infection and disease in PNG, more so than in the other regions.
4.1. CCR5 variation in PNG
−2459A and ORF wt alleles
We found that in the PNG population, the frequency of the −2459A allele was the highest yet reported (0.98). Only one other study, using a total of 49 samples from highland and lowland population groups in PNG (which may be different from those in our study) has reported the frequency of this allele (0.85) (Clark and Dean, 2004). In most populations, the frequency of the −2459A allele ranges from 0.35 to 0.55 (Clark and Dean, 2004; Gonzalez et al., 2001; Mehlotra et al., 2011). We also found that the Δ32 allele was absent in PNG (Clark and Dean, 2004; Martinson et al., 2000; Su et al., 2000).
We, and others, have found that the −2459G and Δ32 alleles were associated with significantly reduced in vitro promoter activity, CCR5 expression, and HIV propagation, compared with the −2459A and ORF wt alleles respectively (Hladik et al., 2005; Kawamura et al., 2003; McDermott et al., 1998; Mummidi et al., 2000; Salkowitz et al., 2003). These findings provide a biologic basis for understanding certain aspects of host genetic susceptibility to HIV-1 propagation and progression to AIDS. Furthermore, they parallel the genetic susceptibility studies performed in large cohorts of HIV-infected individuals, showing that the −2459G and Δ32 alleles were associated with protection against HIV infection and/or delayed disease progression, compared with the −2459A and ORF wt alleles respectively (Dean et al., 1996; Martin et al., 1998; McDermott et al., 1998; Zimmerman et al., 1997). Thus, a very high frequency of the −2459A allele, together with the absence of the Δ32 allele in PNG populations, may indicate a greater predisposition to HIV infection and disease.
HHE haplotype
The most prevalent −2459A allele-carrying CCR5 haplotype is HHE, the frequency of which ranges from 0.2 to 0.32 in most populations (Gonzalez et al., 1999; Gonzalez et al., 2001). We found that in the PNG population, the frequency of HHE was 0.92 and 84% individuals were homozygous for this haplotype.
Along the line that the −2459G/A and ORF wt/Δ32 alleles show differences in phenotypic effects in vitro as well as in HIV/AIDS cohorts, different CCR5 haplotypes influence HIV infection and disease outcomes differently (Gonzalez et al., 1999; Huik et al., 2014; Kostrikis et al., 1999; Mangano et al., 2001; Tang et al., 2002a; Tang et al., 2002b). Among the CCR5 haplotypes, the consistency of the associations of HHE homozygosity (E/E diplotype) with unfavorable outcomes is noteworthy. The E/E diplotype was significantly associated with disease acceleration, particularly an accelerated progression to death in Caucasians (Gonzalez et al., 1999). The E/E diplotype was also significantly associated with HIV-1 seroconversion, higher early HIV-1 RNA levels, and shorter time to AIDS in diverse North American cohorts (Tang et al., 2002a; Tang et al., 2002b). Association between HHE and rapid HIV-1 disease progression was also observed in patients from Rwanda (R.A. Kaslow et al., 8th Conference on Retroviruses and Opportunistic Infections, Chicago, IL, 2001, Abstract #45B), Spain (Li et al., 2005), and Thailand (Nguyen et al., 2004). There was a strong association between the E/E diplotype and susceptibility to perinatal transmission of HIV-1, accelerated rate of progression to AIDS, and a more rapid progression to death in Argentinean children (Mangano et al., 2001). Recently, a study investigating the association between CCR5 haplotypes and HIV tropism in Estonian Caucasians found that HHE was associated with the presence of HIV-1 X4-tropic viruses (Huik et al., 2014). Such consistency across racial/ethnic groups and populations suggest that HHE confers similar effects against distinct genetic backgrounds, and therefore may increase the risk of HIV infection and rate of disease progression in PNG as well.
Twelve percent individuals were HHE/HHF*2 heterozygous (Table S5). HHF*2 carries the CCR2 190A (64Ile) allele, and consistent with the protective effects of the 190A allele on HIV infection and/or disease progression, HHF*2 was found to be protective in various cohort studies (Gonzalez et al., 1999; Mangano et al., 2001; Tang et al., 2002a). However, its effects, when paired with HHE are unclear (Mangano et al., 2001; Tang et al., 2002a).
CCR5 promoter haplotype diversity and population differentiation
Genetic differentiation among populations reflects the action of both random processes (e.g., genetic drift) and natural selection. The 5’ cis-regulatory region of CCR5 has been shown to have undergone balancing selection in humans, under a wide range of demographic histories (Bamshad et al., 2002; Ramalho et al., 2010), whereas there appears to have been a selective sweep in chimpanzee (Wooding et al., 2005). Thus, natural selection may appear to have played a role in the pattern of genetic differentiation observed at CCR5 promoter in the PNG population. To investigate this possibility, the present data is limited, and further analysis of the patterns of genetic variation in and around the CCR5 locus is warranted. Regardless of whether demography or natural selection is responsible for the observed pattern of population differentiation, our results can still be regarded as a starting point in identifying the contribution of variation in CCR5 promoter to HIV infection/disease progression in PNG.
4.2. CXCL12 801G>A in PNG
We found that in the PNG population, the frequency of the 801A allele was 0.64. A previous study has reported that the frequency of this allele in three PNG populations (details of which were not provided) was 0.66–0.72 (Su et al., 2000). Globally, the distribution of the 801A allele varies extensively, from being absent to as high as 0.43 (Su et al., 1999; Su et al., 2000).
CXCL12 801G>A, SDF-1 levels, and HIV infection/disease
The findings regarding the association of CXCL12 801 genotypes with HIV infection/disease progression are complex; the mutant allele, 801A, may be protective (Hendel et al., 1998; Passam et al., 2005; Tiensiwakul, 2004; Winkler et al., 1998) as well as susceptible (Daar et al., 2005; Mummidi et al., 1998; Petersen et al., 2005). That the 801A allele may be protective as well as susceptible may depend in part on the relationship between the allele and SDF-1 protein levels, and between the SDF-1 protein levels and HIV infection/disease progression. The 801 AA genotype may be associated with higher (Tiensiwakul, 2004) or lower (Soriano et al., 2002) protein levels, or there may be no association between the genotype and protein levels (Petersen et al., 2005; Verma et al., 2007). Similarly, there may be a positive (Derdeyn et al., 1999) or negative (Ikegawa et al., 2001) correlation between the SDF-1 protein levels and disease progression, which may depend on the stage of infection/disease (Ikegawa et al., 2001). Thus, the interrelationship of the 801A allele, SDF-1 protein levels, and HIV infection/disease may be population and/or stage specific.
Composite risk scores – role of CXCL12 801G>A vs. CCR5 −2459G>A
In order to further understand how CCR2, CCR5, and CXCL12 variation may influence the risk of HIV infection/disease progression in PNG, we first calculated composite partial risk scores, without including CCR5 −2459G>A and considering only the protective effects of the CXCL12 801A allele. Similar to the previous study (Su et al., 2000), we found that these risk scores were significantly lower in the PNG population compared with any other study population (Figure 1A), indicating high-level protection from AIDS onset, or even HIV-1 infection (Su et al., 1999; Su et al., 2000). However, it seems not to be the case because when CCR5 −2459G>A was included, the composite full risk scores were significantly higher in the PNG population compared with any other study population (Figure 1B). In addition, due to a complex interrelationship of the 801A allele, SDF-1 protein levels, and HIV infection/disease, we calculated composite full risk scores by considering both protective (scenarios I and II) and susceptibility (scenarios III and IV) effects of the 801A allele. Regardless of whether we considered the 801A allele protective or susceptible, the risk scores were significantly higher in the PNG population compared with any other study population (Figure 1B). The only possible explanation for this is that a large majority of the individuals carried CCR5 −2459G>A (−2459 AA), which resulted in most individuals with high risk (Figure 2) and a significantly higher composite simple risk score of the population (Figure 3). Therefore, it is plausible that among the genetic variants included in this study, CCR5 −2459G>A alone may be a significant risk factor for a large majority of the PNG population.
4.3. Limitations
We acknowledge that this study has some limitations. First, PNG samples came from two areas in the same province, and may not truly represent the countrywide CCR5 variation. Therefore, we genotyped all nine CCR2-CCR5 polymorphisms in a total of 41 samples collected from 12 out of 20 total provinces, which were provided by Dr. Mark Stoneking (Max Planck Institute for Evolutionary Anthropology, Germany) for a malaria epidemiological study (Mehlotra et al., 2005). In these samples, the CCR5 −2459A allele frequency was 0.89 and the distribution of CCR5 haplotypes was as follows: HHC, 0.11; HHE, 0.65; and HHF*2, 0.24. Forty-six percent of the samples (19/41) were HHE homozygous, and 32% (13/41) were HHE/HHF*2 heterozygous. These results suggest that the CCR5 −2459A allele and HHE haplotype are quite predominant in PNG.
Second, that we used an arbitrary risk score of 2, 1, or 0 to each of the CCR2 190, CCR5 −2459, CCR5 ORF, and CXCL12 801 genotypes (Table 2), and calculated composite partial/full/simple risk scores (range 2–6, Figures 1-3), could be considered a limitation. Although these arbitrary risk scores are based on the previously reported associations of these genotypes with HIV acquisition and/or disease progression, it is important to recognize that they per se are not indicative of any genetic association. The scoring that we used is unable to take into account differing strengths of genetic effects in the same direction and that is one of its weaknesses. Nevertheless, it does allow for some comparison across populations. On the other hand, the RH values computed in previous studies (Su et al., 1999; Su et al., 2000) also have limitations: they were computed considering only the protective effects of the CXCL12 801 AA genotype (Winkler et al., 1998), and not the susceptibility effects of the genotype. In addition, and importantly, the contribution of CCR5 −2459G>A was not considered in those studies.
Finally, this study was not designed to analyze other host genes potentially important in HIV/AIDS pathogenesis. An example is CCR5 ligands RANTES and macrophage inflammatory protein 1α (Clark and Dean, 2004; Gonzalez et al., 2001; Gonzalez et al., 2005; McDermott et al., 2000). Outside the chemokine receptor-ligand nexus, host genetic factors that are associated with viral load control have been identified by recent genome-wide association studies (van Manen et al., 2012). Variation in immune response genes – human leukocyte antigen complex and killer cell immunoglobulin-like receptor (Bashirova et al., 2011; Martin and Carrington, 2013), β-defensin (Hardwick et al., 2012; Mehlotra et al., 2012), and toll-like receptor (Willie et al., 2014) – has been found to affect the natural history of HIV infection and disease progression. Therefore, future studies that include these additional genetic variants could provide more comprehensive insights regarding HIV/AIDS pathogenesis in PNG, and could potentially alter the interpretation of a risk score as employed here.
4.4. Conclusion and future directions
A very high frequency of the CCR5 −2459A allele and absence of Δ32, resulting in a very high frequency of the HHE haplotype, together with the susceptibility effects of the CXCL12 801A allele suggests that a large majority of the PNG population may be at greater predisposition to HIV infection and disease. Therefore, to better estimate the effects of polymorphisms involved in HIV infection/disease progression in PNG, large-scale, comprehensive genetic association studies are necessary. More specifically, an association between the HHE haplotype and CCR5 expression in vitro, as well as between the HHE haplotype and susceptibility to HIV infection/disease dynamics in clinical cohorts should be analyzed. In addition, further genotyping of CCR5 −1918C>G needs to be performed, and its role in CCR5 transcriptional regulation vis-à-vis HIV/AIDS outcomes needs to be defined. These approaches may strengthen the identification of at-risk individuals, design and implementation of beneficial treatment strategies, and development of strategies to eliminate the threat of HIV/AIDS in PNG and the Asia-Pacific region.
Supplementary Material
Acknowledgements
R.K.M. dedicates this paper to the loving memories of late Dr. Richard Krause (Fogarty International Center/NIH), who was instrumental in changing his research and otherwise life.
We are thankful to Dr. Claire Ryan and Dr. Paul Gorry (Burnet Institute, Australia) for supporting this study by providing funding through the Australian Centre for HIV and Hepatitis Virology Research (to P.A.Z. and R.K.M.). Financial support for this study was also provided by the National Institutes of Health (FIC, D43TW007377, support for B.J.; NHLBI, T32HL007567, support for N.B.H.), a grant from the James B. Pendleton Charitable Trust (to P.A.Z.), and an Infectious Diseases Research Support from a STERIS Corporation grant (to R.K.M.). We sincerely thank Dr Daniel Tisch, Dave McNamara, Barne Willie, Krufinta Bun, Valentine Siba, and Ramalakshmi Janamanchi for helpful discussions and critical evaluation of the manuscript.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Anyiwo CE, Imai M, Igo JD, Ogunbanjo B, Iwamoto A, Babona DV. Genotyping of human immunodefiency virus isolates in Papua New Guinea. Nig. Q. J. Hosp. Med. 2010;20:181–185. [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, Ahuja SS, Dunn DM, Watkins WS, Wooding S, Stone AC, Jorde LB, Weiss RB, Ahuja SK. A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirova AA, Thomas R, Carrington M. HLA/KIR restraint of HIV: surviving the fittest. Annu. Rev. Immunol. 2011;29:295–317. doi: 10.1146/annurev-immunol-031210-101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark VJ, Dean M. Haplotype structure and linkage disequilibrium in chemokine and chemokine receptor genes. Hum. Genomics. 2004;1:255–273. doi: 10.1186/1479-7364-1-4-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan B, Millan J, Malau C, Kaldor J, Toole M. The HIV epidemic in Papua New Guinea. J. Acquir. Immune Defic. Syndr. 2011;58:e48–51. doi: 10.1097/QAI.0b013e3182293417. [DOI] [PubMed] [Google Scholar]
- Daar ES, Lynn HS, Donfield SM, Lail A, O’Brien SJ, Huang W, Winkler CA, Hemophilia G, Development S. Stromal cell-derived factor-1 genotype, coreceptor tropism, and HIV type 1 disease progression. J. Infect. Dis. 2005;192:1597–1605. doi: 10.1086/496893. [DOI] [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien SJ. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Costello C, Kilby JM, Sfakianos G, Saag MS, Kaslow R, Bucy RP. Correlation between circulating stromal cell-derived factor 1 levels and CD4+ cell count in human immunodeficiency virus type 1-infected individuals. AIDS Res. Hum. Retroviruses. 1999;15:1063–1071. doi: 10.1089/088922299310359. [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, Cabrera S, McBride M, Cao XH, Merrill G, O’Connell P, Bowden DW, Freedman BI, Anderson SA, Walter EA, Evans JS, Stephan KT, Clark RA, Tyagi S, Ahuja SS, Dolan MJ, Ahuja SK. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Dhanda R, Bamshad M, Mummidi S, Geevarghese R, Catano G, Anderson SA, Walter EA, Stephan KT, Hammer MF, Mangano A, Sen L, Clark RA, Ahuja SS, Dolan MJ, Ahuja SK. Global survey of genetic variation in CCR5, RANTES, and MIP-1α: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O’Connell R,J, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Hardwick RJ, Amogne W, Mugusi S, Yimer G, Ngaimisi E, Habtewold A, Minzi O, Makonnen E, Janabi M, Machado LR, Viskaduraki M, Mugusi F, Aderaye G, Lindquist L, Hollox EJ, Aklillu E. beta-defensin genomic copy number is associated with HIV load and immune reconstitution in sub-saharan Africans. J. Infect. Dis. 2012;206:1012–1019. doi: 10.1093/infdis/jis448. [DOI] [PubMed] [Google Scholar]
- Hendel H, Henon N, Lebuanec H, Lachgar A, Poncelet H, Caillat-Zucman S, Winkler CA, Smith MW, Kenefic L, O’Brien S, Lu W, Andrieu JM, Zagury D, Schachter F, Rappaport J, Zagury JF. Distinctive effects of CCR5, CCR2, and SDF1 genetic polymorphisms in AIDS progression. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1998;19:381–386. doi: 10.1097/00042560-199812010-00009. [DOI] [PubMed] [Google Scholar]
- Hladik F, Liu H, Speelmon E, Livingston-Rosanoff D, Wilson S, Sakchalathorn P, Hwangbo Y, Greene B, Zhu T, McElrath MJ. Combined effect of CCR5-Δ32 heterozygosity and the CCR5 promoter polymorphism -2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J. Virol. 2005;79:11677–11684. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huik K, Avi R, Uibopuu H, Pauskar M, Margus T, Karki T, Krispin T, Kool P, Ruutel K, Talu A, Abel-Ollo K, Uuskula A, Carrillo A, He W, Ahuja SK, Lutsar I. Association Between HIV-1 Tropism and CCR5 Human Haplotype E in a Caucasian Population. J. Acquir. Immune Defic. Syndr. 2014;66:239–244. doi: 10.1097/QAI.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa M, Yuan J, Matsumoto K, Herrmann S, Iwamoto A, Nakamura T, Matsushita S, Kimura T, Honjo T, Tashiro K. Elevated plasma stromal cell-derived factor 1 protein level in the progression of HIV type 1 infection/AIDS. AIDS Res. Hum. Retroviruses. 2001;17:587–595. doi: 10.1089/088922201300119680. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Rosenberg PS, Goedert JJ, Ashton LJ, Benfield TL, Buchbinder SP, Coutinho RA, Eugen-Olsen J, Gallart T, Katzenstein TL, Kostrikis LG, Kuipers H, Louie LG, Mallal SA, Margolick JB, Martinez OP, Meyer L, Michael NL, Operskalski E, Pantaleo G, Rizzardi GP, Schuitemaker H, Sheppard HW, Stewart GJ, Theodorou ID, Ullum H, Vicenzi E, Vlahov D, Wilkinson D, Workman C, Zagury JF, O’Brien TR. International Meta-Analysis of, H.I.V.H.G., 2001. Effects of CCR5-Δ32, CCR2-64I, and SDF-1 3′A alleles on HIV-1 disease progression: An international meta-analysis of individual-patient data. Ann. Intern. Med. 135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- Kasehagen LJ, Mueller I, Kiniboro B, Bockarie MJ, Reeder JC, Kazura JW, Kastens W, McNamara DT, King CH, Whalen CC, Zimmerman PA. Reduced Plasmodium vivax erythrocyte infection in PNG Duffy-negative heterozygotes. PLoS One. 2007;2:e336. doi: 10.1371/journal.pone.0000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Gulden FO, Sugaya M, McNamara DT, Borris DL, Lederman MM, Orenstein JM, Zimmerman PA, Blauvelt A. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8401–8406. doi: 10.1073/pnas.1432450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrikis LG, Huang Y, Moore JP, Wolinsky SM, Zhang L, Guo Y, Deutsch L, Phair J, Neumann AU, Ho DD. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat. Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- Kostrikis LG, Neumann AU, Thomson B, Korber BT, McHardy P, Karanicolas R, Deutsch L, Huang Y, Lew JF, McIntosh K, Pollack H, Borkowsky W, Spiegel HM, Palumbo P, Oleske J, Bardeguez A, Luzuriaga K, Sullivan J, Wolinsky SM, Koup RA, Ho DD, Moore JP. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J. Virol. 1999;73:10264–10271. doi: 10.1128/jvi.73.12.10264-10271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Song R, Masciotra S, Soriano V, Spira TJ, Lal RB, Yang C. Association of CCR5 human haplogroup E with rapid HIV type 1 disease progression. AIDS Res. Hum. Retroviruses. 2005;21:111–115. doi: 10.1089/aid.2005.21.111. [DOI] [PubMed] [Google Scholar]
- Mangano A, Gonzalez E, Dhanda R, Catano G, Bamshad M, Bock A, Duggirala R, Williams K, Mummidi S, Clark RA, Ahuja SS, Dolan MJ, Bologna R, Sen L, Ahuja SK. Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus. J. Infect. Dis. 2001;183:1574–1585. doi: 10.1086/320705. [DOI] [PubMed] [Google Scholar]
- Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol. Rev. 2013;254:245–264. doi: 10.1111/imr.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, Lee B, Doms RW, Margolick J, Buchbinder S, Goedert JJ, O’Brien TR, Hilgartner MW, Vlahov D, O’Brien SJ, Carrington M. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- Martinson JJ, Hong L, Karanicolas R, Moore JP, Kostrikis LG. Global distribution of the CCR2-64I/CCR5-59653T HIV-1 disease-protective haplotype. AIDS. 2000;14:483–489. doi: 10.1097/00002030-200003310-00003. [DOI] [PubMed] [Google Scholar]
- McDermott DH, Beecroft MJ, Kleeberger CA, Al-Sharif FM, Ollier WE, Zimmerman PA, Boatin BA, Leitman SF, Detels R, Hajeer AH, Murphy PM. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS. 2000;14:2671–2678. doi: 10.1097/00002030-200012010-00006. [DOI] [PubMed] [Google Scholar]
- McDermott DH, Zimmerman PA, Guignard F, Kleeberger CA, Leitman SF, Murphy PM. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- Mehlotra RK, Cheruvu VK, Blood Zikursh MJ, Benish RL, Lederman MM, Salata RA, Gripshover B, McComsey GA, Lisgaris MV, Fulton S, Subauste CS, Jurevic RJ, Guillemette C, Zimmerman PA, Rodriguez B. Chemokine (C-C motif) receptor 5 -2459 genotype in patients receiving highly active antiretroviral therapy: race-specific influence on virologic success. J. Infect. Dis. 2011;204:291–298. doi: 10.1093/infdis/jir262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Dazard JE, John B, Zimmerman PA, Weinberg A, Jurevic RJ. Copy Number Variation within Human beta-Defensin Gene Cluster Influences Progression to AIDS in the Multicenter AIDS Cohort Study. J. AIDS Clin. Res. 2012;3 doi: 10.4172/2155-6113.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Kasehagen LJ, Baisor M, Lorry K, Kazura JW, Bockarie MJ, Zimmerman PA. Malaria infections are randomly distributed in diverse holoendemic areas of Papua New Guinea. Am. J. Trop. Med. Hyg. 2002;67:555–562. doi: 10.4269/ajtmh.2002.67.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK, Lorry K, Kastens W, Miller SM, Alpers MP, Bockarie M, Kazura JW, Zimmerman PA. Random distribution of mixed species malaria infections in Papua New Guinea. Am. J. Trop. Med. Hyg. 2000;62:225–231. doi: 10.4269/ajtmh.2000.62.225. [DOI] [PubMed] [Google Scholar]
- Mehlotra RK, Mattera G, Bhatia K, Reeder JC, Stoneking M, Zimmerman PA. Insight into the early spread of chloroquine-resistant Plasmodium falciparum infections in Papua New Guinea. J. Infect. Dis. 2005;192:2174–2179. doi: 10.1086/497694. [DOI] [PubMed] [Google Scholar]
- Mulherin SA, O’Brien TR, Ioannidis JP, Goedert JJ, Buchbinder SP, Coutinho RA, Jamieson BD, Meyer L, Michael NL, Pantaleo G, Rizzardi GP, Schuitemaker H, Sheppard HW, Theodorou ID, Vlahov D, Rosenberg PS, International Meta-Analysis of, H.I.V.H.G. Effects of CCR5-Δ32 and CCR2-64I alleles on HIV-1 disease progression: the protection varies with duration of infection. AIDS. 2003;17:377–387. doi: 10.1097/01.aids.0000050783.28043.3e. [DOI] [PubMed] [Google Scholar]
- Mummidi S, Ahuja SS, Gonzalez E, Anderson SA, Santiago EN, Stephan KT, Craig FE, O’Connell P, Tryon V, Clark RA, Dolan MJ, Ahuja SK. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat. Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- Mummidi S, Bamshad M, Ahuja SS, Gonzalez E, Feuillet PM, Begum K, Galvis MC, Kostecki V, Valente AJ, Murthy KK, Haro L, Dolan MJ, Allan JS, Ahuja SK. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J. Biol. Chem. 2000;275:18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Li M, Chaowanachan T, Hu DJ, Vanichseni S, Mock PA, van Griensven F, Martin M, Sangkum U, Choopanya K, Tappero JW, Lal RB, Yang C. CCR5 promoter human haplogroups associated with HIV-1 disease progression in Thai injection drug users. AIDS. 2004;18:1327–1333. doi: 10.1097/00002030-200406180-00012. [DOI] [PubMed] [Google Scholar]
- Passam AM, Zafiropoulos A, Miyakis S, Zagoreos I, Stavrianeas NG, Krambovitis E, Spandidos DA. CCR2-64I and CXCL12 3′A alleles confer a favorable prognosis to AIDS patients undergoing HAART therapy. J. Clin. Virol. 2005;34:302–309. doi: 10.1016/j.jcv.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Petersen DC, Glashoff RH, Shrestha S, Bergeron J, Laten A, Gold B, van Rensburg EJ, Dean M, Hayes VM. Risk for HIV-1 infection associated with a common CXCL12 (SDF1) polymorphism and CXCR4 variation in an African population. J. Acquir. Immune Defic. Syndr. 2005;40:521–526. doi: 10.1097/01.qai.0000186360.42834.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho RF, Santos EJ, Guerreiro JF, Meyer D. Balanced polymorphism in bottlenecked populations: the case of the CCR5 5′ cis-regulatory region in Amazonian Amerindians. Hum. Immunol. 2010;71:922–928. doi: 10.1016/j.humimm.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Ryan CE, Gare J, Crowe SM, Wilson K, Reeder JC, Oelrichs RB. The heterosexual HIV type 1 epidemic in Papua New Guinea is dominated by subtype C. AIDS Res. Hum. Retroviruses. 2007;23:941–944. doi: 10.1089/aid.2007.0043. [DOI] [PubMed] [Google Scholar]
- Salkowitz JR, Bruse SE, Meyerson H, Valdez H, Mosier DE, Harding CV, Zimmerman PA, Lederman MM. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro. Clin. Immunol. 2003;108:234–240. doi: 10.1016/s1521-6616(03)00147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, Goedert JJ, O’Brien TR, Jacobson LP, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner MW, O’Brien SJ. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- Soriano A, Martinez C, Garcia F, Plana M, Palou E, Lejeune M, Arostegui JI, De Lazzari E, Rodriguez C, Barrasa A, Lorenzo JI, Alcami J, del Romero J, Miro JM, Gatell JM, Gallart T. Plasma stromal cell-derived factor (SDF)-1 levels, SDF1-3′A genotype, and expression of CXCR4 on T lymphocytes: their impact on resistance to human immunodeficiency virus type 1 infection and its progression. J. Infect. Dis. 2002;186:922–931. doi: 10.1086/343741. [DOI] [PubMed] [Google Scholar]
- Su B, Jin L, Hu F, Xiao J, Luo J, Lu D, Zhang W, Chu J, Du R, Geng Z, Qiu X, Xue J, Tan J, O’Brien SJ, Chakraborty R. Distribution of two HIV-1-resistant polymorphisms (SDF1-3′A and CCR2-64I) in East Asian and world populations and its implication in AIDS epidemiology. Am. J. Hum. Genet. 1999;65:1047–1053. doi: 10.1086/302568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Sun G, Lu D, Xiao J, Hu F, Chakraborty R, Deka R, Jin L. Distribution of three HIV-1 resistance-conferring polymorphisms (SDF1-3′A, CCR2-641, and CCR5-Δ32) in global populations. Eur. J. Hum. Genet. 2000;8:975–979. doi: 10.1038/sj.ejhg.5200568. [DOI] [PubMed] [Google Scholar]
- Tang J, Shelton B, Makhatadze NJ, Zhang Y, Schaen M, Louie LG, Goedert JJ, Seaberg EC, Margolick JB, Mellors J, Kaslow RA. Distribution of chemokine receptor CCR2 and CCR5 genotypes and their relative contribution to human immunodeficiency virus type 1 (HIV-1) seroconversion, early HIV-1 RNA concentration in plasma, and later disease progression. J. Virol. 2002a;76:662–672. doi: 10.1128/JVI.76.2.662-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Wilson CM, Schaen M, Myracle A, Douglas SD, Kaslow RA, Group RS. CCR2 and CCR5 genotypes in HIV type 1-infected adolescents: limited contributions to variability in plasma HIV type 1 RNA concentration in the absence of antiretroviral therapy. AIDS Res. Hum. Retroviruses. 2002b;18:403–412. doi: 10.1089/088922202753614164. [DOI] [PubMed] [Google Scholar]
- Tiensiwakul P. Stromal cell-derived factor (SDF) 1-3′A polymorphism may play a role in resistance to HIV-1 infection in seronegative high-risk Thais. Intervirology. 2004;47:87–92. doi: 10.1159/000077831. [DOI] [PubMed] [Google Scholar]
- Vallely A, Page A, Dias S, Siba P, Lupiwa T, Law G, Millan J, Wilson DP, Murray JM, Toole M, Kaldor JM. The prevalence of sexually transmitted infections in Papua New Guinea: a systematic review and meta-analysis. PLoS One. 2010;5:e15586. doi: 10.1371/journal.pone.0015586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallely A, Ryan CE, Allen J, Sauk JC, Simbiken CS, Wapling J, Kaima P, Kombati Z, Law G, Fehler G, Murray JM, Siba P, Kaldor JM. High prevalence and incidence of HIV, sexually transmissible infections and penile foreskin cutting among sexual health clinic attendees in Papua New Guinea. Sexual health. 2014;11:58–66. doi: 10.1071/SH13197. [DOI] [PubMed] [Google Scholar]
- van Manen D, van ’t Wout AB, Schuitemaker H. Genome-wide association studies on HIV susceptibility, pathogenesis and pharmacogenomics. Retrovirology. 2012;9:70. doi: 10.1186/1742-4690-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Gupta RB, Singh K, Bhasin R, Anand Shukla A, Chauhan SS, Luthra K. Distribution of CCR5Δ32, CCR2-64I and SDF1-3′A and plasma levels of SDF-1 in HIV-1 seronegative North Indians. J. Clin. Virol. 2007;38:198–203. doi: 10.1016/j.jcv.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Willie B, Hall NB, Stein CM, Jurevic RJ, Weinberg A, Mehlotra RK, Zimmerman PA. Association of Toll-like receptor polymorphisms with HIV status in North Americans. Genes Immun. 2014;15:569–577. doi: 10.1038/gene.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Modi W, Smith MW, Nelson GW, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, Buchbinder S, Vittinghoff E, Goedert JJ, O’Brien TR, Jacobson LP, Detels R, Donfield S, Willoughby A, Gomperts E, Vlahov D, Phair J, O’Brien SJ. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- Wooding S, Stone AC, Dunn DM, Mummidi S, Jorde LB, Weiss RK, Ahuja S, Bamshad MJ. Contrasting effects of natural selection on human and chimpanzee CC chemokine receptor 5. Am. J. Hum. Genet. 2005;76:291–301. doi: 10.1086/427927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy PE, Kumaraswami V, Giorgi JV, Detels R, Hunter J, Chopek M, Berger EA, Fauci AS, Nutman TB, Murphy PM. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol. Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- Zimmerman PA, Dadzie KY, De Sole G, Remme J, Alley ES, Unnasch TR. Onchocerca volvulus DNA probe classification correlates with epidemiologic patterns of blindness. J. Infect. Dis. 1992;165:964–968. doi: 10.1093/infdis/165.5.964. [DOI] [PubMed] [Google Scholar]
- Zimmerman PA, Woolley I, Masinde GL, Miller SM, McNamara DT, Hazlett F, Mgone CS, Alpers MP, Genton B, Boatin BA, Kazura JW. Emergence of FY*A(null) in a Plasmodium vivax-endemic region of Papua New Guinea. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13973–13977. doi: 10.1073/pnas.96.24.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.